Abstract
Background
The ability to identify patients with Crohn’s disease (CD) at highest risk of surgery would be invaluable in guiding therapy. Genome-wide association (GWA) studies have identified multiple IBD loci with unknown phenotypic consequences.
Objectives
1) to identify associations between known and novel CD loci with early resective CD surgery2) to develop the best predictive model for time to surgery using a combination of phenotypic, serologic and genetic variables.
Methods
Genotyping was performed on 1115 subjects using Illumina-based Genome-wide technology. Univariate and multivariate analyses tested genetic associations with need for surgery within 5 years. Analyses were performed by testing known CD loci (n=71) and by performing a GWA study. Time to surgery was analyzed using Cox regression modeling. Clinical and serologic variables were included along with genotype to build predictive models for time to surgery.
Results
Surgery occurred within 5 years in 239 subjects at a median time of 12 months. Three CD susceptibility loci were independently associated with surgery within 5 years (IL12B, IL23R, C11orf30). GWA identified novel putative loci associated with early surgery; 7q21 (CACNA2D1) and 9q34 (RXRA, COL5A1). The most predictive models of time to surgery included genetic and clinical risk factors. More than a 20% difference in frequency of progression to surgery was seen between the lowest and highest risk groups.
Conclusion
Progression to surgery is faster in CD patients with both genetic and clinical risk factors. IL12B is independently associated with need and time to early surgery in CD patients and justifies the investigation of novel and existing therapies that affect this pathway.
Introduction
Crohn’s disease (CD) is a disease of diverse clinical phenotypes and patients with complicated disease phenotypes such as those with stricturing and/or internal penetrating disease behaviors often require surgical intervention. Over a 10 year period, approximately 80% of patients can expect to develop a complication and close to 50% of CD patients will progress to surgery (1, 2). This is in contrast to the small percentage of patients presenting with a complication, and requiring surgery at time of diagnosis. One of the biggest challenges facing clinicians today is predicting which patients will progress to complication resulting in intestinal resection. The identification of these “rapidly progressive” patients early in the course of their disease may assist in the decision regarding the early introduction of effective intervention strategies and customizing therapies based on risk of underlying disease. Those at highest risk of disease complication may well benefit most from early use of immunomodulators and/or biologic therapies. Clinical factors, such as extensive small bowel and perianal disease location, earlier age at diagnosis, and use of corticosteroids have been shown to be associated with “disabling” CD (3). Antibodies to microbial antigens have been shown repeatedly to be associated with complicated disease behaviors and need for surgery. Both the number of antibodies as well as the magnitude of the antibody response has been reported to be associated with surgical intervention in both the pediatric and adult CD populations (4–11). Recently, a risk prediction tool demonstrated that the use of biologics within the first 90 days of diagnosis in a child predicted to be at high risk of disease progression resulted in a 75% absolute risk reduction in disease complication rate as compared to children not treated with biologics within this time frame (12). This is the first study to demonstrate a change in the natural history in a predicted at risk population of CD patients. Despite major advances in genome discovery, there have only been a few studies that have considered the role of genetics in determining disease behavior. Variation in NOD2/CARD15 has been shown to be associated with stricturing CD, need for surgery as well as surgical recurrence (13–18). Henckaerts et al reported on over 30 loci known to be associated with CD and changes in disease behavior and need for surgery (19). The rs1363670 G allele near the IL12B gene was found to be independently associated with both stricturing disease and more rapid development of strictures. Recent studies have extended the number of CD loci achieving association at a genome-wide level to 71 (20–22). In looking for genes that predict natural history in CD, a logical place to start would be with the known susceptibility loci, but it is also quite possible that non-susceptibility loci contribute to disease behavior, as has been demonstrated in coronary artery disease (23, 24). These concepts justify testing associations with known CD loci and identifying novel loci using a whole genome approach for finding loci that influence the development of complications and surgery. The aims of this study were to identify associations between known CD susceptibility loci and novel genetic loci with need for resective surgery in CD patients and to develop the best predictive model for resective CD surgery using a combination of clinical, serologic and genetic variables. The identification of CD patients at highest risk for early resective surgery would be helpful to patients and clinicians alike in guiding initial therapy and identifying patients who would benefit most from early effective intervention.
Methods
Patient Population
A total of 1115 well characterized CD patients from both the Adult and Pediatric IBD Centers at CSMC, The Western Regional Research Alliance for Pediatric IBD and Wisconsin were included in this study. This study was approved by the IRB at all participating sites.
Phenotyping
All data was collected by chart review and stored in a secured database. For the purpose of this study, phenotype was defined as all variables that were not genetic.
Clinical Phenotype
These included demographic and clinical variables: age, gender, disease duration, age at diagnosis, date of diagnosis, disease location, type of disease complication (stricturing and/or internal penetrating disease), date of disease complication, type of CD surgery, time from diagnosis to CD surgery or time from diagnosis to last follow up in those patients who had not required surgery.
Phenotype Definitions
Surgery was defined as intestinal resection only for penetrating or stricturing CD.
