Abstract
Release of gonadotropins in adult rodents is sex specific and dependent upon kisspeptin (Kiss1) neurons. This crucial pathway within the hypothalamic-pituitary-gonadal (HPG) axis is profoundly influenced by neonatal estrogens, which induce a male-like phenotype. Classically, estrogen activity is mediated via the estrogen receptors a and β (ERα and ERβ), but the relative roles each plays in organizing the sex-specific ontogeny of kisspeptin signaling pathways remain unresolved. Thus, the present study used in situ hybridization histochemistry (ISHH) to map the temporal and sexually dimorphic neonatal mRNA expression profiles of ERα, ERβ, and Kiss1 in the anterioventral periventricular nucleus (AVPV), medial preoptic area (MPOA), ventromedial nucleus (VMN), and arcuate nucleus (ARC), all regions critical for kisspeptin regulation of gonadotropin secretion. In general, females had higher levels of ERα, in all regions examined, a sex difference that persisted until postnatal day (PND) 19 except in the ARC. Males had significantly more ERβ expression in the AVPV at birth, but this sex difference was lost and then re-emerged on PND 19, with females having more than males. VMN ERβ levels were higher in females until PND 19. Kiss1 was not detectable until PND 11 in the anterior hypo-thalamus, but expression levels were equivalent at birth in the ARC. By PND 2, ARC ERα and Kiss1 levels were abundant, sexually dimorphic (higher in females), and, respectively, showed a U- and a bell-shaped pattern with age. Sex differences in ARC Kiss1 expression provide evidence that Kiss1 may play a role in the sexual dimorphic organization of the neonatal brain. These detailed profiles of neonatal Kiss1 and ERs mRNA levels will help elucidate the relative roles each plays in the sex-specific, estrogen-dependent organization of gonadotropin signaling pathways.
INDEXING TERMS: development, anteroventral periventricular nucleus (AVPV), medial preoptic area (MPOA), arcuate nucleus (ARC), kisspeptin, sex differences
It is well established that the neuroendocrine pathways that regulate gonadotropin release in rodents are sexually dimorphic and profoundly influenced by neonatal estrogens. In males, circulating androgen levels are high in the first few days after birth, and estrogens, aromatized from this testicular androgen, masculinize the system such that gonadotropin surges cannot be elicited in adulthood. In females, the opposite occurs, and low circulating levels of estrogens permit the ontogeny of the female-typical hypothalamic-pituitary-gonadal (HPG) axis. Administration of estrogens or aromatizable androgens to females during this critical period is sufficient to defeminize the system such that a surge cannot be elicited during adulthood (Arai and Gorski, 1968; Simerly, 2002) and to reduce the number of preoptic kisspeptin (Kiss1) cells, required to stimulate the surge, to male-typical levels (Kauffman et al., 2007b; Bateman and Patisaul, 2008; Patisaul et al., 2009). Classically, the organizing effects of estrogens are mediated through two nuclear estrogen receptor subtypes, ERα (Green et al., 1986) and ERβ (Kuiper et al., 1996), but the relative mechanistic roles each play in the estrogen-dependent organization of sexually dimorphic neuroendocrine pathways remain unclear.
To better elucidate the mechanisms by which neonatal estrogens sexually differentiate the neuroendocrine pathways that mediate gonadotropin secretion, we mapped the neonatal mRNA expression of the two primary ER subtypes, ERα and ERβ, and the gonadotropin-stimulating peptide Kiss1 in the preoptic area (POA) and mediobasal hypothalamus of male and female rats from postnatal day zero (PND 0; the day of birth) through 19. Although the hypothalamic distribution of all three genes within the adult rodent brain of both sexes has previously been described in detail (Simerly et al., 1990; Lauber et al., 1991; Shughrue et al., 1996, 1997a,b; Shughrue and Merchenthaler, 2001b; Zhang et al., 2002; Ikeda et al., 2003; Mitra et al., 2003, Nomura et al., 2003; Kauffman et al., 2007b; Mikkelsen and Simonneaux, 2009), sex-specific expression levels (mRNA or protein) in the early postnatal hypothalamus are less well characterized. Only a few studies to date have made identification of sex differences a primary focus (Yokosuka et al., 1997; Perez et al., 2003), whereas others did not obtain samples from the first few days of life (Orikasa et al., 2002; Orikasa and Sakuma, 2003), the developmental window classically considered the neonatal critical period (Simerly, 2002).
In addition, although a few studies have reported fluctuations in mRNA expression levels across early development, the results are inconsistent across studies, and early investigations largely employed techniques that assess ER density indirectly and could not discriminate between ERα and ERβ (MacLusky et al., 1979a,b; White et al., 1979; Gerlach et al., 1983; DonCarlos and Handa, 1994; Kuhnemann et al., 1994; DonCarlos, 1996; Yokosuka et al., 1997). Many also quantified expression within large hypothalamic regions rather than discrete nuclei, an approach that may fail to uncover region-specific sex differences. The resulting need for a more detailed investigation of neonatal expression patterns was the critical data gap addressed by the present set of experiments. Regions of interest included the anteroventral periventricular nucleus (AVPV), the medial preoptic area (MPOA), the arcuate nucleus (ARC), and the ventromedial hypothalamic nucleus (VMN) because all are well established to be sexually dimorphic and important for the neuroendocrine control of gonadotropin release as well as other aspects of female reproductive physiology and behavior (Pfaff and Keiner, 1973; Colpaert and Wiep-kema, 1976; Nance, 1976; Pfaff et al., 1994; Simerly, 2002)
We also characterized the sex-specific ontogeny of Kiss1 expression in the rat hypothalamus because rapidly emerging evidence reveals that these ER-expressing neurons are critical “gatekeepers” of gonadotropin release (Oakley et al., 2009). In the adult rodent, there are two major populations of Kiss1-expressing neurons. The first population is located in an anterior hypothalamic region comprising the rostral and caudal aspects of the preoptic periventricular nucleus and the AVPV, which is now collectively referred to as the rostral periventricular area of the third ventricle (RP3V) (Herbison, 2008; Clarkson et al., 2009). The second population resides in the ARC (Kauffman et al., 2007b). The expression of Kiss1 mRNA and the number of kisspeptin-immunoreactive (-ir) cells is sexually dimorphic in the adult rodent RP3V, with females having more of both than males (Kauffman et al., 2007b; Clarkson et al., 2009), but no sex differences in expression levels or cell number have been found in the peripu-bertal (Losa et al., 2011) or adult (Kauffman et al., 2007b) ARC.
By using dual-labeled in situ hybridization, it has been shown that almost all Kiss1-expressing neurons in the RP3V and ARC co-express ERα (Smith et al., 2005a, 2006), and Kiss1 mRNA expression is under the control of sex hormones. In females, it is upregulated by estrogens in the RP3V, but downregulated in the ARC (Smith et al., 2005b, 2006; Kauffman et al., 2007b), suggesting that the anterior population participates in steroid-positive feedback on gonadotropin release, whereas the posterior population plays a role in steroid-negative feedback (Smith and Davidson, 1974; Scott et al., 1997; Hoffman et al., 2005; Smith et al., 2005a,b, 2006; Dungan et al., 2006; Kauffman et al., 2007a; Kauffman, 2009; Smith, 2009).
Evidence obtained by RT-PCR using dissected blocks of whole hypothalamus revealed evidence of persistent Kiss1 expression in both sexes from PND 1 to adulthood (Navarro et al., 2004). More recently, another group reported that kisspeptin-ir neurons are visible in the mouse RP3V from PND 25 to PND 61,with females having more kisspeptin-ir cells than males at all time points (Clarkson and Herbison, 2006). In a subsequent study, this group found that these cells are present as early as PND 15, but did not look at the ontogeny of Kiss1 expression or kisspeptin-ir cell numbers in the ARC (Clarkson et al., 2009), leaving open the possibility that the hypo-thalamic Kiss1 expression identified previously as early as PND 1 (Navarro et al., 2004) may be localized to the ARC. Therefore, we sought to fill this data gap by quantifying Kiss1 expression in the RP3V and ARC across postnatal development from PND 0 through 19 in male and female rats using in situ hybridization.
For these experiments, we elected to maintain the animals on a soy-free diet because soy-based chow contains phytoestrogens, endocrine disrupting compounds known to disrupt the sexual differentiation of the rodent brain (Brown and Setchell, 2001; Degen et al., 2002; Patisaul, 2005). Prior studies exploring postnatal expression of ERs and Kiss1 used soy-rich diets, thus potentially “masking” or confounding existing sex differences. The present study is the first to characterize the sexual dimorphic expression of ERs and Kiss1 in animals maintained in an environment where exposure to phytoestrogens and other endocrine disrupting compounds is deliberately minimized. Determining precisely when and where in postnatal developmental sex differences in ERα, ERβ and Kiss1 gene expression emerge in the rat brain will help elucidate the mechanistic roles each play in the sex specific organization and maturation of the HPG axis.
MATERIALS AND METHODS
Animal care and tissue collection
Timed pregnant Long Evans (LE) rats were purchased from Charles River (n = 7; Charles River, Raleigh, NC), and were individually housed in a temperature- and light-controlled room (14:10-hour light/dark cycle; lights on at 0700 hours) at the Biological Resource Facility of North Carolina State University (NCSU). All dams were housed in thoroughly washed polysulfone (Bisphenol A [BPA] free) caging and fed a semipurified, phytoestrogen-free diet ad libitum (AIN-93G, Test Diet, Richmond, IN) to minimize exposure to endocrine-disrupting compounds. Three LE females from our existing colony were used to obtain tissue (brain and ovary) for probe generation and testing. All animals in these studies were maintained according to the applicable portions of the Animal Welfare Act and the U.S. Department of Health and Human Services Guide for the Care and Use of Laboratory Animals, and the study was approved by the Institutional Animal Care and Use Committee of NCSU.
Beginning on the day of birth, designated postnatal day 0 (PND 0), female and male pups (n = 5–7 per group) were obtained from litters born to the timed pregnant rats and sacrificed by rapid decapitation at PND 0 (within 12 hours after littering took place), PND 2, PND 4, PND 7, PND 11 (only used to assess Kiss1 expression), and PND 19. To minimize potential litter effects, for each time point, only one animal of each sex was selected from a single litter, and the rest were obtained from other litters. For the PND 0, 2, and 4 animals, the whole head was rapidly frozen on powdered dry ice and stored at −80°C until cryosectioning. For the older animals, the brains were rapidly removed and directly frozen on powdered dry ice.
Gene cloning and plasmid construction for ERα, ERβ, and Kiss1 mRNA templates
Probe generation for ERα and ERβ was guided by previously described procedures (Shughrue et al., 1997a; Shughrue and Merchenthaler, 2001b). Total RNA was extracted from the LE rat ovary with the Ultraspec-II RNA isolation system (Biotecx, Houston, TX) according to the manufacturer’s instructions. Synthesis of cDNA was performed in a 25-µl reaction volume containing 1X M-MLV reverse transcriptase reaction buffer, 0.5 mM dNTPs, 25 U recombinant RNasin ribonuclease inhibitor, 0.5 µg random primers, 200 U M-MLV reverse transcriptase (Promega, Madison, WI), and 2.5 µg total RNA. The reaction conditions were as follows: 1 cycle of 25°C for 5 minutes, 1 cycle of 42°C for 30 minutes, 1 cycle of 95°C for 5 minutes. For long-term storage, the cDNA was kept at –20°C.
The primers (Sigma-Genosys, The Woodlands, TX) used for the polymerase chain reaction (PCR) are listed in Table 1. cDNA of ERα, ERβ and Kiss1 mRNA was amplified by using 0.4 µM of each primer, 0.2 mM each dNTP, 2.5 mM MgCl2, and 5 U taq DNA polymerase (Promega). The PCR conditions were: 1 cycle of 95°C for 3 minutes, 40 cycles of 95°C for 1 minute, 60°C for 1 minute, 72°C for 30 seconds, and a final cycle at 72°C for 10 minutes.
TABLE 1.
Primer Sets for PCR
| Genes | Primers | 5’ →3’ sequence | Amplicon size (bp) | GenBank no. |
|---|---|---|---|---|
| ERα | FW | AATTCTGACAATCGACGCCAG | 345 | NM_012689 |
| RV | GTGCTTCAACATTCTCCCTCC | |||
| ERβ | FW | TTCCCGGCAGCACCAGTAAC | 501 | NM_012754 |
| RV | CACACCGTTCTCTCCTGGATCC | |||
| Kiss1 | FW | TCTCCTCTGTGTGGCCTCTT | 318 | NM_181692 |
| RV | AGGCCAAAGGAGTTCCAGTT |
For abbreviations, see list.
The PCR fragments were then cloned into the PCRII-TOPO vector, and transformed into One Shot TOP10 chemically competent Escherichia coli cells according to the user manual accompanying TOPO TA Cloning Kit (Invi-trogen, Carlsbad, CA). Colonies were selected, the plas-mid was isolated, and verification of the sequence, size, and orientation was accomplished by DNA sequencing (GENEWIZ, South Plainfield, NJ). The plasmid was isolated from the bacterial cultures by using a midi-plasmid preparation kit (Qiagen, Valencia, CA), and the resulting plasmid was digested with HindIII and EcoRV restriction enzymes (New England BioLabs, Boston, MA) to linearize the plasmid for preparation of the antisense or sense transcription templates.
ERα in situ hybridization histochemistry (ISHH)
Each brain was cryosectioned (Leica CM1900, Nus-sloch, Germany) into three serial sets of 12-µm coronal sections (each containing all regions of interest), mounted onto Superfrost plus slides (Fisher Scientific, Pittsburgh, PA), and stored at −80°C until ISHH processing. One set of sections from each animal was used for ISHH of ERα mRNA. All sections containing the region of interest (e.g., RP3V or mediobasal hypothalamus), from both sexes at all time points, were processed and analyzed simultaneously as a large batch. The transcriptional template for the ERα antisense and sense-strand probes was a 345-bp cDNA fragment (Table 1). NEG 35S-labeled UTP (15 µl, PerkinElmer, Boston, MA) was dried down in a vacuum centrifuge, and the following reagents were added to it: 2.0 µl 5X transcription buffer, 1.0 µl 100 mM dithioxytocinhreitol (DTT), 1.5 µl 3.3 mM ATP, GTP, CTP mix (Promega, Madison, WI) and 1.0 µg linearized template. Finally, nuclease-free water was added to obtain a final volume of 10 µl. The contents were mixed well and the following was added: 0.5 µl RNAasin and 0.5 µl Sp6 or T7 RNA polymerase (Promega) for antisense and sense probes, respectively. After a brief vortexing and centrifu-gation, the mixture was incubated at 37°C for 30 minutes, and then another 0.5 µl RNA polymerase was added, followed by an additional 30-minute incubation. Following this, 0.5 µl RNAasin and 2.0 µl RQ1 RNase-Free DNase (Promega) were added to each of the tubes, which were incubated for 30 minutes at 37°C. Finally, the cRNA probe was purified with the Qiaquick Nucleotide Remove Kit (Qiagen) according the user manual.
On the day of the ISHH, the appropriate cryosections were thawed at room temperature for 10 minutes. The slides were prepared for hybridization with a series of pre-hybridization washes as described previously (Petersen et al., 1996). Briefly, air-dried slides were fixed with 4% formalin/phosphate-buffered saline (formalin/PBS, pH 7.4) for 15 minutes, and then treated with 0.25% acetic anhydride in 0.1 M triethanolamine (pH 8.0). Slides were subsequently dehydrated in a series of ethanol washes, dilapidated in chloroform for 5 minutes, and then rinsed in 100% and 95% ethanol. After the sections were dried, the cRNA probes (∼0.5 × 106 cpm) were applied to each tissue section in 25 µl of hybridization buffer. The hybridization buffer contained 2X standard saline citrate solution (2X SSC; 1X SSC = 0.15 M NaCl and 0.015 M sodium citrate, pH 7.2), 50% (v/v) formamide, 10% (w/v) dextran sulfate, 250 µg/ml tRNA, 1X Denhardt’s solution (Sigma-Aldrich, St. Louis, MO), and 200 mM DTT. Sections were covered with glass coverslips and incubated in humid chambers overnight at 50°C.
After hybridization, the coverslips were removed, and the slides were washed for 10 minutes in 1X SSC (pH 7.0) at room temperature. The slides were then washed according to a standard protocol as described previously (Petersen et al., 1996). Briefly, they were bathed for 20 minutes in 50% (v/v) formamide/2X SSC at 50°C, rinsed for 10 minutes in 2X SSC, incubated in RNAse buffer with 25 µg/ml RNAse A (Sigma, St. Louis, MO) at 37°C for 30 minutes, rinsed twice for 10 minutes in 2X SSC, and then incubated in 50% formamide/2X SSC (v/v) for 20 minutes at 50°C. Finally, the slides were rinsed in 2X SSC, and then dehydrated in a series of increasing concentrations of ethanol. Dried slides were apposed to Kodak Biomax MR X-ray film (Eastman Kodak, Rochester, NY) for 17 days (AVPV and MPOA) or 14 days (VMN and ARC). After exposure, films were developed by using a Konica SRX-101A film processor (Konica, Tokyo, Japan).
To confirm the specificity of the labeling as well as the results obtained from the films, the slides were dipped in NTB3 emulsion (Kodak) to detect ERα mRNA signal, kept at 4°C for 50 days (AVPV and MPOA) or 42 days (VMN and ARC), and then developed in Dektol developer and Kodak fixer (Kodak) according to the user manual for quantification of the resulting silver grains.
ERβ ISHH
To determine the expression level of ERβ in neonatal rat brain, we performed single-label ISHH as described above on a second set of sections. 35S-UTP-labeled cRNA antisense and sense probes for ERβ mRNA were prepared by using 501-bp cDNA templates (Table 1) with Sp6 and T7 RNA polymerase, respectively. In vitro transcription and ISHH procedures were performed as described above. Slides were exposed to film for 36 days (AVPV and MPOA) or 25 days (VMN and ARC). Because transcript abundance was relatively low, the slides were then dipped in NTB3 emulsion to ensure the specificity of the labeling. Slides were exposed for 105 days (AVPV and MPOA) or 73 days (VMN and ARC) and then developed.
All of the emulsion-dipped slides were then counter-stained with Mayer’s hematoxylin (Sigma) to help visualize the cell-specific silver grain clusters. Slides were dipped in staining solution for 3 minutes, rinsed in tap water for 10 minutes, and then dehydrated by a series of increasing concentrations of ethanol. Finally, the slides were dipped in xylene (Sigma) for 30 minutes and mounted with DPX mountant (Sigma).
Kiss1 ISHH
The expression level of Kiss1 was identified in a third set of coronal sections. The probe was generated from a 318-bp fragment (Table 1), and T7 and Sp6 RNA polymerase were used for antisense and sense probes synthesis, respectively. Probe synthesis and ISHH procedures were as described above. Because a recent publication showed that kisspeptin protein in mouse RP3V is not detectable at PND 10 (Clarkson and Herbison, 2006), we did not use the PND 4 brains for these experiments, but rather added a group of PND 11 males and females to quantify labeling in the anterior hypothalamus. For the assessment of labeling in the VMN and ARC, only brains obtained from PND 0, 2, 7, and 19 animals were used. Films were exposed to the labeled sections for 30 days (AVPV and MPOA) or 24 days (VMN and ARC). After exposure to film, slides were emulsion-dipped and exposed 106 days (AVPV and MPOA) or 91 days (VMN and ARC), and then developed. All of the emulsion-dipped slides were counterstained with Mayer’s hematoxylin as described above.
Landmark identification, image production, and analysis
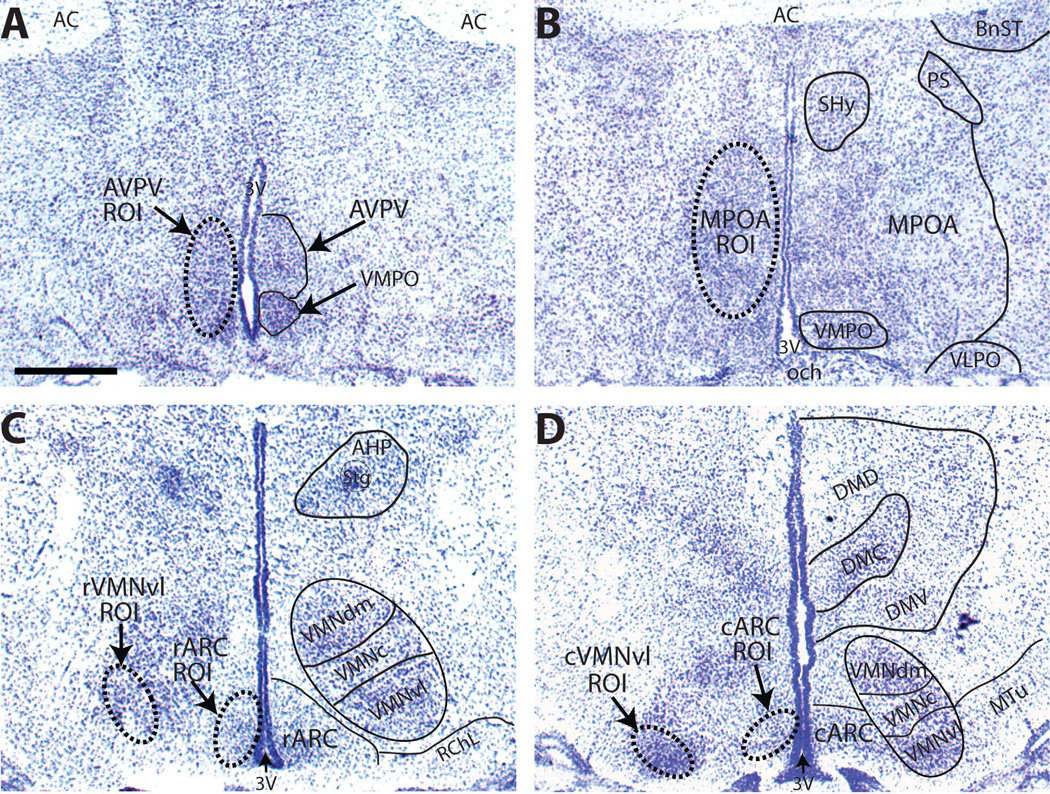
All measurements were made by an investigator blind to the treatment groups. Identification of each region of interest across neonatal development was conducted with the aid of a standard rat brain atlas (Paxinos and Watson, 2007), an in-house library of age matched, Nissl-stained hypothalamic sections (one for each sex), and adjacent counterstained sections when available (Fig. 1). The AVPV (Fig. 1A) abuts the third ventricle (3V), just rostral to the anterior portion of the optic chiasm (och), and contains more darkly stained cells with higher packing density than the surrounding area, which is relatively cell sparse (Gu and Simerly, 1997). For quantification of gene expression in the AVPV, three unilateral sections per animal were obtained, one from the more rostral aspect (defined by the presence of the optic chiasm and the size and shape of the 3V), one midlevel section, and one from the more caudal aspect (defined as the region just anterior to the convergence of the anterior commissure). The MPOA (Fig. 1B) was identified by using conventional landmarks (Hines et al., 1985; Ju and Swanson, 1989) including the shape of the 3V, anterior commissure, and optic chiasm; three unilateral sections per animal were used for the analysis. The subdivisions of this region are not well defined in neonates so they were not considered in the analysis. The VMN and ARC were identified by using well-defined landmarks (Chronwall, 1985; Sa and Madeira, 2005),as we have done previously (Patisaul et al., 2008) including the presence of the median eminence, the shape of the 3V, and the location of the fornix.
Figure 1.
Representative NISSL-stained sections from PND 2 females depicting the anatomical landmarks (right side of each panel) used to identify each brain area of interest and the corresponding sampling template created to define the region of interest (ROI; dashed oval; left side of each panel) and used to quantify the autoradiographic signal within that brain area. Because the size and position of each brain area change with age, an ROI was developed for each age and then used for all sections, regardless of sex, within that age group. Depicted is a midlevel section of the AVPV (A), and MPOA (B), a representative section containing both the rVMNvl and the rARC (C), and a section containing both the cVMNvl and cARC (D). All sections were obtained from animals in our existing colony to match against the hybridized tissue used for the present studies. For abbreviations, see list. Scale bar = 500 µm in A (applies to A–D). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Cells within the VMNvl and ARC are more densely packed than in the surrounding regions,and both are reasonably well defined by the presence of ERα labeling because the surrounding region is devoid of ERα (Shughrue et al., 1997b; Patisaul et al., 2008). Because these regions are larger than the AVPV and MPOA, the rostral and caudal aspects were analyzed separately and designated the rostral VMN (rVMN) and rostral ARC (rARC), and the caudal VMN (cVMN) and caudal ARC (cARC), respectively (Fig. 1C,D). For both the VMN and ARC, the rostral region was defined by the emergence of the median eminence, the height of the 3V, and the slight elongation of the optic tract. The anterior edge of the caudal region was defined by the loss of the lateral part of the retrochiasmatic area (RChL) and thus the more ventral position of the VMN. Five anatomically matched, unilateral sections were used for the rARC, rVMN, cARC, and cVMN.
X-ray film images were viewed by using a monochrome QICAM 1394 12-bit camera (QImaging, Surry, BC, Canada) mounted above a light-box (Northern Lights; Bert-hold, Australia). Relative levels of ERα, ERβ, and Kiss1 mRNA were first assessed by optical density from the film autoradiograms by using the digital densitometry application of the MCID Core Image software program (InterFocus Imaging, Cambridge, UK) and following procedures similar to what we and others have described previously (Kuhar et al., 1986; Patisaul et al., 1999). For each brain area, a sampling template encompassing the region of interest (ROI) was created and used for all sections to standardize the area examined (Fig. 1A–D, left panel in each). As described above, ROIs were defined with the aid of a standard rat brain atlas (Paxinos and Watson, 2007), an in-house library of age-matched, Nissl-stained hypothalamic sections females only), and adjacent counterstained sections (when available). Female sections were used to define the ROI because the AVPV is larger in females than males by peripuberty (Simerly, 2002).
Because the size of each region changed with age (being larger in the older animals), the ROI was slightly different for each age group. For the AVPV, for example, an oval encompassing this region (defined by using Nissl-stained female sections) was created for each age and then used to analyze all sections in that age group (both sexes; Fig. 1A). Because ERβ expression was primarily confined to the periventricular region of the AVPV and MPOA, the ROI was narrowed slightly to encapsulate just this region. ROI and background levels were measured unilaterally from anatomically matched sections. For quantification of ERα and ERβ three serial sections per animal were used for the AVPV and MPOA analysis, and five serial sections per animal were used for each subregion of the VMN and ARC. For quantification of Kiss1, the R3PV and ARC were analyzed; three serial sections per animal were used for the RP3V, and four sections per animal were used to quantify signal in the ARC. The resulting values for each brain section were then averaged to obtain a representative measurement (for that region) for each animal. Average background levels were then subtracted to obtain a final value. Optical densities were converted to nCi/g tissue equivalents by using a “best fit” curve (5th degree polynomial) generated from autoradiographic 14C microscales (Amersham Life Sciences, Arlington Heights, IL). In all cases, signal was within the limits of the curve.
To verify the results observed on the autoradiograms, the counterstained, emulsion-dipped slides (both sexes, all ages) containing ERα label in the AVPV and MPOA were imaged by using a Retiga 2000R color camera (QImaging) under the 20× plan apo objective of our Leica 5000DM microscope, and the silver grains within each subregion were counted by using the grain counter tool of the MCID Image software program. To ensure consistency, unilateral measurements were taken from three sections per animal using the same sized ROI as was used for the densitometry analysis. The same landmarks and anatomical criteria were used to define the sampling area. For all other dipped slides (e.g., ERβ labeling in the AVPV), silver grain analysis was only used to confirm the presence and location of specific labeling (but not quantified). The presence of dense clusters confined within discreetly labeled nuclei was considered to be confirmation of label specificity. These slides were also used to confirm that the region selected for the autoradiographic analysis was appropriate and to ensure that the sex differences observed on the autoradiograms were also apparent on the dipped slides. This was particularly critical for regions in which the density of cells expressing the gene of interest was low or diffuse.
To obtain suitable images for publication, all selected images were cropped, and the brightness and contrast were adjusted slightly only to improve the visibility of the landmarks by using the 2010 version of Microsoft Picture Manage. Silver grain images were similarly adjusted by using Microsoft Picture Manager, to highlight silver deposition and counterstaining. All adjustments were minor so as to provide a representative overview of the observations.
Statistical analysis
Only sections that were intact and had consistent labeling were used for the statistical analysis (n = 3–7; sample sizes indicated in the figures). All datasets were first tested for homogeneity of variance and normality by using GraphPad Prism (Graphpad Software, La Jolla, CA), and all met these criteria. For each region, and each gene of interest, the data were first analyzed by two-way analysis of variance (ANOVA) with sex and age as factors. A significant interaction was found in nearly all cases. Thus the temporal pattern of expression levels was then analyzed within each sex by one-way ANOVA, with age as the factor, and followed up with Dunnett’s multiple comparison post hoc test, with PND 0 as the control group, to identify specific differences between age groups. A t-test was then used at each time point to determine whether a significant sex difference in mRNA expression was present. When a significant interaction was not identified by two-way ANOVA, the significant main effects were followed up individually. For age, the data were analyzed by one-way ANOVA with Dunnett’s multiple comparison post hoc test. For sex, a t-test was used at each age to test for sex differences in expression. Similar to optical density analysis, AVPV and MPOA ERα silver grain labeling was first compared by two-way ANOVA, then by one-way ANOVA within each sex, with age as the factor, followed by Dunnett’s post hoc test when appropriate to identify age-specific differences. A t-test was then performed at each age to identify sex differences. All analyses were two-tailed, and results were considered significant when P ≤ 0.05.
RESULTS
Confirmation of the specificity of the ERα, ERβ, and Kiss1 cRNA probes
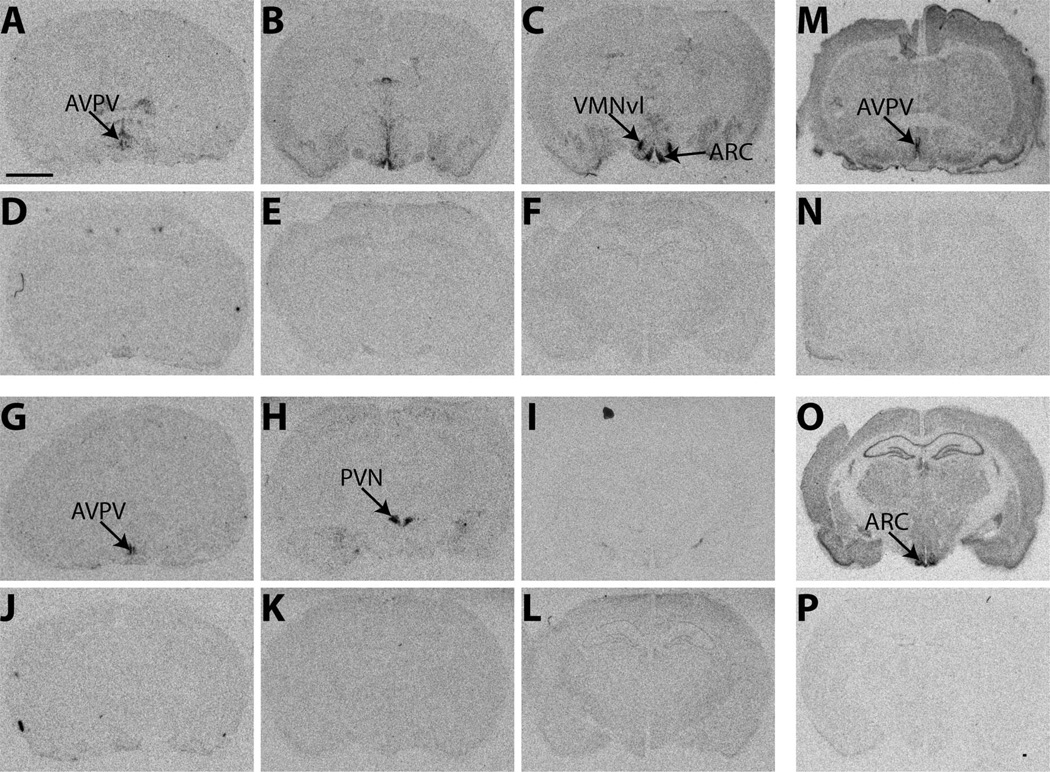
Coronal brain sections obtained previously from three adult LE rats (20 µm) were used for an initial ISHH test. As anticipated, all three sense strand probes produced only background levels of signal (Fig. 2D–F,J-L,N,P). Specificity of the antisense labeling of ERα was confirmed by the presence of robust signal in the AVPV (Fig. 2A), ARC, and VMN (Fig. 2C), but not in the paraventricular hypothalamic nucleus (PVN), a region known to be largely devoid of ERα, except in the parvocellular subdivisions (Shughrue et al., 1996; Hrabovszky et al., 1998; Shughrue and Merchenthaler, 2001a). The ERβ antisense probe produced strong labeling in the AVPV (Fig. 2G) and PVN (Fig. 2H), but no obvious signal in the ARC (Fig. 2I), a region previously shown to be nearly devoid of ERβ (Shughrue et al., 1996). The Kiss1 antisense cRNA probe showed strong signal in the RP3V and ARC (Fig. 2M,O), but not elsewhere, an expression pattern that is consistent with what has been described previously in rodents (Smith et al., 2006; Kauffman et al., 2007b). Importantly, it did not produce signal in the dorsomedial nucleus of the hypothalamus (DMH), a region that contains related proteins, but not Kiss1.