Early surgery was considered need for surgery within 60 months of diagnosis. Perianal surgery and stricturoplasty were excluded.
Age at diagnosis was defined as equal to or less than 16 years of agevs. greaterthan 16.
Disease location was defined as disease of maximal extent at last follow up prior to surgery or at time of last follow up prior to data extraction if they have not undergone surgery. Disease extent was based on endoscopic, histologic and radiographic evidence of inflammation. There were 3 disease locations that patients were categorized into: 1) Small bowel only: disease of the small bowel proximal to the cecum and distal to the ligament of treitz; 2) Large bowel only: any colonic location between cecum and rectum with no small bowel disease; 3) Small and large bowel: disease of the small bowel and any location between cecum and rectum.
Stricturing disease (S): was defined as the occurrence of constant luminal narrowing demonstrated by radiologic, endoscopic or surgical examination combined with pre-stenotic dilatation and/or obstructive signs or symptoms. Internal penetrating disease (IP): was defined if patients had evidence of entero-enteric or entero-vesicular fistulae, intra-abdominal abscesses or intestinal perforation (4, 5, 10, 12, 14).
Immune Phenotype
Serum was collected on all patients and analyzed at CSMC. Serum immune responses: anti-Saccharomyces Cereviciae antibodies (ASCA IgG and IgA), perinuclear anti-nuclear cytoplasmic antibody (pANCA), anti-flagellin (anti-CBir1), anti-outer membrane porin C (anti-OmpC) and anti-Pseudomonas fluorescens-associated sequence I2 (anti-I2) were analyzed blinded to disease outcome by ELISA as previously described (5, 25). Antibody quartile score which is the quartile score for each antibody level was calculated(<25% = 1; 25%–50% = 2; 51%–_75% = 3; 75%–100% = 4). Quartile sum score is the sum of quartile scores for all 3 antibodies (ASCA IgA or IgG, anti-OmpC, and anti-CBir1). Minimum score is 4 (all antibodies had a quartile score of 1), and maximum score is 16 (all antibodies had a quartile score of 4). The quartile sum scores were grouped in order to minimize the number of patient subsets: quartile sum score 4–−7, group 1; 8–10, group 2; 11–12, group 3; and 13–16, group 4 (5). ANCA was analyzed separately given that ANCA has been shown to be negatively associated with CD resective surgery (26)
Genotype
Genotyping was performed at the Medical Genetics Institute at CSMC using the Illumina Human610 platform (n=887) and Children’s Hospital of Philadelphia (CHOP) using the Illumina Human 550 platform (n=228) (27). Genotyping for the 3 common CD associated NOD2 SNPs (SNPs 8, 12 and 13) was performed using TaqMan MGB platform (28). We performed principal components analysis (PCA) separately for our 2 primary outcomes; need for surgery and time to surgery given the sample size difference in the 2 outcome groups included for analysis.
Need for surgery
For the purpose of quality control, SNPs with a minor allele frequency (MAF) <0.01, genotype failure rate > 0.02, HWE p-value < 10−6 and Heterozygosity> 0.53 were excluded from the analysis. Following quality control, 483,359 SNPs were available in all data sets for analysis. We observed 2 Individuals with Missing rate > 0.02 and 2 individuals with cryptic relatedness (π̂>0.125). Principal components (PC) analysis (using EIGENSTRAT) was conducted to examine population stratification (29). As we examined the first 10 PCs with the default options provided by EIGENSTRAT, we observed that 23 patients were identified as outliers and the direction of the first 2 PCs shown in multidimensional scatter plot were separate (Fig., Supplemental Digital Content 1, http://links.lww.com/IBD/A154). Thus, we excluded the 27 patients identified by QC and PCA procedures, and corrected for population stratification by adding the first 2 PCs as covariates in the model of association analysis.
Time to Surgery
Following quality control, 484,724 SNPs were available in all data sets for analysis. We observed 3 Individuals with Missing rate > 0.02 and 3 individuals with cryptic relatedness (π̂>0.125). Principal components (PC) analysis (using EIGENSTRAT) was conducted to examine population stratification (29). As we examined the first 10 PCs with the default options provided by EIGENSTRAT, we observed that 64 patients were identified as outliers and the direction of the first 2 PCs shown in multidimensional scatter plot were separate (Fig., Supplemental Digital Content 1, http://links.lww.com/IBD/A154). Thus, we excluded the 70 patients identified by QC and PCA procedures, and corrected for population stratification by adding the first 2 PCs as covariates in the model of association analysis.