Figure 2.
Representative autoradiographic images depicting the specificity of the 35S-labeled cRNA probes of ERα, ERβ, and Kiss1 in the adult female rat brain. A–C: Labeling for ERα, was visible in the AVPV (A), ARC (C), and VMNvl (C), all regions known to contain ERα, but not the PVN (B), a region known to be devoid of ERα. G–I: Labeling for ERβ was robust in the AVPV (G) and PVN (H) but not the VMNvl (I), a region devoid of ERβ in adulthood. M,O: Kiss1 labeling was confined to the AVPV (M) and, caudally, within the RP3V, along with the ARC (O), as expected. No signal above background was present on any of the autoradiographs generated with the sense probes. D–F: ERα sense labeling. J–L: ERβ sense labeling. N,P: Kiss1 sense labeling. For abbreviations, see list. Scale bar = 2 µm in A (applies to A–P).
Sexually dimorphic expression levels of ERα mRNA in the POA
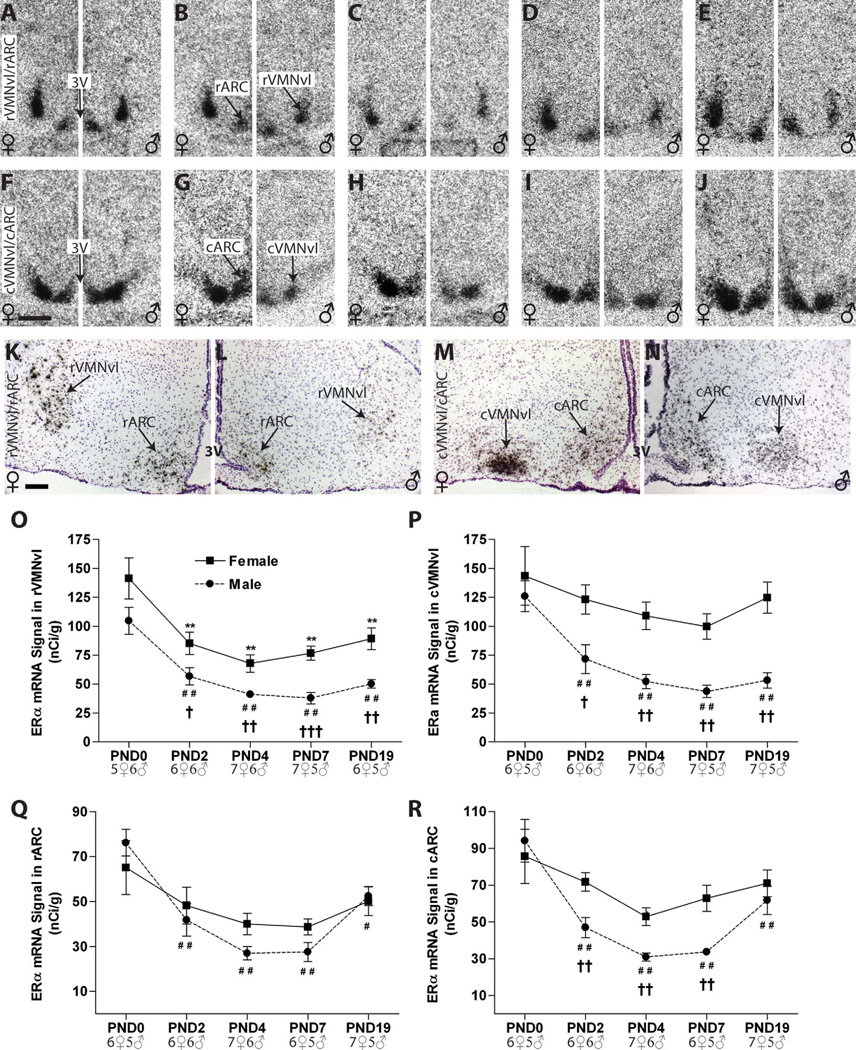
All results are summarized in Table 2. In both sexes (age PND0–19), ERα expression was detected in all sub- regions examined, and the intensity of ERα labeling was generally higher in the MPOA than the AVPV (Figs. 3, 4), but in both subregions a clearly defined, sexually dimorphic temporal pattern of expression was observed on both the autoradiograms (Fig. 3) and the emulsion-dipped slides (Fig. 4), with neonatal females having higher expression levels than males.
TABLE 2.
Sex Differences in the Expression of ERα, ERβ, and Kiss1 mRNA in the Postnatal Rat Hypothalamus
| Preoptic area |
Mediobasal hypothalamus |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Age (PND) | AVPV | MPOA | rARC | cARC | rVMNvl | cVMNvl | |||
| ERα | 0 | F >M | F = M | F = M | F = M | F = M | F = M | |||
| 2 | F >M | F >M | F = M | F>M | F>M | F>M | ||||
| 4 | F>M | F>M | F = M | F>M | F>M | F>M | ||||
| 7 | F = M | F>M | F = M | F>M | F>M | F>M | ||||
| 19 | F = M | F>M | F = M | F = M | F>M | F>M | ||||
| ERβ | 0 | F<M | F = M | ND | ND | F>M | F>M | |||
| 2 | F = M | F = M | ND | ND | F>M | F>M | ||||
| 4 | F = M | F = M | ND | ND | F>M | F>M | ||||
| 7 | F = M | F = M | F = M | F = M | F = M** | F>M | ||||
| 19 | F = M* | F>M | F = M | F = M | F>M | F = M | ||||
| rRP3V | cRP3V | ARC | VMN | |||||||
| Kiss1 | 0 | ND | ND | F = M | ND | |||||
| 2 | ND | ND | F>M | ND | ||||||
| 7 | ND | ND | F>M | NQ | ||||||
| 11 | ND | F = M | NA | NA | ||||||
| 19 | Female only | F>M | F = M | NQ | ||||||
ND, not detectable; NQ, not quantified (very weak signal); NA, not available. For other abbreviations, see list.
P = 0.09.
P = 0.06.
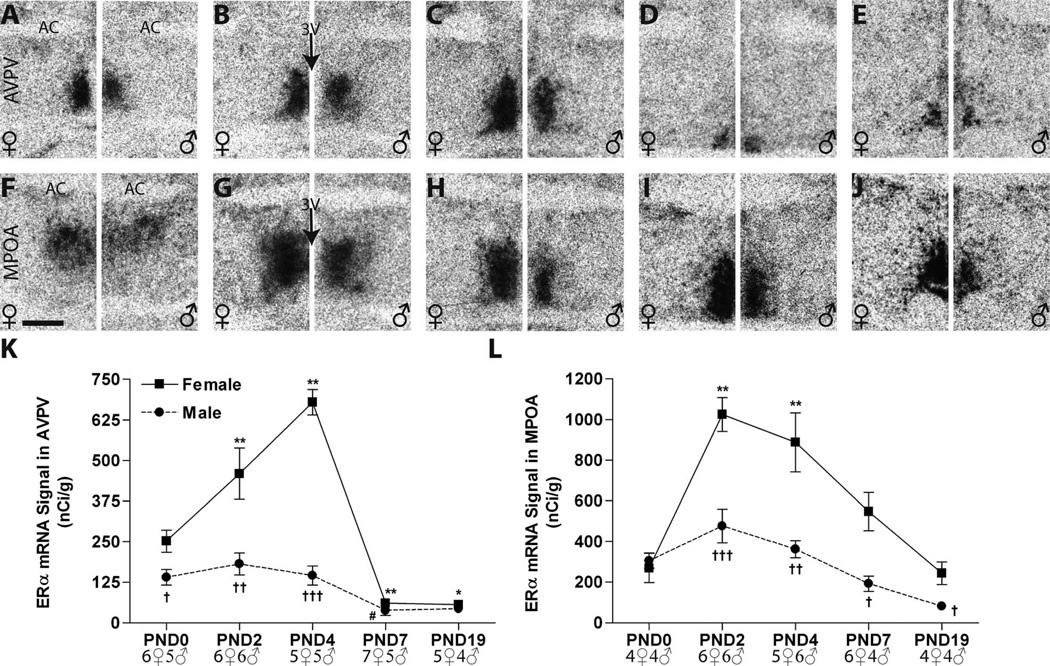
Figure 3.
Representative autoradiographic images depicting ERα mRNA signal in the neonatal rat anterior hypothalamus of both sexes (females on the left side of each panel, males on the right). A–E: Within the AVPV, labeling intensified in the females (left sides) from birth (A) through PNDs 2 (B) and 4 (C), then appreciably declined by PND 7 (D), and remained low on PND 19 (E). In males (right sides), signa was also robust, but intensity remained relatively flat across the neonatal period until PND 7. F–J: Signal was readily detectable at all ages examined in the MPOA but was appreciably more intense within the first 4 days of life (F–H). A pronounced sex difference was observable on all days except PND 0 (G–J). K,L: Expression intensity, as measured by optical density, was sexually dimorphic in both the AVPV and MPOA by PND 2 but declined in magnitude after PND 4 to where the sex difference was abolished by PND 7 in the AVPV but remained significant in the MPOA through PND 19. Significant differences in expression compared with PND 0 levels are represented by (*) P ≤ 0.05, (**) P ≤ 0.01 for the females, and (#) P ≤ 0.05, for the males. Statistically significant sex differences in expression at each age are represented by (†) P ≤ 0.05, (††) P ≤ 0.01, and (†††) P ≤ 0.001. The sample size for each group is presented in the graphs, and the data points represent mean ± SEM. For abbreviations, see list. Scale bar = 500 µm in F (applies to A-J).
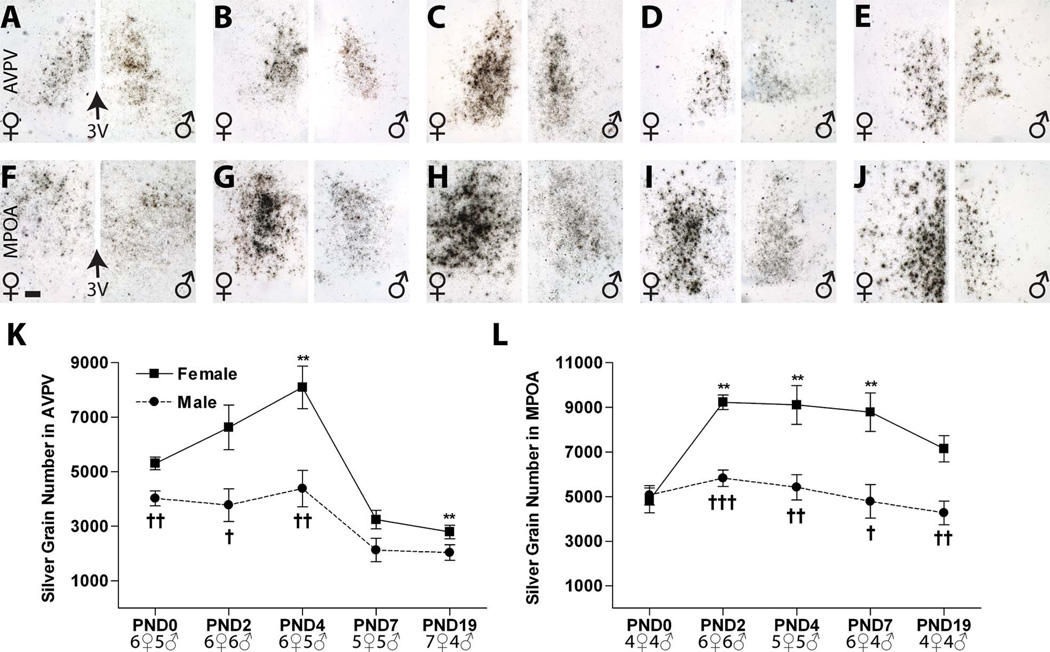
Figure 4.
Deposition of silver grain labeling for ERα mRNA in the neonatal rat anterior hypothalamus (females on the left side of each panel, males on the right). A–J: The representative photomicrographs depict labeling in the (A–E) AVPV and (F–J) MPOA from PND 0–19. In all cases, labeling consisted of discrete clusters of silver grains with very little background labeling. Notably (F), on PND 0, label within the MPOA was largely confined to the dorsal portion of the MPN. K,L: Quantification of silver grain deposition revealed a significant sex difference in ERα mRNA levels by PND 2 in both regions. This sex difference was eliminated by PND 7 in the AVPV, but persisted through PND 19 in the MPOA. Significant differences in expression compared with PND 0 levels are represented by (**) P ≤ 0.01, for the females. (No significant change with age was observed in the males.) Significant sex differences in expression at each age are represented by (†) P ≤ 0.05, (††) P ≤ 0.01, and (†††) P ≤ 0.001. The graphs depict mean ± SEM, and the sample size at each age is provided at the bottom. For abbreviations, see list. Scale bar = 100 µm in F (applies to A–J). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
AVPV. The distribution of ERα mRNA in the AVPV was robust and surrounded the third ventricle (3V, Fig. 3)
Intense labeling for ERα was observed in the AVPV from PND 0 to PND 4 in both males and females (Fig. 3A–C). Two-way ANOVA identified a significant effect of sex (F(1,44) = 65.39, P ≤ 0.0001) and age (F(4,44) = 37.40, P ≤ 0.0001) plus a significant interaction (F(4,44) = 17.11, P ≤ 0.0001). Follow-up one-way ANOVA revealed a significant effect of age in females(F(4,24) = 37.00, P ≤ 0.001) and males (F(4,20) = 6.10, P ≤ 0.002) (Fig. 3K). In females, (Fig. 3A–E, left panels) ERα mRNA levels were significantly increased on PNDs 2 and 4, compared with PND 0, and then sharply declined to significantly lower levels compared with PND 0, on PNDs 7 and 19. In males (Fig. 3A–E, right panels), levels did not statistically differ from PND 0 through PND 4, and then dropped, but levels were only significantly lower compared with PND 0 on PND 7. Thus, ERα expression levels in the AVPV were sexually dimorphic from PNDs 0 to 4 (P ≤ 0.04, P ≤ 0.01, P ≤ 0.0001, respectively), with significantly higher levels in females, but this sex difference disappeared by PND 7 and remained absent on PND 19 (Fig. 3K, Table 2).
Quantification of deposited silver grain labeling following emulsion dipping confirmed the results obtained from the autoradiograms (Fig. 4A–E,K). ERα signal in the AVPV was broadly distributed around the third ventricle in both sexes, and the temporal pattern of ERα mRNA levels was also similar. As with the film analysis, two-way ANOVA revealed a significant effect of age (F(4,45) = 18.6, P ≤ 0.0001) and sex (F(1,45) = 32.3, P ≤ 0.0001) as well as a significant interaction (F(4,45) = 2.80 , P ≤ 0.04). In females (Fig. 4A–E, left panels), a significant effect of age was again found (F(4,25) = 17.11, P ≤ 0.0001), with levels rising through PND 4 and then dropping to significantly lower levels than PND 0 by PND 19. In the males (Fig. 4A–E, right panels), there was also a significant effect of age (F(4,20) = 4.45, P ≤ 0.01), with levels relatively flat and then significantly decreased compared with PND 0 on PNDs 7 and 19. As observed in the autoradiograms, quantification of the silver grains revealed sexually dimorphic expression of ERα on PNDs 0, 2, and 4 (P ≤ 0.001, P ≤ 0.02, P ≤ 0.006, respectively; Fig. 4K, Table 2).
MPOA. The autoradiographs revealed that the distribution of ERα mRNA within the MPOA changed with age in both sexes (Fig. 3F–J,L)
On PND 0 it was largely restricted to the dorsal part of the medial preoptic nucleus (MPN; Fig. 3F). By PNDs 2 and 4, ERα labeling was readily observed in both the medial and lateral compartments of the MPN (Fig. 3G,H). On PNDs 7 and 19, ERα signal was mainly restricted to the medial part of the MPN (Fig. 3I,J). A significant effect of age (F(4,39)= 16.80, P ≤ 0.0001) and sex (F(1,39) = 34.43, P ≤ 0.0001) as well as a significant interaction (F(4,39) = 4.38, P ≤ 0.005) was identified by two-way ANOVA. In females, age was a significant main effect (F(4,20) = 12.17, P ≤ 0.0001), with significantly elevated levels on PNDs 2 (P ≤ 0.01) and 4 (P ≤ 0.01) (Fig. 3K). In males, there was also a significant effect of age (F(4,19) = 7.12, P ≤ 0.001) the magnitude of which was considerably less pronounced than in females (Fig. 3K) and was largely driven by the difference between expression levels on PNDs 2 and 19. A significant sex difference in ERα mRNA levels was observed on PNDs 2, 4, 7, and 19 (P ≤ 0.001, P ≤ 0.004, P ≤ 0.02, and P ≤ 0.04, respectively), with the magnitude of this sex difference diminishing with age, largely attributable to markedly declined expression levels in females (Fig. 3K, Table 2).
Analysis of the dipped slides confirmed the autoradiographic finding that ERα mRNA is mainly confined to the dorsal MPN on PND 0 in both sexes (Fig. 4F). Moreover, silver grain quantification revealed a similar, but not identical, pattern of ERα mRNA signal in both sexes (Fig. 4F–J,L; Table 2). Consistent with the autoradiographic results, two-way ANOVA identified a significant effect of age (F(4,38) = 5.82, P ≤ 0.001) and sex (F(1,38) = 46.47, P ≤ 0.0001) as well as a significant interaction (F(4,38) = 3.62, P ≤ 0.014). One-way ANOVA revealed a significant effect of age in females (F(4,20) = 6.32, P ≤ 0.002, Fig. 3F) with elevated levels on PNDs 2 and 4, but also 7 (P ≤ 0.01 compared with PND 0), and then a gradual decline to PND 0 levels by PND 19 (Fig. 4L). In contrast, no significant main effect of age on ERα mRNA levels in the male MPOA was detected with this method, because PND 2 levels were not found to be as high, although the temporal expression pattern was similar to that observed on the autoradiograms (Figs. 3L, 4L). The sex difference in expression on PNDs 2–19 (P ≤ 0.04 or below; Fig. 3F), however, was confirmed. Collectively, the results from both analytical assessments indicate that, during the early neonatal period, ERα expression is higher in females compared with males, and then levels decline in both sexes, becoming less dimorphic by PND 19.
Sexually dimorphic expression levels of ERα mRNA in the postnatal mediobasal hypothalamus
In the mediobasal hypothalamus, ERα mRNA was found in both the VMN and ARC (Fig. 5). In the VMN, ERα mRNA was mainly confined to the ventrolateral (vl) division. In the ARC, ERα signal was readily detectable but was less pronounced than that in the VMN. In nearly all mediobasal regions explored here, the intensity of ERα labeling changed with age (Fig. 5O–R) with females having significantly more labeling than males at nearly all ages examined.
Figure 5.
Autoradiographs depicting ERα mRNA labeling in the ARC and VMNvl of male and female neonatal rats (females on the left side of each panel, males on the right). A–J: Labeling in the (A–E) rVMN and rARC and (F–J) cVMN and cARC was robust and tightly confined to the regions of interest in both sexes at all ages examined. K–N: Representative hemotoxylin-counterstained sections depicting deposition of silver grain labeling for ERα in the mediobasal hypothalamus of PND 19 females (K,M) and males (L,N). Signal in the VMN was appreciably more robust than in the ARC. Label was visible as dark, discrete clusters of silver grains localized on counterstained nuclei. Qualitative assessment revealed that the temporal and sexual dimorphic pattern of labeling was congruous to that observed on the autoradiographs. O–R: A significant sex difference in ERα expression was present in all regions except the rARC by PND 2. By PND 19, the sex difference in the VMN remained but was lost in the ARC. Expression intensity of ERα mRNA in the rVMNvl of both sexes decreased between PNDs 0 and 2 then remained relatively stable. In the cVMNvl, levels declined with age in males but remained comparatively flat in females. In the male ARC, temporal expression of ERα was “U-shape” with the nadir on or near PND 7. In contrast, expression levels remained stable in females, with no significant effect of age in either subregion. A main effect of sex was identified in the cARC, but not the rARC, with females having higher levels of expression than males on PNDs 2, 4, and 7 in the cARC. Significant differences in expression compared with PND 0 levels are represented by (**) P≤ 0.01 for the females, and (#) P≤ 0.05, (###) P≤ 0.01, for the males. Significant sex differences in expression are represented by (†) P≤ 0.05, (††) P≤ 0.01, and (†††) P ≤ 0.001 at each age. The graphs depict mean ± SEM, and the sample size is provided for each age. For abbreviations, see list. Scale bar = 500 µm in F (applies to A–J); 100 µm in K (applies to K–N). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
VMN: ERα mRNA signal was mainly confined to the VMNvl and was highest on PND 0 in both sexes
Two-way ANOVA of rVMNvl labeling revealed a significant effect of age (F(4,49) = 19.63, P ≤ 0.0001) and sex (F(1,49) = 36.68, P ≤ 0.0001) but no significant interaction (P = 0.9). Compared with the day of birth, expression levels were significantly lower from PND 2 to 19 in both sexes (Fig. 5B–E; females on the left, males on the right). Sexually dimorphic expression was observed from PND 2 through PND 19, with significantly higher levels in females compared with age-matched males (P ≤ 0.04, P ≤ 0.01, P ≤ 0.001, and P ≤ 0.005, respectively; Fig. 5O, Table 2). Silver grain deposition on the emulsion-dipped slides revealed that the distribution of label was in discrete clusters, but was more diffusely distributed in the males, and qualitatively confirmed the autoradiographic results (Fig. 5K,L).
Temporal expression levels were similar in the cVMNvl, with two-way ANOVA revealing a significant effect of age (F(4,50) = 6.54, P ≤ 0.0003) and sex (F(1,50) = 35.49, P ≤ 0.0001) but no significant interaction (P = 0.4). In females, expression levels were robust and remained relatively stable, with no significant main effect of age (Fig. 5F – J). In males, however, ERα mRNA levels decreased with age (Fig. 4F–J, right panels) (F(4,22) = 15.32, P ≤ 0.0001), with levels becoming statistically lower than PND 0 levels by PND 2 (P ≤ 0.01). These divergent temporal expression patterns resulted in a significant sex difference in expression from PNDs 2 through 19 (P ≤ 0.02, P ≤ 0.002, P ≤ 0.003, and P ≤ 0.002, respectively), with levels significantly higher in females (Fig. 5P, Table 2). Visual inspection of silver grain deposition within the ARC confirmed these findings (Fig. 5K – N).
ARC. ERα expression was less robust in the ARC compared with the VMN, but the temporal pattern of expression was similar in both sexes (Fig. 5, Table 2)
Two-way ANOVA revealed a significant effect of age (F(4,49) = 10.83, P ≤ 0.0001) but not sex (F(1,49) = 0.68, P = 0.410) in the rARC and no significant interaction (F(4,49) = 1.14, P = 0.35). In females, expression levels remained relatively unchanged with age, but there was a significant effect of age (F(4,22) = 14.54, P ≤ 0.0001) in males, with levels declining overtime and then recovering slightly by PND 19 (Fig. 5Q). Levels were significantly lower compared with the day of birth beginning on PND 2 (P ≤ 0.01). In the cARC, however, both age (F(4,49) = 11.72, P ≤ 0.0001) and sex (F(1,49) = 9.89, P ≤ 0.003) were significant factors but there was only a trend for a significant interaction (F(4,49) = 1.95, P ≤ 0.12). In females, there was only a trend for an effect of age in the cARC (F(4,27) = 2.13, P = 0.1; Fig. 5F–J, right panels). In contrast, there was a significant effect of age in the males (F(4,22) = 15.32, P ≤ 0.0001), with levels significantly lower from PND 2 through PND 7 compared with PND 0 but slightly recovering by PND 19. In the cARC, a sex difference emerged on PND 2 (P ≤ 0.01) and remained on PNDs 4 (P ≤ 0.003) and 7 (P ≤ 0.005), but disappeared by PND 19, due largely to increased signal in the males (Fig. 4R, Table 2). Visual inspection of the deposited silver grains confirmed the overall expression patterns observed on the autoradiographs (Fig. 5K–N).
Sexually dimorphic expression levels of ERβ mRNA in the postnatal preoptic area
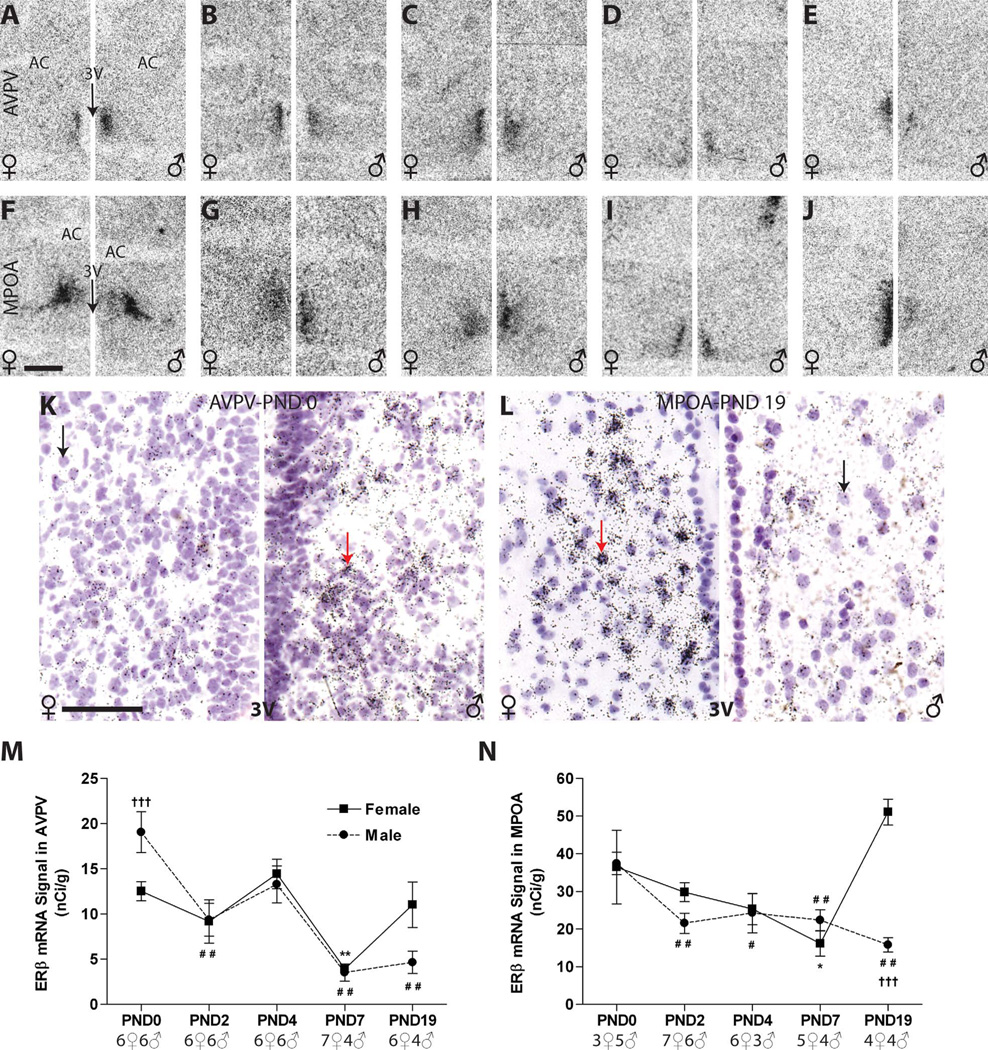
ERβ signal was appreciably lower than ERα in the anterior hypothalamus, but the distribution of ERβ was similar in both sexes at all ages examined (Fig. 6). ERβ mRNA signal in the AVPV (Fig. 6A – E) and MPOA (Fig. 6F–J) was found mainly in the periventricular part except on PND 0, when ERβ was distributed in the dorsal MPN (Fig. 6F). This distribution pattern is similar to what was observed for ERα (Figs. 3F, 4F). Examination of silver grain deposition confirmed the distribution and signal intensity observed on the autoradiograms, but was not quantified (Fig. 6K,L).
Figure 6.
A–J: Autoradiographs showing ERβ mRNA expression in the RP3V of male and female neonatal rats (females on the left side of each panel, males on the right). Signal was relatively weak at all ages examined but distinguishable from background and quantifiable by optical density in the AVPV (A–E) and MPOA (F–J). The pattern of autoradiographic labeling was confirmed by observing silver grain deposition on the emulsion-dipped, counterstained slides. K,L: Label appeared as tight clusters (gray arrows) over the counterstained nuclei (black arrows). ERβ expression was sexually dimorphic at birth in the AVPV (A,K,M) with males (right panel of A,K) having significantly higher expression levels than females (left panel of A,K). This sex difference was rapidly lost, and no sex difference was observed until PND 19 (M,N), at which point females had significantly more ERβ signal than males in the MPOA (J,L), but only a trend for a difference was observed in the AVPV (E; P = 0.09). M,N: In both sexes, ERβ expression was notably lower on PND 7, compared with PND 0, but levels recovered in both regions by PND 19 in females. For the females, significant differences in temporal expression levels are represented by (*) P ≤ 0.05 and (**) P ≤ 0.01, compared with PND 0. For the males, significant differences in expression from PND 0 are indicated by (#) P ≤ 0.05 and (###) P ≤ 0.01. Significant sex differences are represented by (†) P ≤ 0.05, (††) P ≤ 0.01, and (†††) P ≤ 0.001. The sample size for each age is indicated on the graphs, and the data points represent mean ± SEM. For abbreviations, see list. Scale bar = 500 µm in F (applies to A–J); 50 µm in K (applies toK,L). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
AVPV. ER β mRNA signal was detectible but less intense, compared with ERα expression, in the postnatal AVPV, with the intensity of staining generally weakest in the anterior region and increasing caudally
Two-way ANOVA revealed a significant effect of age (F(4,47) = 13.57, P ≤ 0.0001) and a significant interaction of age and sex (F(4,47) = 3.08, P ≤ 0.02) but no main effect of sex (P = 0.8). There was a statistically significant effect of age in both males (F(4,21) = 10.55, P ≤ 0.0001) and females (F(4,26) = 5.74, P ≤ 0.002), with levels declining to near the limit of detection in both sexes by PND 7 and remaining low thereafter (Fig. 6M). The only statistically significant sex difference observed was on PND 0 (P ≤ 0.005), with higher levels in males (Fig. 6A,K,M, Table 2), a pattern opposite to what was found for ERα. A trend for a sex difference on PND 19 (P = 0.09) was observed, with females having higher expression than males (Fig. 6M).
MPOA
Two-way ANOVA revealed a significant effect of age (F(4,37) = 6.83, P ≤ 0.0003) and sex (F(1,37) = 9.46, P ≤ 0.004) as well a significant interaction (F(4,37) = 9.06, P ≤ 0.0001). At birth, MPOA ERβ mRNA levels were nearly identical in both sexes (Fig. 6F,N), and there was a statistically significant effect of age in males (F(4,17) = 7.43, P ≤ 0.001) and females (F(4,20) = 8.82, P ≤ 0.0003), with levels generally decreasing over early postnatal development through PND 7 (Fig. 6F–I,N). In males, this drop from PND 0 levels was slight but statistically significant beginning on PND 2 (P ≤ 0.01). In females, by contrast, levels were relatively stable but were significantly lower on PND 7 compared with the day of birth (P ≤ 0.05). Levels then markedly increased (Fig. 6J,N, Table 2), resulting in a significant sex difference on PND 19 (P ≤ 0.001), a difference that was particularly pronounced on the emulsion-dipped slides, which revealed numerous dense clusters of silver grains in females (Fig. 6L, left panel), but not males (Fig. 6L, right panel).
Sexually dimorphic expression levels of ERβ mRNA in the postnatal mediobasal hypothalamus
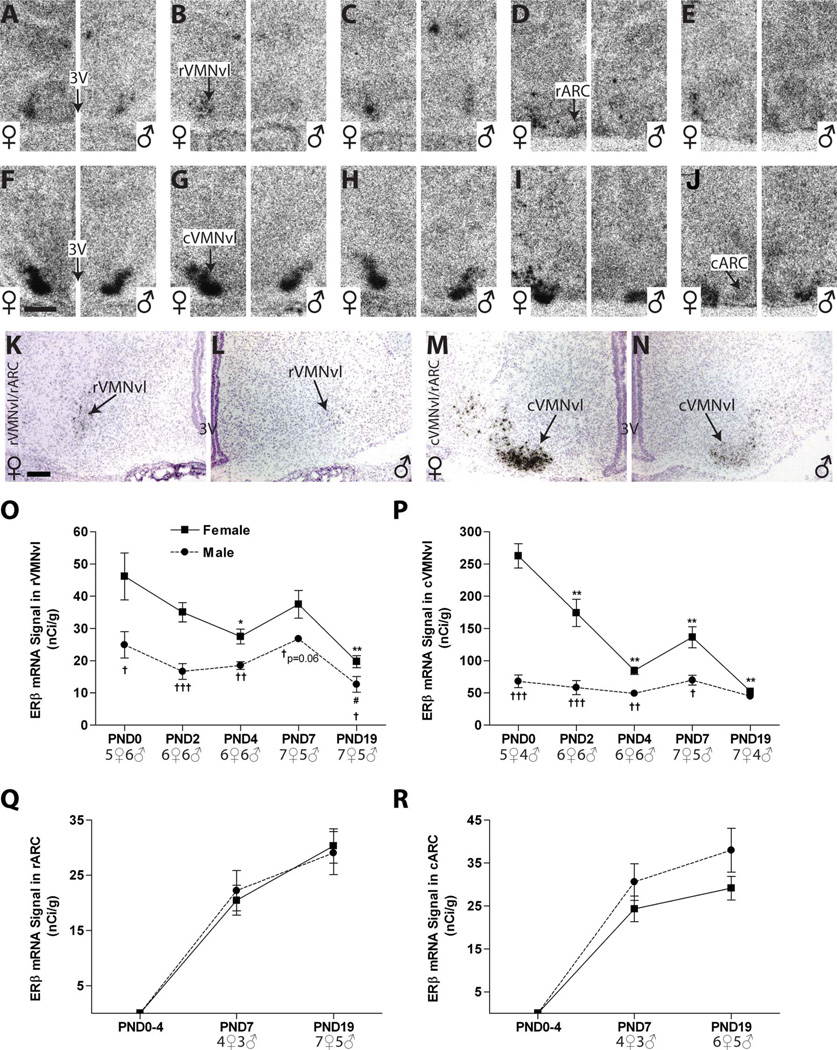
In the VMN, ERβ signal was largely restricted to the VMNvl, and was more intense in the cVMNvl than in the rVMNvl (Fig. 7). No obvious signal on either the autoradiograms or the emulsion-dipped slides was detected in the ARC from PND 0 to PND 4 (Fig. 7A–C,F–H), and only a very weak signal was detected on PNDs 7 and 19 (Fig. 7D,E,I,J).
Figure 7.
Representative autoradiographic and photomicrographs depicting ERβ labeling in the mediobasal hypothalamus (females on the left side of each panel, males on the right). A–J: ERβ signal on the autoradiographs was appreciably weaker in the (A–E) rVMNvl compared with the (F–J) cVMNvl and was virtually absent in the ARC (A–J). K–N: Silver grain deposition following emulsion dipping revealed few, but prominent, silver grain clusters in the rVMNvl on PND 2 and more intense labeling in the cVMNvl (K–N), particularly in females (K,M). O,P: Quantification by optical density revealed a significant sex difference in ERβ expression on PND 0 in both VMNvl subregions, the magnitude of which diminished overtime in the cVMNvl, and was eliminated in both subregions by PND 19. Expression intensity remained relatively flat in the males across neonatal development throughout the VMNvl but declined significantly and steadily in the female cVMNvl. Q,R: No signal was detected in the ARC on the autoradiograms until PND 7. Although expression was very low between PND 7 and 19, it was discernable above background (D,E,I,J) and quantification by optical density found no appreciable sex difference. Significant differences in expression, as measured by optical density, compared with PND 0 are represented by (*) P ≤ 0.05 and (**) P ≤ 0.01 for the females and (#) P ≤ 0.05 for the males. Significant sex differences at each age are indicated by (†) P ≤ 0.05, (††) P ≤ 0.01, and (†††) P ≤ 0.001. The sample size for each age is provided on the graphs, and the data points represent mean ± SEM. For abbreviations, see list. Scale bar = 500 µm in F (applies to A –J); 100 µm in K (applies to K–N). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
VMN
A low level of ERβ mRNA was found in the rVMNvl. Two-way ANOVA revealed a significant effect of age (F(4,49) = 10.15, P ≤ 0.0001) and sex (F(1,49) = 38.79, P ≤ 0.0001), but no significant interaction. In both sexes, expression levels were relatively flat across postnatal development, but there was a significant main effect of age in females (F(4,26) = 6.57, P ≤ 0.001) and in males (F(4,23) = 4.91, P ≤ 0.01) largely attributable to the significant drop in expression on PND 19 (P ≤ 05 in both sexes; Fig. 7E,O). ERβ mRNA levels in the rVMNvl were sexually dimorphic at birth (Fig. 7A), with levels higher in females (P ≤ 0.03), and this sex difference remained through PNDs 2 (Fig. 7K,L; P ≤ 0.001) and 4 (P ≤ 0.001) but was reduced on PNDs 7 (P ≤ 0.06) and 19 (P ≤ 0.04) as expression levels dropped to near the limit of detection in both sexes (Fig. 7O, Table 2).
ERβ mRNA expression levels were considerably more pronounced in the cVMNvl (Fig. 7F – J) than in the rVMNvl, and the temporal pattern of expression was similar, with a more robust sex difference on PNDs 0–7. Two-way ANOVA revealed a significant effect of age (F(4,46) = 24.92, P ≤ 0.0001) and sex (F(1,46) = 106.30, P ≤ 0.0001) as well as a significant interaction (F(4,46) = 16.32, P ≤ 0.0001). There was a significant effect of age (F(4,26) = 29.05, P ≤ 0.0001) in females, with levels decreasing over time, but levels remained stable in males, and no significant impact of age was detected (P = 0.2; Fig. 7P). There was a marked sex difference at birth, with females having significantly higher levels than males through PND 7 (P ≤ 0.0001 on PND 0, P ≤ 0.001 on PND 2, P ≤ 0.0001 on PND 4, and P ≤ 0.01 on PND 7), but this sex difference was abolished by PND 19 (Fig. 7P, Table 2) as expression levels in female diminished. The pronounced sex difference on PND 2 was also observable on the emulsion-dipped slides (Fig. 7M,N).
ARC
In the ARC, there was no visible ERβ mRNA signal on either the autoradiograms or the emulsion-dipped slides from PND 0 to PND 4 in either subregion (Fig. 7A–C,F–H). A very weak signal was observed on PND 7 and PND 19 (Fig. 7D,E,I,J), but was quantifiable in only a small cohort of animals. There was no appreciable sex difference (Fig. 7Q,R, Table 2), and visual inspection of silver grain deposition confirmed that only a few cells contained signal for ERβ mRNA. (Fig. 7K – N depicts the absence of signal in either sex on PND 2.)
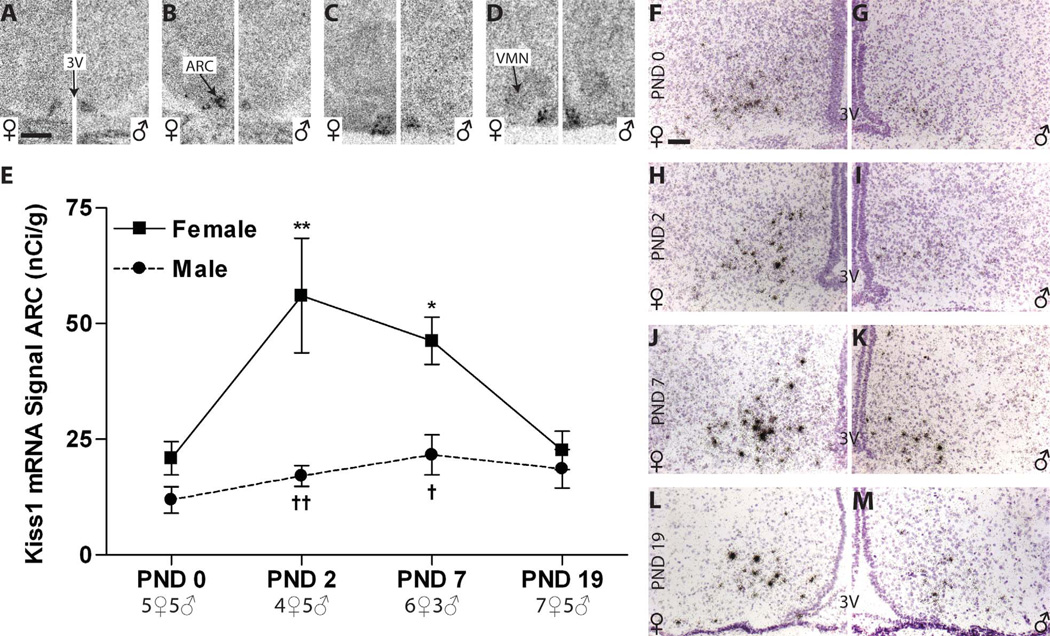
Sexually dimorphic expression levels of Kiss1 mRNA in the postnatal RP3V
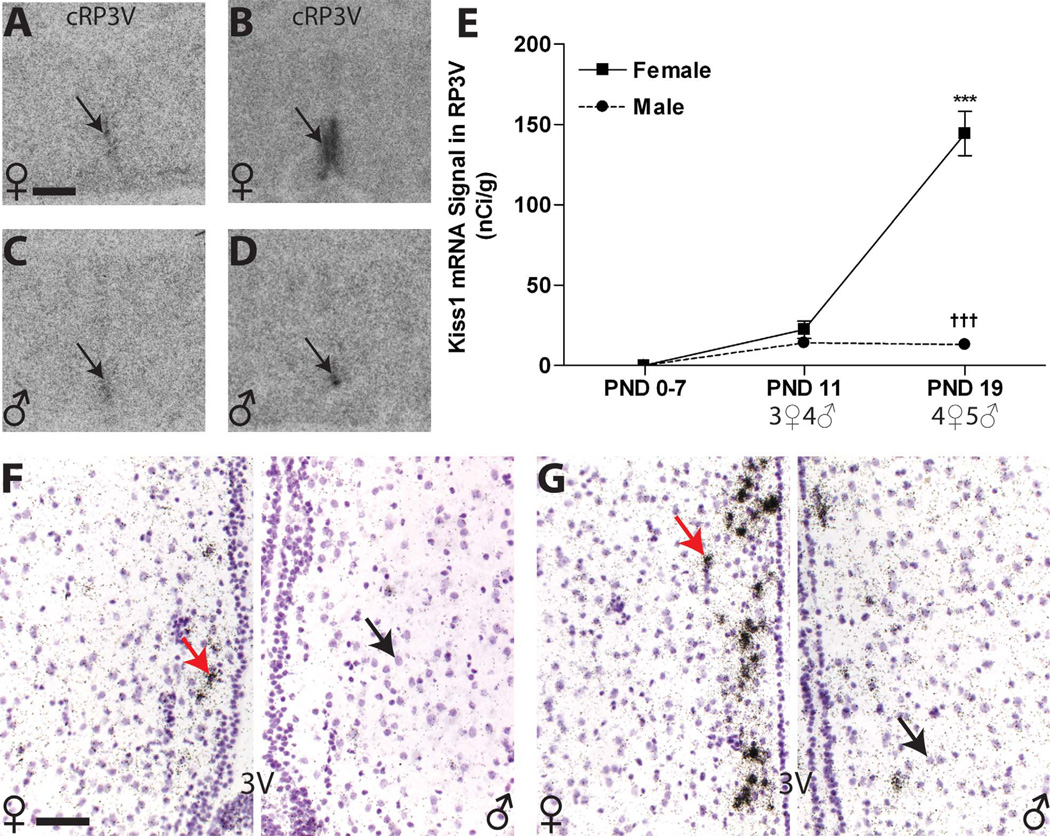
AVPV
Because previous studies had indicated that RP3V Kiss1 levels are not appreciable until around PND 15 (Clarkson and Herbison, 2006), we eliminated the PND 4 time point and added PND 11 (Fig. 8A,C). Additionally, because signal was weak in the AVPV, especially in the most rostral sections, Kiss1 mRNA was quantified in the RP3V as a whole. Kiss1 mRNA signal was not observed from PNDs 0 to 7 in either the females or the males (Fig. 8E), but signal was apparent in both sexes on PND 11 (Fig. 8A,C,E). Thus, expression was only compared between PNDs 11 and 19 (Fig. 8B,D). Two-way ANOVA revealed a significant effect of age (F(1,12) = 66.12, P ≤ 0.0001) and sex (F(1,12) = 87.91, P ≤ 0.0001) as well as a significant interaction (F(1,12) = 68.16, P ≤ 0.0001). By PND 19, females had significantly higher levels than males (P ≤ 0.001), but male levels remained very weak (Fig. 8B,D,E, Table 2). Within the AVPV, silver grain deposition revealed that expression was localized to a relatively small number of cells in both sexes, with females having more clusters than males. The most intense labeling was observed in the periventricular portion of the caudal aspect, along the borders of the third ventricle (Fig. 8F,G). Silver grain deposition was not observed in the rostral portion of the RP3V until PND 19, and only in the females (Fig. 8F). Within the caudal aspect of the RP3V, silver grains were largely confined to the periventricular nucleus and were more densely clustered in females compared with males (Fig. 8G). Thus silver grain deposition confirmed the results obtained from the auto-radiograms and revealed that both the number of Kiss1 cells and the intensity of expression is lower males compared with females
Figure 8.
A–D: Kiss1 signal in the RP3V autoradiographs was not detectable until PND 11 (black arrow; A,C) and sexually dimorphic at PND 19 (B,D,E). E: Quantification of signal by optical density revealed a significant sex difference in expression on PND 19 across the entire RP3V due to a significant increase in female expression. F,G: Qualitative assessment of silver grain deposition on the emulsion-dipped, counterstained slides revealed that, on PND 19, labeling was clustered (arrows on the left sides) around the labeled nuclei (arrows on the right sides) and primarily confined to a narrow region adjacent to the third ventricle (3V) but less robust in the rostral portion of the RP3V (F) in both sexes (female on the left, male on the right) compared with the caudal portion (G; female on the left, male on the right) . Significant differences in expression, compared with PND 0 are represented by (***) P ≤ 0.001 for the females. Significant sex differences at P19 are indicated by (†††) P ≤ 0.001. The sample size for each age is shown on the graph, and the data points represent mean ± SEM. For abbreviations, see list. Scale bar = 500 µm in A (applies to A–D); 50 µm in F (applies to F,G). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Sexually dimorphic expression levels of Kiss1 mRNA in the postnatal mediobasal hypothalamus
VMN
No signal was found on PND 0 or PND 2 (Fig. 9A,B) within the VMN of either sex, but a very weak signal was observed on PNDs 7 and 19 (Fig. 9C,D). It was not strong enough, however, to be quantifiable.
Figure 9.
Kiss1 expression in postnatal rat mediobasal hypothalamus. A–D, F–M: Representative autoradiographs (A–D) and photomicrographs (F–M) revealed discrete signal on all days examined in both sexes. In addition, a weak signal can be seen in the VMN on PNDs 7 and 19 (C,D). E: ARC expression levels increased in females between PND 0 and 7 but then dipped on PND 19. Levels in males remained comparatively unchanged across all days examined. A significant sex difference in Kiss1 expression was present on PNDs 2 and 7 but disappeared, due to declining levels in females by PND 19. Significant differences in expression, compared with PND 0, are represented by (*) P ≤ 0.05 and (**) P ≤ 0.01 for the females. No effect of age was found in the males. Significant sex differences at each age are indicated by (†) P ≤ 0.05 and (††) P ≤ 0.01. The sample size for each age is indicated on the graphs, and the data points represent mean ± SEM. For abbreviations, see list. Scale bar = 500 µm in A (applies to A–D); 50 µm in F (applies to F–M). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
ARC
Expression levels were consistent throughout the ARC so analysis of Kiss1 expression was conducted in the ARC as a whole (rather than as rARC and cARC). Kiss1 mRNA expression on the autoradiograms was visible in both males (Fig. 9A–D, right panels) and females (Fig. 9A–D, left panels) at all ages examined. A significant effect of age (F(3,32) = 6.84, P ≤ 0.0001) and sex (F(1,32) = 25.67, P ≤ 0.0001; Fig. 9E) as well as a significant interaction (F(3,32) = 4.33, P ≤ 0.01) was detected by two-way ANOVA. In females there was a main effect of age (F(3,18) = 7.69, P ≤ 0.002), with levels rising above PND 0 levels across PNDs 2 (P ≤ 0.01) through 7 (P ≤ 0.02) and then declining somewhat by PND 19 (Fig. 9A – E). No effect of age was detected in males (P = 0.3). No sex difference in expression levels was apparent at birth, but by PND 2 and also on PND 7, females had significantly more Kiss1 mRNA than males (P ≤ 0.01, P ≤ 0.02). This sex difference was largely lost by PND 19 (P = 0.08) largely due to declining expression levels in the females (Fig. 9E, Table 2). Visual inspection of silver grain deposition revealed numerous discrete clusters throughout the ARC in both sexes at all ages (Fig. 9F – M).
DISCUSSION
The present study used in situ hybridization to produce the most comprehensive analysis to date of the temporal and sexually dimorphic expression patterns of ERα, ERβ , and Kiss1 mRNA in the anterior and mediobasal hypothalamus during the neonatal critical period and early peri-puberty (PND-19). Overall, ERα expression was more extensive and robust than ERβ expression during the neonatal period, an observation that is consistent with and expands upon what has previously been reported for the rat (MacLusky et al., 1979a,b; White et al., 1979; DonCarlos and Handa, 1994; Kuhnemann et al., 1994; DonCarlos, 1996; Yokosuka et al., 1997; Orikasa et al., 2002; Perez et al., 2003). Moreover, the results highlight that sexually dimorphic ER expression patterns can be transient and region specific, but robust in neonatal life. As expected, based on prior reports obtained in mice (Clarkson and Herbison, 2006; Clarkson et al., 2009), Kiss1 and ER expression largely overlapped in the postnatal rat brain, but their ontogeny differed somewhat, particularly in the anterior hypothalamus.
The results also provide the first description of Kiss1 mRNA expression in the postnatal ARC. ERα and Kiss1 levels were abundant on the day of birth in the ARC, but the general pattern of ERα and Kiss1 expression diverged, with the ontogeny of ERα levels resembling a “U-shape” and Kiss1 ontogeny resembling the opposite. This expression pattern likely results from the sexually dimorphic levels of brain estradiol present in the postnatal period. In the adult, estradiol decreases ARC Kiss1 expression in both sexes, a process that requires ERα (Smith et al., 2005a). In neonatal males, estradiol levels are higher, which may simultaneously drive down ERα expression via autoregulation, and Kiss1 expression. This hypothesis is supported by the observation that, for both ERα and Kiss1, a pronounced sex difference emerged by PND 2 (females > males). This difference was lost by PND 19, a finding consistent with previous studies indicating that ARC Kiss1 levels are not sexually dimorphic post puberty (Kauffman et al., 2007b; Losa et al., 2011). In the VMNvl, females had higher expression levels of both ERs, but temporal expression patterns were sex specific, with ERα levels more notably decreasing with age in males, and ERβ levels decreasing more dramatically with age in females. Moreover, evidence for trace levels of Kiss1 expression in the VMNvl was also found, but this observation will have to be replicated and explored in more detail in future studies. Collectively, the results provide evidence that Kiss1 may play a role in the sexual dimorphic organization of the neonatal brain and suggest that the expression of Kiss1 and ERs might be related and influenced by each other during the neonatal critical period.
Developmental and sexual dimorphic expression of ERα, ERβ, and Kiss1 mRNA in the anterior hypothalamus
The AVPV and MPOA are both sexually dimorphic brain regions that are highly estrogen sensitive (Simerly, 2002). The AVPV is crucial for regulating the preovulatory lutei-nizing hormone (LH) surge, and lesions to this region result in anovulation in female rats (Wiegand et al., 1980; Wiegand and Terasawa, 1982; Ronnekleiv and Kelly, 1986; Petersen and Barraclough, 1989). The MPOA has long been recognized to play an important role in regulating female parental behavior (Numan, 1994; Numan and Stolzenberg, 2009) and male sexual behavior (Arnold and Gorski, 1984; Coolen and Wood, 1999). More recently, the sexually dimorphic expression of Kiss1 within the RP3V, a region encompassing the AVPV and the preoptic periventricular region of the MPOA (Herbison, 2008; Clarkson et al., 2009), indicates that this nucleus is also critical for coordinating gonadotropin-releasing hormone (GnRH) secretion (Kauffman et al., 2007b; Oakley et al., 2009). In the present study, we found that temporal expression levels of, and sex differences in, ERα, ERβ, and Kiss1 gene expression were similar in the AVPV and MPOA.
Prior work using dual-labeled ISHH revealed that almost all Kiss1 neurons within the adult female rat AVPV co-express ERα, whereas only approximately 21% co-express ERβ (Smith et al., 2006). Moreover, knockout studies have shown that activation of ERα appears to be essential to mediate the estrogen-induced increase in Kiss1 expression required to initiate the preovulatory GnRH surge (Smith et al., 2005a). What remains less clear, however, is the relative roles ERα and ERβ play in organizing the sex-specific RP3V Kiss1 neuron signaling pathways. Neonatal administration of androgen (Kauffman et al., 2007b), estrogen, or an ERα agonist (Bateman and Patisaul, 2008; Patisaul et al., 2009) to females results in a male-typical expression pattern of Kiss1 in the adult AVPV, results that emphasize the mechanistic importance of ERα in the sexual differentiation process. Although the expression of ERα and ERβ in the preoptic area was higher prior to PND 7 compared with later ages, Kiss1 did not become detectable until PND 11, at which point it rapidly increased in females, resulting in a 3-fold sex difference in expression in the AVPV, and a nearly 11-fold difference in the MPOA by PND 19. A concomitant rise in female RP3V ERβ expression accompanied the emergence of Kiss1 expression and, similarly, females had higher levels of ERβ on PND 19 than males. These observations suggest that the expression of peripubertal Kiss1 and ERβ is related and, perhaps, interdependent. The ontogeny of ERα expression did not follow this pattern, a result that was somewhat unexpected given that ERα appears to be required for estrogen-dependent regulation of Kiss1 expression (Smith et al., 2005a; Oakley et al., 2009).
The mechanism by which neonatal estrogen induces the sexually dimorphic expression of Kiss1 more than a week later remains unclear. One possibility is that input from other sexual dimorphic nuclei may play an important role in the initiation of peripubertal Kiss1 expression. For example, many neuronal populations known to play a role in the regulation of sex-appropriate gonadotropin secretion, such as GABA neurons (Ottem et al., 2004) and do-paminergic neurons (Orikasa et al., 2002), are frequently co-localized with ERα and/or ERβ in the AVPV. A study published while the present manuscript was in review tested the hypothesis that BAX-dependent apoptosis might contribute to sex differences in Kiss1 expression but found that it only regulated Kiss1 cell numbers in the ARC, not the RP3V (Semaan et al., 2010). Thus the mechanism by which this sex difference emerges remains to be determined.
In both the AVPV and the MPOA, ERα expression was more extensive and robust than ERβ expression during the neonatal period, an observation that is consistent with and expands upon what has previously been reported for the rat. A pair of studies conducted prior to the discovery of ERβ (DonCarlos and Handa, 1994; Kuhnemann et al., 1994) provide some of the earliest evidence that preoptic levels of ER mRNA (presumably ERα) are sexually dimorphic at birth (females > males). The present data are largely congruous with DonCarlos and Handa (1994) in that we observed that this sex difference is abolished as early as PND 7 in the AVPV (Fig. 2), whereas Kuhnemann et al. (1994) reported that it persists through early adulthood. Our results differ, however, from a recent study using nonradioactive in situ hybridization, which found that ERα mRNA levels are higher in females than males on PND 14 (Orikasa et al., 2002). Sex differences in ERα expression across the neonatal period likely result primarily from increased mRNA production in females rather than differences in the number of ERα-producing neurons because immunohistochemical data reveal that ERα-ir cells are present in the rat MPOA from PND 1 to PND 35, with sex differences in cell number only appreciable on PNDs 10 and 21 (Yokosuka et al., 1997).
In contrast to ERα, AVPV ERβ expression was found to be sexually dimorphic at birth but, in this case, males had higher expression levels than females. This difference rapidly disappeared, and then reemerged by PND 19, with females now having higher expression than males. This sex difference on PND 0 has previously been identified in mice (Wolfe et al., 2005) and, additionally, a pair of studies has reported that this sex difference subsequently flips again, with males having higher expression levels than females in adulthood (Zhang et al., 2002; Kudwa et al., 2004). Our findings are consistent with a prior mouse study using semiquantitative RT-PCR that found detectable levels of ERβ mRNA in the preoptic region throughout perinatal development. In males, levels of mRNA expression increased until the day of birth and remained high through PND 15, but no concomitant increase was seen in females (Karolczak and Beyer, 1998). Our results differ, however, from two related studies reporting that rat females have higher levels of both ERβ mRNA and ERβ immunoreactivity in the AVPV from PND 7 to adulthood (day of birth defined as PND 1, compared with PND 0 in the present study) (Orikasa et al., 2002; Orikasa and Sakuma, 2003). This group did not, however, examine animals younger than PND 7. Another recent study failed to detect ERβ immunoreactivity in the rat AVPV from PND 3 to PND 14 (Perez et al., 2003), but did not look earlier or at control for sex, making the results difficult to interpret.
These inconsistent observations between research groups may result from several factors. First, the relatively low expression level of ERβ makes it difficult to detect. Second, historically, labeling obtained from ERβ antibodies has been problematic and has produced discordant results. Subtle strain differences in expression may also be a factor (Fisher 344 rats were used by Perez et al. [2003], Sprague-Dawley rats were used by Orikasa et al. [2002], and LE rats were used for the present study). Finally, the angle of cryosectioning and the methods used to define anatomical borders differ slightly between studies. Orikasa et al. (2003) performed a topographic analysis and found a greater density of expression near the ependymal lining of the third ventricle in females compared with males, a quantitative approach that was not used in the present study. Qualitatively, however, we also found that ERβ expression was largely confined to this region.
The functional significance of the transient sex difference in AVPV ERβ mRNA expression observed on the day of birth remains to be established, but it may be related to the sex-specific organization of the HPG axis. It has long been recognized that estrogens, aromatized from testicular androgens, are required to organize the male rodent HPG axis such that no surge of GnRH can be elicited with hormone priming (Corbier et al., 1978, 1992; Weisz and Ward, 1980; MacLusky and Naftolin, 1981; DonCarlos et al., 1995; Simerly, 2002). A recent study found that the androgen 5α-androstan-3β, 17β-diol (3β-diol) activates estrogen response element (ERE)-depend-ent ERβ-induced transcription equivalent to that of 17β-estradiol in neural tissue (Pak et al., 2005), further supporting the hypothesis that neonatal ERβ may facilitate estrogen-dependent male-typical hypothalamic development. Our observations are also compatible with the hypothesis that ERβ may play an important role in the “defeminization” of the male hypothalamus (Kudwa et al., 2005).
Notably, within the MPOA, both ERs were confined mainly to the dorsal aspect on the day of birth (PND 0), and were then uniformly distributed at later ages. Thus, this group of ER-expressing cells may still be in the process of migrating. This observation is consistent with an immunohistochemical study conducted in perinatal mice (Wolfe et al., 2005) but contrasts with an earlier study that found that ERα was distributed evenly within the MPOA on the day of birth (Yokosuka et al., 1997).
Developmental and sexual dimorphic expression of ERα, ERβ, and Kiss1 mRNA in the mediobasal hypothalamus
A different relationship among the three genes emerged in the mediobasal hypothalamus. Evidence that estrogen administration in the ARC suppresses plasma LH levels suggests that the ARC may mediate the negative feedback of sex steroids on GnRH secretion (Scott et al., 1997). This negative feedback action has been hypothesized to be mediated by Kiss1 neurons (Smith et al., 2005a,b, 2006; Dungan et al., 2006; Kauffman et al., 2007a,b; Maeda et al., 2007; Kauffman, 2009). The present study is the first to examine the expression of Kiss1, ERα, and ERβ in early neonatal life, as this feedback system undergoes sex-specific organization. In the ARC, ERβ expression levels were minimal across postnatal development and only rose slightly with age in both sexes, with no apparent sex difference in expression levels at any age examined. In contrast, ERα and Kiss1 levels were readily detectable on the day of birth, but the general pattern of ERα and Kiss1 expression diverged, with the ontogeny of female ERα levels resembling a “U-shape” and Kiss1 ontogeny resembling the opposite.
Collectively, these results suggest that their expression might be related to and influenced by each other, a hypothesis supported by prior studies showing that nearly all Kiss1 neurons in the adult ARC co-express ERα (Smith et al., 2005a,b, 2006). Our data provide evidence that Kiss1 in the developing ARC may also be under the control of sex hormones. In males, estradiol, aromatized from testicular androgens, is likely driving down the expression of both ERα and Kiss1 in this region, whereas in females the opposite is occurring and, in the absence of estradiol, Kiss1 expression is unrestrained until peri-puberty, when estradiol levels rise.
An RT-PCR study using whole hypothalamus detected persistent expression of Kiss1 in rat hypothalamus from PND 1 to adulthood in both males and females (Navarro et al., 2004). The results obtained from the present study reveal that this expression is confined to the ARC. Collectively, the findings are congruous with parallel work from our lab showing that the number of Kiss1-ir cells is not dimorphic at either PND 20 or PND 30 (Losa et al., 2011), an additional study indicating that ARC Kiss1 mRNA levels are not sexually dimorphic in adulthood (Kauffman et al., 2007b), and data that emerged while this paper was under review revealing a sex difference in the number of ARC neurons immunoreactive for Kiss1 PND 7 (Iijima et al., 2011).
Interestingly, however, we and numerous others have noted that the density of Kiss1-ir neuronal fibers within the ARC is significantly higher in male rodents compared with females (Clarkson et al., 2009; Patisaul et al., 2009; Losa et al., 2011). It remains unresolved whether this dense plexus of fibers within the ARC originates from the ARC population of Kiss1 neurons, or the more rostral RP3V population, but our findings are more supportive of the latter. Moreover, although it has been established that RP3V Kiss1 neurons make direct projections to GnRH neurons (Clarkson and Herbison, 2011), whether the ARC population also does so remains undetermined. It is clear, however, that the ARC population regulates GnRH release from the ME (Navarro et al., 2009).
Previous studies using different techniques have also observed a similar pattern of ERα and ERβ expression in the ARC (Simerly et al., 1990; Kuhnemann et al., 1994; Yokosuka et al., 1997), but our findings are the first to describe the ERα mRNA expression levels in the early postnatal stage. Moreover, although immunolabeling failed to detect ARC ERβ within the first 2 weeks of life (Perez et al., 2003), here ISHH revealed a weak signal on PNDs 7 and 19, but no appreciable sex difference. Collectively, these findings demonstrate that neonatal ERβ expression in the ARC is minimal at most, and remains low throughout life in both sexes (Shughrue et al., 1997a; Shughrue and Merchenthaler, 2001b).
It had previously been shown that VMN ERα expression is sexually dimorphic from PND 10 to adulthood (Kuhnemann et al., 1994; Yokosuka et al., 1997), and the results from the present study both support and enhance this finding by demonstrating that sex-specific ERα expression exists as early as PND 2 and persists throughout the neonatal period. Thus, a detailed map of lifetime VMN ERα expression patterns is now complete. Our results are also compatible with the well-established observation that the adult rodent VMN expresses primarily ERα (Shughrue et al., 1996, 1997b; Zhang et al., 2002). Moreover, we found that the temporal expression pattern of ERα was different in the cVMNvl than in the rVMNvl, a result suggesting that the rVMNvl may be functionally distinct from cVMNvl. This hypothesis is supported by a behavioral study in which higher levels of lordosis were observed in animals with estrogen implants in the rVMN rather than the cVMN (Pleim et al., 1989).
In addition, we are the first to provide a detailed temporal profile of ERβ expression in the VMN, and we show that ERβ expression is sexually dimorphic as early as PND 0, with more than threefold higher levels in females. These results expand upon prior work showing that ERβ immunoreactivity is present in the cVMN of postnatal rats (Perez et al., 2003) and adult mice (Mitra et al., 2003; Nomura et al., 2003). Interestingly, the ontogeny of the sex difference in ERα expression is opposite to that of ERβ in the cVMNvl, data that provides evidence that ERα and ERβ play functionally different roles in mediating brain sexual dimorphisms within this region and are potentially influencing each other’s expression levels and activity (Ogawa et al., 1998).
CONCLUSIONS
The detailed profile of neonatal Kiss1 and ER mRNA levels provided here will help elucidate the relative roles each plays in the sex-specific, estrogen-dependent organization of gonadotropin signaling pathways. Although sex differences in endogenous steroid hormone levels likely contribute to the sex differences in ERs and Kiss1 expression observed here, particularly during the early neonatal period, not all of our observations are consistent with this mechanism. It is well established that hypothala-mic aromatase activity and local estrogen levels are higher in neonatal males than females (Lauber et al., 1997; Amateau et al., 2004), and it has previously been purported that, because estrogen downregulates its own receptors through negative feedback, this sex difference in local estrogen levels accounts for sex differences in hypothalamic ER expression (DonCarlos et al., 1995; Kuhnemann et al., 1995; Ikeda et al., 2003). This postulate cannot explain, however, why ERα and ERβ had different temporal expression patterns across postnatal life within the same hormonal environment. Thus, other unknown mechanisms could be involved, at least partially, in the regulation of the two ER subtypes and the two populations of Kiss1 (Semaan et al., 2010).
Emerging evidence suggests that promoter methylation may be a possibility (Nugent et al., 2011). Research on maternal behavior has revealed that increased methylation of the ERα promoter decreases ERα expression, resulting in lower levels of maternal behavior (Champagne et al., 2006). Moreover, a recent study found significantly higher methylation of exon 1b of the ERα promoter in males and females neonatally exposed to estradiol benzoate (Kurian et al., 2010). It has not yet been established whether epigenetic mechanisms might account for the region-specific expression of Kiss1 or its emergence in the RP3V just prior to puberty. Exploration of how the environment, (hormonal, behavioral, or otherwise), shapes the sex-specific ontogeny of gonadotropin signaling pathways is the theme of our ongoing work.
ACKNOWLEDGMENTS
We thank Emily Merritt for her careful work in quantifying a portion of the ARC ERβ autoradiograms as part of her undergraduate research project, and Karina Todd for assistance with animal care and her critical reading of this manuscript.
Grant sponsor: National Institutes of Environmental Health Sciences (NIEHS); Grant number: RO1 ES016001 (to H.B.P.).
Abbreviations
- 3V
third ventricle
- AC
anterior commissure
- AHP
anterior hypothalamic area, posterior part
- ANOVA
analysis of variance
- ARC
arcuate nucleus
- rARC
rostral arcuate nucleus
- cARC
caudal arcuate nucleus
- AVPV
anteroventral periventricular nucleus
- BPA
bisphenol A
- C
caudal
- DMC
dorsomedial hypothalamic nucleus, compact part
- DMD
dorsomedial hypothalamic nucleus, dorsal part
- DMN
dorsomedial nucleus of the hypothalamus
- DMV
dorsomedial hypothalamic nucleus, ventral part
- DTT
dithioxytocinhreitol
- ER
estrogen receptor
- ERα
estrogen receptor α
- ERβ
estrogen receptor β
- ERE
estrogen response element
- Fx
fornix
- GnRH
onadotropin-releasing hormone
- GABA
γ-aminobutyric acid
- HPG
hypothalamic-pituitary-gonadal
- ir
immunoreactive
- ISHH
in situ hybridization histochemistry
- Kiss1
kisspeptin
- LE
Long Evans
- LH
luteinizing hormone
- LHA
anterior lateral hypothalamus
- MPN
medial preoptic nucleus
- MPOA
medial preoptic area
- MTu
medial tuberal nucleus
- Och
optic chiasm
- PCR
polymerase chain reaction
- PND
postnatal day
- POA
preoptic area
- PVN
paraventricular hypothalamic nucleus
- r
rostral
- RChL
retrochiasmatic area, lateral part
- ROI
regionofinterest
- RP3V
rostral periventricular area of the third ventricle
- RT-PCR
reverse transcription-polymerase chain reaction
- SD
Sprague-Dawley
- SSC
standard saline citrate solution
- Stg
stigmoid hypothalamic nucleus
- vl
ventrolateral
- VLPO
ventrolateral preoptic nucleus
- VMN
ventromedial hypothalamic nucleus
- rVMN
rostral ventromedial hypothalamic nucleus
- cVMN
caudal ventromedial hypothalamic nucleus
- VMNc
ventromedial hypothalamic nucleus, central part
- VMNdm
ventromedial hypothalamic nucleus, dorsomedial part
- VMNvl
ventrolateral division of the ventromedial hypothalamic nucleus
- cVMNvl
caudal ventrolateral division of the ventromedial hypothalamic nucleus
- rVMNvl
rostral ventrolateral division of the ventromedial hypothalamic nucleus
- VMPO
posterior part of the ventral medial nucleus
LITERATURE CITED
- Amateau SK, Alt JJ, Stamps CL, McCarthy MM. Brain estradiol content in newborn rats: sex differences, regional heterogeneity, and possible de novo synthesis by the female telencephalon. Endocrinology. 2004;145:2906–2917. doi: 10.1210/en.2003-1363. [DOI] [PubMed] [Google Scholar]
- Arai Y, Gorski RA. Critical exposure time for androgeni-zation of the developing hypothalamus in the female rat. Endocrinology. 1968;82:1010–1014. doi: 10.1210/endo-82-5-1010. [DOI] [PubMed] [Google Scholar]
- Arnold AP, Gorski RA. Gonadal steroid induction of structural sex differences in the central nervous system. Ann Rev Neurosci. 1984;7:413–442. doi: 10.1146/annurev.ne.07.030184.002213. [DOI] [PubMed] [Google Scholar]
- Bateman HL, Patisaul HB. Disrupted female reproductive physiology following neonatal exposure to phytoestro-gens or estrogen specific ligands is associated with decreased GnRH activation and kisspeptin fiber density in the hypothalamus. Neurotoxicology. 2008;29:988–997. doi: 10.1016/j.neuro.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NM, Setchell KD. Animal models impacted by phytoestrogens in commercial chow: implications for pathways influenced by hormones. Lab Invest. 2001;81:735–747. doi: 10.1038/labinvest.3780282. [DOI] [PubMed] [Google Scholar]
- Champagne FA, Weaver IC, Diorio J, Dymov S, Szyf M, Meaney MJ. Maternal care associated with methylation of the estrogen receptor-alpha1b promoter and estrogen receptor-alpha expression in the medial preoptic area of female offspring. Endocrinology. 2006;147:2909–2915. doi: 10.1210/en.2005-1119. [DOI] [PubMed] [Google Scholar]
- Chronwall BM. Anatomy and physiology of the neuroen-docrine arcuate nucleus. Peptides. 1985;6(suppl 2):1–11. doi: 10.1016/0196-9781(85)90128-7. [DOI] [PubMed] [Google Scholar]
- Clarkson J, Herbison AE. Postnatal development of kiss-peptin neurons in mouse hypothalamus; sexual dimorphism and projections to gonadotropin-releasing hormone neurons. Endocrinology. 2006;147:5817–5825. doi: 10.1210/en.2006-0787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson J, Herbison AE. Dual phenotype kisspeptin-do-pamine neurones of the rostral periventricular area of the third ventricle project to GnRH neurones. J Neuroendocri-nol. 2011;152:1551–1561. doi: 10.1111/j.1365-2826.2011.02107.x. [DOI] [PubMed] [Google Scholar]
- Clarkson J, Boon WC, Simpson ER, Herbison AE. Postnatal development of an estradiol-kisspeptin positive feedback mechanism implicated in puberty onset. Endocrinology. 2009;150:3214–3220. doi: 10.1210/en.2008-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colpaert FC, Wiepkema PR. Effects of ventromedial hypothalamic lesions on spontaneous intraspecies aggression in male rats. Behav Biol. 1976;16:117–125. doi: 10.1016/s0091-6773(76)91225-6. [DOI] [PubMed] [Google Scholar]
- Coolen LM, Wood RI. Testosterone stimulation of the medial preoptic area and medial amygdala in the control of male hamster sexual behavior: redundancy without amplification. Behav Brain Res. 1999;98:143–153. doi: 10.1016/s0166-4328(98)00063-1. [DOI] [PubMed] [Google Scholar]
- Corbier P, Kerdelhue B, Picon R, Roffi J. Changes in tes-ticular weight and serum gonadotropin and testosterone levels before, during, and after birth in the perinatal rat. Endocrinology. 1978;103:1985–1991. doi: 10.1210/endo-103-6-1985. [DOI] [PubMed] [Google Scholar]
- Corbier P, Edwards DA, Roffi J. The neonatal testosterone surge: a comparative study. Arch Int Physiol Biochim Biophys. 1992;100:127–131. doi: 10.3109/13813459209035274. [DOI] [PubMed] [Google Scholar]
- Degen GH, Janning P, Diel P, Bolt HM. Estrogenic isofla-vones in rodent diets. Toxicol Lett. 2002;128:145–157. doi: 10.1016/s0378-4274(02)00009-7. [DOI] [PubMed] [Google Scholar]
- DonCarlos LL. Developmental profile and regulation of estrogen receptor (ER) mRNA expression in the preoptic area of prenatal rats. Brain Res Dev Brain Res. 1996;94:224–233. doi: 10.1016/0165-3806(96)00067-3. [DOI] [PubMed] [Google Scholar]
- DonCarlos LL, Handa RJ. Developmental profile of estrogen receptor mRNA in the preoptic area of male and female neonatal rats. Brain Res Dev Brain Res. 1994;79:283–289. doi: 10.1016/0165-3806(94)90133-3. [DOI] [PubMed] [Google Scholar]
- DonCarlos LL, McAbee M, Ramer-Quinn DS, Stancik DM. Estrogen receptor mRNA levels in the preoptic area of neonatal rats are responsive to hormone manipulation. Brain Res. 1995;84:253–260. doi: 10.1016/0165-3806(94)00179-4. [DOI] [PubMed] [Google Scholar]
- Dungan HM, Clifton DK, Steiner RA. Minireview: kiss-peptin neurons as central processors in the regulation of gonadotropin-releasing hormone secretion. Endocrinology. 2006;147:1154–1158. doi: 10.1210/en.2005-1282. [DOI] [PubMed] [Google Scholar]
- Gerlach JL, McEwen BS, Toran-Allerand CD, Friedman WJ. Perinatal development of estrogen receptors in mouse brain assessed by radioautography, nuclear isolation and receptor assay. Brain Res. 1983;313:7–18. doi: 10.1016/0165-3806(83)90197-9. [DOI] [PubMed] [Google Scholar]
- Green S, Walter P, Kumar V, Krust A, Bornert JM, Argos P, Chambon P. Human oestrogen receptor cDNA: sequence, expression and homology to v-erb-A. Nature. 1986;320:134–139. doi: 10.1038/320134a0. [DOI] [PubMed] [Google Scholar]
- Greene GL, Gilna P, Waterfield M, Baker A, Hort Y, Shine J. Sequence and expression of human estrogen receptor complementary DNA. Science. 1986;231:1150–1154. doi: 10.1126/science.3753802. [DOI] [PubMed] [Google Scholar]
- Gu GB, Simerly RB. Projections of the sexually dimorphic anteroventral periventricular nucleus in the female rat. J Comp Neurol. 1997;384:142–164. [PubMed] [Google Scholar]
- Herbison AE. Estrogen positive feedback to gonadotro-pin-releasing hormone (GnRH) neurons in the rodent: the case for the rostral periventricular area of the third ventricle (RP3V) Brain Res Rev. 2008;57:277–287. doi: 10.1016/j.brainresrev.2007.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines M, Davis FC, Coquelin A, Goy RW, Gorski RA. Sexually dimorphic regions in the medial preoptic area and the bed nucleus of the stria terminalis of the guinea pig brain: a description and an investigation of their relationship to gonadal steroids in adulthood. J Neurosci. 1985;5:40–47. doi: 10.1523/JNEUROSCI.05-01-00040.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman GE, Le WW, Schulterbrandt T, Legan SJ. Estrogen and progesterone do not activate Fos in AVPV or LHRH neurons in male rats. Brain Res. 2005;1054:116–124. doi: 10.1016/j.brainres.2005.06.082. [DOI] [PubMed] [Google Scholar]
- Hrabovszky E, Kallo I, Hajszan T, Shughrue PJ, Merchenthaler I, Liposits Z. Expression of estrogen receptor-beta messenger ribonucleic acid in oxytocin and vasopressin neurons of the rat supraoptic and paraventricular nuclei. Endocrinology. 1998;139:2600–2604. doi: 10.1210/endo.139.5.6024. [DOI] [PubMed] [Google Scholar]
- Iijima N, Takumi K, Sawai N, Ozawa H. An immunohisto-chemical study on the expressional dynamics of kisspeptin neurons relevant to GnRH neurons using a newly developed anti-kisspeptin antibody. J Mol Neurosci. 2011;43:146–154. doi: 10.1007/s12031-010-9433-y. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Nagai A, Ikeda MA, Hayashi S. Sexually dimorphic and estrogen-dependent expression of estrogen receptor beta in the ventromedial hypothalamus during rat postnatal development. Endocrinology. 2003;144:5098–5104. doi: 10.1210/en.2003-0267. [DOI] [PubMed] [Google Scholar]
- Ju G, Swanson LW. Studies on the cellular architecture of the bed nuclei of the stria terminalis in the rat: I. Cytoarchitecture. J Comp Neurol. 1989;280:587–602. doi: 10.1002/cne.902800409. [DOI] [PubMed] [Google Scholar]
- Karolczak M, Beyer C. Developmental sex differences in estrogen receptor-beta mRNA expression in the mouse hypothalamus/preoptic region. Neuroendocrinology. 1998;68:229–234. doi: 10.1159/000054370. [DOI] [PubMed] [Google Scholar]
- Kauffman AS. Sexual differentiation and the Kiss1 system: hormonal and developmental considerations. Peptides. 2009;30:83–93. doi: 10.1016/j.peptides.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman AS, Clifton DK, Steiner RA. Emerging ideas about kisspeptin- GPR54 signaling in the neuroendocrine regulation of reproduction. Trends Neurosci. 2007a;30:504–511. doi: 10.1016/j.tins.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Kauffman AS, Gottsch ML, Roa J, Byquist AC, Crown A, Clifton DK, Hoffman GE, Steiner RA, Tena-Sempere M. Sexual differentiation of Kiss1 gene expression in the brain of the rat. Endocrinology. 2007b;148:1774–1783. doi: 10.1210/en.2006-1540. [DOI] [PubMed] [Google Scholar]
- Kudwa AE, Gustafsson JA, Rissman EF. Estrogen receptor beta modulates estradiol induction of progestin receptor immunoreactivity in male, but not in female, mouse medial preoptic area. Endocrinology. 2004;145:4500–4506. doi: 10.1210/en.2003-1708. [DOI] [PubMed] [Google Scholar]
- Kudwa AE, Bodo C, Gustafsson JA, Rissman EF. A previously uncharacterized role for estrogen receptor beta: defeminization of male brain and behavior. Proc Natl Acad Sci U S A. 2005;102:4608–4612. doi: 10.1073/pnas.0500752102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhar MJ, De Souza EB, Unnerstall JR. Neurotransmitter receptor mapping by autoradiography and other methods. Annu Rev Neurosci. 1986;9:27–59. doi: 10.1146/annurev.ne.09.030186.000331. [DOI] [PubMed] [Google Scholar]
- Kuhnemann S, Brown TJ, Hochberg RB, MacLusky NJ. Sex differences in the development of estrogen receptors in the rat brain. Horm Behave. 1994;28:483–491. doi: 10.1006/hbeh.1994.1046. [DOI] [PubMed] [Google Scholar]
- Kuhnemann S, Brown TJ, Hochberg RB, MacLusky NJ. Sexual differentiation of estrogen receptor concentrations in the rat brain: effects of neonatal testosterone exposure. Brain Res. 1995;691:229–234. doi: 10.1016/0006-8993(95)00640-c. [DOI] [PubMed] [Google Scholar]
- Kuiper GG, Enmark E, Pelto-Huikko M, Nilsson S, Gustafsson JA. Cloning of a novel receptor expressed in rat prostate and ovary. Proc Natl Acad Sci U S A. 1996;93:5925–5930. doi: 10.1073/pnas.93.12.5925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian JR, Olesen KM, Auger AP. Sex differences in epi-genetic regulation of the estrogen receptor-alpha promoter within the developing preoptic area. Endocrinology. 2010;151:2297–2305. doi: 10.1210/en.2009-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber AH, Mobbs CV, Muramatsu M, Pfaff DW. Estrogen receptor messenger RNA expression in rat hypothala-mus as a function of genetic sex and estrogen dose. Endocrinology. 1991;129:3180–3186. doi: 10.1210/endo-129-6-3180. [DOI] [PubMed] [Google Scholar]
- Lauber ME, Sarasin A, Lichtensteiger W. Sex differences and androgen-dependent regulation of aromatase (CYP19) mRNA expression in the developing and adult rat brain. J Steroid Biochem Mol Biol. 1997;61:359–364. [PubMed] [Google Scholar]
- Losa SM, Todd KL, Sullivan AW, Cao J, Mickens JA, Patisaul HB. Neonatal exposure to genistein adversely impacts the ontogeny of hypothalamic kisspeptin signaling pathways and ovarian development in the peripubertal female rat. Reprod Toxicol. 2011;31:280–289. doi: 10.1016/j.reprotox.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLusky NJ, Naftolin F. Sexual differentiation of the central nervous system. Science (New York) 1981;211:1294–1302. doi: 10.1126/science.6163211. [DOI] [PubMed] [Google Scholar]
- MacLusky NJ, Chaptal C, McEwen BS. The development of estrogen receptor systems in the rat brain and pituitary: postnatal development. Brain Res. 1979a;178:143–160. doi: 10.1016/0006-8993(79)90094-5. [DOI] [PubMed] [Google Scholar]
- MacLusky NJ, Lieberburg I, McEwen BS. The development of estrogen receptor systems in the rat brain: perinatal development. Brain Res. 1979b;178:129–142. doi: 10.1016/0006-8993(79)90093-3. [DOI] [PubMed] [Google Scholar]
- Maeda K, Adachi S, Inoue K, Ohkura S, Tsukamura H. Metastin/kisspeptin and control of estrous cycle in rats. Rev Endocr Metab Disord. 2007;8:21–29. doi: 10.1007/s11154-007-9032-6. [DOI] [PubMed] [Google Scholar]
- Mikkelsen JD, Simonneaux V. The neuroanatomy of the kisspeptin system in the mammalian brain. Peptides. 2009;30:26–33. doi: 10.1016/j.peptides.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Mitra SW, Hoskin E, Yudkovitz J, Pear L, Wilkinson HA, Haya-shi S, Pfaff DW, Ogawa S, Rohrer SP, Schaeffer JM, McE-wen BS, Alves SE. Immunolocalization of estrogen receptor beta in the mouse brain: comparison with estrogen receptor alpha. Endocrinology. 2003;144:2055–2067. doi: 10.1210/en.2002-221069. [DOI] [PubMed] [Google Scholar]
- Nance DM. Sex differences in the hypothalamic regulation of feeding behavior in the rat. Adv Psychobiol. 1976;3:75–123. [PubMed] [Google Scholar]
- Navarro VM, Castellano JM, Fernandez-Fernandez R, Barreiro ML, Roa J, Sanchez-Criado JE, Aguilar E, Dieguez C, Pinilla L, Tena-Sempere M. Developmental and hormonally regulated messenger ribonucleic acid expression of KiSS-1 and its putative receptor, GPR54, in rat hypothalamus and potent luteinizing hormone-releasing activity of KiSS-1 pep-tide. Endocrinology. 2004;145:4565–4574. doi: 10.1210/en.2004-0413. [DOI] [PubMed] [Google Scholar]
- Navarro VM, Gottsch ML, Chavkin C, Okamura H, Clifton DK, Steiner RA. Regulation of gonadotropin-releasing hormone secretion by kisspeptin/dynorphin/neurokinin B neurons in the arcuate nucleus of the mouse. J Neurosci. 2009;29:11859–11866. doi: 10.1523/JNEUROSCI.1569-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura M, Korach KS, Pfaff DW, Ogawa S. Estrogen receptor beta (ERbeta) protein levels in neurons depend on estrogen receptor alpha (ERalpha) gene expression and on its ligand in a brain region-specific manner. Brain Res Mol Brain Res. 2003;110:7–14. doi: 10.1016/s0169-328x(02)00544-2. [DOI] [PubMed] [Google Scholar]
- Nugent BM, Schwarz JM, McCarthy MM. Hormonally mediated epigenetic changes to steroid receptors in the developing brain: implications for sexual differentiation. Horm Behav. 2011;59:338–344. doi: 10.1016/j.yhbeh.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numan M. A neural circuitry analysis of maternal behavior in the rat. Acta Paediatr Suppl. 1994;397:19–28. doi: 10.1111/j.1651-2227.1994.tb13261.x. [DOI] [PubMed] [Google Scholar]
- Numan M, Stolzenberg DS. Medial preoptic area interactions with dopamine neural systems in the control of the onset and maintenance of maternal behavior in rats. Front Neuroendocrinol. 2009;30:46–64. doi: 10.1016/j.yfrne.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Oakley AE, Clifton DK, Steiner RA. Kisspeptin signaling in the brain. Endocr Rev. 2009;30:713–743. doi: 10.1210/er.2009-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Inoue S, Watanabe T, Orimo A, Hosoi T, Ouchi Y, Muramatsu M. Molecular cloning and characterization of human estrogen receptor betacx: a potential inhibitor ofestrogen action in human. Nucleic Acids Res. 1998;26:3505–3512. doi: 10.1093/nar/26.15.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orikasa C, Sakuma Y. Possible involvement of preoptic estrogen receptor beta positive cells in luteinizing hormone surge in the rat. Domest Anim Endocrinol. 2003;25:83–92. doi: 10.1016/s0739-7240(03)00047-x. [DOI] [PubMed] [Google Scholar]
- Orikasa C, Kondo Y, Hayashi S, McEwen BS, Sakuma Y. Sexually dimorphic expression of estrogen receptor beta in the anteroventral periventricular nucleus of the rat pre-optic area: implication in luteinizing hormone surge. Proc Natl Acad Sci U S A. 2002;99:3306–3311. doi: 10.1073/pnas.052707299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottem EN, Godwin JG, Krishnan S, Petersen SL. Dual-phenotype GABA/glutamate neurons in adult preoptic area: sexual dimorphism and function. J Neurosci. 2004;24:8097–8105. doi: 10.1523/JNEUROSCI.2267-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak TR, Chung WC, Lund TD, Hinds LR, Clay CM, Handa RJ. The androgen metabolite, 5alpha-androstane-3beta, 17beta-diol, is a potent modulator of estrogen receptor-beta1-mediated gene transcription in neuronal cells. Endocrinology. 2005;146:147–155. doi: 10.1210/en.2004-0871. [DOI] [PubMed] [Google Scholar]
- Patisaul HB. Phytoestrogen action in the adult and developing brain. J Neuroendocrinol. 2005;17:57–64. doi: 10.1111/j.1365-2826.2005.01268.x. [DOI] [PubMed] [Google Scholar]
- Patisaul HB, Whitten PL, Young L. Regulation of estrogen receptor beta mRNA in the brain: opposite effects of 17b–estradiol and the phytoestrogen, coumestrol. Brain Res Mol Brain Res. 1999;67:165–171. doi: 10.1016/s0169-328x(99)00058-3. [DOI] [PubMed] [Google Scholar]
- Patisaul HB, Fortino AE, Polston EK. Sex differences in serotonergic but not {gamma>-aminobutyric acidergic (GABA) projections to the rat ventromedial nucleus of the hypothalamus. Endocrinology. 2008;149:397–408. doi: 10.1210/en.2007-0666. [DOI] [PubMed] [Google Scholar]
- Patisaul HB, Todd KL, Mickens JA, Adewale HB. Impact of neonatal exposure to the ERalpha agonist PPT, bisphenol-A or phytoestrogens on hypothalamic kisspeptin fiber density in male and female rats. Neurotoxicology. 2009;30:350–357. doi: 10.1016/j.neuro.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Watson C. London: Academic Press; 2007. The rat brain in stereotaxic coordinates. [DOI] [PubMed] [Google Scholar]
- Perez SE, Chen EY, Mufson EJ. Distribution of estrogen receptor alpha and beta immunoreactive profiles in the postnatal rat brain. Brain Res Dev Brain Res. 2003;145:117–139. doi: 10.1016/s0165-3806(03)00223-2. [DOI] [PubMed] [Google Scholar]
- Petersen SL, Barraclough CA. Suppression of spontaneous LH surges in estrogen-treated ovariectomized rats by microimplants of antiestrogens into the preoptic brain. Brain Res. 1989;484:279–289. doi: 10.1016/0006-8993(89)90371-5. [DOI] [PubMed] [Google Scholar]
- Petersen SL, Gardner E, Adelman J, McCrone S. Examination of steroid-induced changes in LHRH gene transcription using 33P–and 35S–labeled probes specific for intron 2. Endocrinology. 1996;137:234–239. doi: 10.1210/endo.137.1.8536618. [DOI] [PubMed] [Google Scholar]
- Pfaff D, Keiner M. Atlas of estradiol-concentrating cells in the central nervous system of the female rat. J Comp Neurol. 1973;151:121–158. doi: 10.1002/cne.901510204. [DOI] [PubMed] [Google Scholar]
- Pfaff DW, Schwanzel-Fukuda M, Parhar IS, Lauber AH, McCarthy LM, Kow LM. GnRH neurons and other cellular and molecular mechanisms for simple mammalian reproductive behaviors. Rec Prog Horm Res. 1994;49:1–25. doi: 10.1016/b978-0-12-571149-4.50005-2. [DOI] [PubMed] [Google Scholar]
- Pleim ET, Brown TJ, MacLusky NJ, Etgen AM, Barfield RJ. Dilute estradiol implants and progestin receptor induction in the ventromedial nucleus of the hypothalamus: correlation with receptive behavior in female rats. Endocrinology. 1989;124:1807–1812. doi: 10.1210/endo-124-4-1807. [DOI] [PubMed] [Google Scholar]
- Ronnekleiv OK, Kelly MJ. Luteinizing hormone-releasing hormone neuronal system during the estrous cycle of the female rat: effects of surgically induced persistent estrus. Neuroendocrinology. 1986;43:564–576. doi: 10.1159/000124583. [DOI] [PubMed] [Google Scholar]
- Sa SI, Madeira MD. Estrogen modulates the sexually dimorphic synaptic connectivity of the ventromedial nucleus. J Comp Neurol. 2005;484:68–79. doi: 10.1002/cne.20451. [DOI] [PubMed] [Google Scholar]
- Scott CJ, Kuehl DE, Ferreira SA, Jackson GL. Hypothalamic sites of action for testosterone, dihydrotestosterone, and estrogen in the regulation of luteinizing hormone secretion in male sheep. Endocrinology. 1997;138:3686–3694. doi: 10.1210/endo.138.9.5401. [DOI] [PubMed] [Google Scholar]
- Semaan SJ, Murray EK, Poling MC, Dhamija S, Forger NG, Kauffman AS. BAX-dependent and BAX-independent regulation of Kiss1 neuron development in mice. Endocrinology. 2010;151:5807–5817. doi: 10.1210/en.2010-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shughrue P, Merchenthaler I. Distribution of estrogen receptor beta immunoreactivity in the rat central nervous system. J Comp Neurol. 2001a;436:64–81. [PubMed] [Google Scholar]
- Shughrue PJ, Merchenthaler I. Distribution of estrogen receptor beta immunoreactivity in the rat central nervous system. J Comp Neurol. 2001b;436:64–81. [PubMed] [Google Scholar]
- Shughrue PJ, Komm B, Merchenthaler I. The distribution of estrogen receptor-β mRNA in the rat hypothalamus. Steroids. 1996;61:678–681. doi: 10.1016/s0039-128x(96)00222-x. [DOI] [PubMed] [Google Scholar]
- Shughrue PJ, Lane MV, Merchenthaler I. Comparative distribution of estrogen receptor-alpha and -beta mRNA in the rat central nervous system. J Comp Neurol. 1997a;388:507–525. doi: 10.1002/(sici)1096-9861(19971201)388:4<507::aid-cne1>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Shughrue PL, Lane MV, Merchenthaler I. Comparative distribution of estrogen receptor α and β mRNA in the rat central nervous system. J Comp Neurol. 1997b;388:507–525. doi: 10.1002/(sici)1096-9861(19971201)388:4<507::aid-cne1>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Simerly RB. Wired for reproduction: organization and development of sexually dimorphic circuits in the mammalian forebrain. Annu Rev Neurosci. 2002;25:507–536. doi: 10.1146/annurev.neuro.25.112701.142745. [DOI] [PubMed] [Google Scholar]
- Simerly RB, Chang C, Muramatsu M, Swanson LW. Distribution of androgen and estrogen receptor mRNA-con-taining cells in the rat brain: an in situ hybridization study. J Comp Neurol. 1990;294:76–95. doi: 10.1002/cne.902940107. [DOI] [PubMed] [Google Scholar]
- Smith ER, Davidson JM. Location of feedback receptors: effects of intracranially implanted steroids on plasma LH and LRF response. Endocrinology. 1974;95:1566–1573. doi: 10.1210/endo-95-6-1566. [DOI] [PubMed] [Google Scholar]
- Smith JT. Sex steroid control of hypothalamic Kiss1 expression in sheep and rodents: comparative aspects. Peptides. 2009;30:94–102. doi: 10.1016/j.peptides.2008.08.013. [DOI] [PubMed] [Google Scholar]
- Smith JT, Cunningham MJ, Rissman EF, Clifton DK, Steiner RA. Regulation of Kiss1 gene expression in the brain of the female mouse. Endocrinology. 2005a;146:3686–3692. doi: 10.1210/en.2005-0488. [DOI] [PubMed] [Google Scholar]
- Smith JT, Dungan HM, Stoll EA, Gottsch ML, Braun RE, Eacker SM, Clifton DK, Steiner RA. Differential regulation of KiSS-1 mRNA expression by sex steroids in the brain of the male mouse. Endocrinology. 2005b;146:2976–2984. doi: 10.1210/en.2005-0323. [DOI] [PubMed] [Google Scholar]
- Smith JT, Popa SM, Clifton DK, Hoffman GE, Steiner RA. Kiss1 neurons in the forebrain as central processors for generating the preovulatory luteinizing hormone surge. J Neurosci. 2006;26:6687–6694. doi: 10.1523/JNEUROSCI.1618-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisz J, Ward IL. Plasma testosterone and progesterone titers of pregnant rats, their male and female fetuses, and neonatal offspring. Endocrinology. 1980;106:306–316. doi: 10.1210/endo-106-1-306. [DOI] [PubMed] [Google Scholar]
- White JO, Hall C, Lim L. Developmental changes in the content of oestrogen receptors in the hypothalamus of the female rat. Biochem J. 1979;184:465–468. doi: 10.1042/bj1840465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand SJ, Terasawa E. Discrete lesions reveal functional heterogeneity of suprachiasmatic structures in regulation of gonadotropin secretion in the female rat. Neuroendocrinology. 1982;34:395–404. doi: 10.1159/000123335. [DOI] [PubMed] [Google Scholar]
- Wiegand SJ, Terasawa E, Bridson WE, Goy RW. Effects of discrete lesions of preoptic and suprachiasmatic structures in the female rat. Alterations in the feedback regulation of gonadotropin secretion. Neuroendocrinology. 1980;31:147–157. doi: 10.1159/000123066. [DOI] [PubMed] [Google Scholar]
- Wolfe CA, Van Doren M, Walker HJ, Seney ML, McClellan KM, Tobet SA. Sex differences in the location of immunochemically defined cell populations in the mouse pre-optic area/anterior hypothalamus. Brain Res Dev Brain Res. 2005;157:34–41. doi: 10.1016/j.devbrainres.2005.03.001. [DOI] [PubMed] [Google Scholar]
- Yokosuka M, Okamura H, Hayashi S. Postnatal development and sex difference in neurons containing estrogen receptor-alpha immunoreactivity in the preoptic brain, the diencephalon, and the amygdala in the rat. J Comp Neurol. 1997;389:81–93. doi: 10.1002/(sici)1096-9861(19971208)389:1<81::aid-cne6>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Zhang JQ, Cai WQ, Zhou DS, Su BY. Distribution differences of estrogen receptor beta immunoreactivity in the brain of adult male female rats. Brain Res. 2002;935:73–80. doi: 10.1016/s0006-8993(02)02460-5. [DOI] [PubMed] [Google Scholar]