Statistical Analysis
A. Need for Early Surgery
Genome-wide association studies
In order to identify genetic factors that influence the need for early surgery within 60 months, we performed genome-wide single SNP association on patients with at least 60 months of follow up. For each SNP, an additive model was assumed. With a multivariate logistic regression model having 2 PCs and a SNP, PLINK was used to evaluate the association between the need for surgery and each single SNP. Genomic inflation factor λ of 0.9899 reveals no significant population stratification after exclusion of the 27 outliers. Although we recognize that accepted genome-wide level of significance is at p < 10−8 we, a priori, considered SNP association with a p-value < 10−5 from the GWAS as significant for this outcome. We believe that it is justified to show these SNPs so that other researchers may be able to utilize these data in interpreting their own data in future studies of similar design. For the 71 CD susceptibility loci (referred below as “top hits”) that were reported in the latest published CD meta-analysis (Table, Supplemental Digital Content 2, http://links.lww.com/IBD/A155), the SNPs that had a p-value < 0.05 in the analyses above were considered “significant” for association with need for surgery (20). In addition, we decided a priori, to assess a third level of significance for this hypothesis (namely that disease associated variants may also influence disease behavior) driven analyses by applying a bonferroni correction to any significant findings at these 71 loci ((p < 0.0007). The role of NOD2, (SNPs 8, 12 and 13) as a consequence of multiple disease predisposition alleles and prior literature, was tested separately using the chi-square test. Odds Ratios (OR) and 95% Confidence intervals (95% CI) were calculated by comparing the odds of surgery in the patients with a specific genotype versus those without the genotype. Genotype counts for all significant genetic associations are shown in Table, Supplemental Digital Content 3, http://links.lww.com/IBD/A156.
B. Time to surgery
Survival genome-wide association analysis
In order to determine genetic variation associated with time to surgery, survival genome- wide single SNP association was performed by assuming an additive genetic model for each SNP. The survival GWAS employed Cox regression modeling, utilizing 2PCs and SNPs as independent variables, and was performed by R package ‘survival’ (30). The SNPs with a p-value < 10−5 from the GWAS were considered as “significant” for this outcome and carried forward for additional analyses. All hazard rations (HR) were expressed as a point estimate with 95% confidence interval. For the known 71 CD ’SNPs’, association with time to surgery was considered significant if the p-value was ≤ 0.05.
Predictive models of time to surgery
Models to predict time to surgery were built using cox regression modeling, combining the following variables:A: Genetics; Top Hits p < 0.05, Genome-wide SNPs p < 10−5, and NOD2 status (any NOD2 SNP with a p ≤ 0.05 was considered positive), B: Serologies; Quartile Sum Score Groups 1,2,3 or 4, and ANCA status (positive or negative) C: Clinical: age at diagnosis (< 16 vs. > 16 yrs), disease duration, gender, and small bowel location. CD complications were not included in these models as this could bias the model to increase time to surgery. Using these variables, 5 model strategies were constructed:
Genetics only (excluding NOD 2 status)
Clinical variables only
Clinical + Serology
Genetics + Clinical
Genetics (including NOD2 status) + Clinical + Serology
Patients who had surgery at diagnosis or within 3 months of diagnosis were excluded from this particular analysis. The final predictive model of time to surgery for each strategic model was determined by stepwise model selection method.
The goal of these analyses were to first determine, using Cox regression modeling, the variables that were independently associated with progression to surgery and then to use log rank testing to compare the survival curves for different risk strata within each model in order to determine which patients progressed to surgery faster. The 3 risk strata were derived based on the sum of the variables in the final predictive models: strata 1 = ≤ the 25% quartile of the sum of variables, strata 2 = between 25% and 75 % quartiles of the sum of variables and strata 3 = ≥ 75% quartile of the sum of variables. Each clinical and serology variable was attributed a score of 1 or 0 and an additive genetic model was adopted. In order to validate the AUCs of each of the models, we used 5-fold cross-validation that randomly partitioned the original sample into 5 subsamples. Of the 5 subsamples, 4 subsamples were used as training data to develop the models, and the remaining subsample was served as the validation data for measuring AUC of the model developed on the training data. Then the cross-validation process was repeated 5 times by utilizing each of the 5 subsamples exactly once as the validation data. The 5 AUCs from the folds were averaged to produce a single estimation of AUC. The cross-validation was performed using the surv AUC R package. The data analyses were generated using SAS/STAT software, Version 9.2 of the SAS System for Windows. Copyright © 2008 SAS Institute Inc. SAS and all other SAS Institute Inc. product or service names are registered trademarks or trademarks of SAS Institute Inc., Cary, NC, USA.
Results
Patient demographics
Table 1 illustrates the key demographic data for all patients. The mean age at diagnosis for this cohort was 15 years of age and more than 50% of the patients were diagnosed at 16 years of age or younger. Approximately 75% of patients had small bowel involvement and just over 50% had complicated disease behaviors as of last follow up or at time of surgery. Four hundred and forty-four of the 1115 (40%) had undergone intestinal resection surgery in our cohort with a median (range) time to surgery of 45.5 (0–516.8) months. A total of 239 of these patients underwent surgery between diagnosis and 60 months of follow up with a median (range) of 12 months. Of these 239 patients, 65, 56, and 115 underwent surgery within 3 months, 3 to 12 months and 12 to 60 months from diagnosis, respectively. The remaining 169 had surgery after 60 months. NOD2 ‘positivity’ (carrier for at least 1 of the 3 common CD associated NOD2 risk alleles) was found in 33% of CD patients consistent with previously published data (13–18). ANCA status was negative in 79% of patients, and Quartile Sum Score groups 1, 2,3 and 4were observed in 8%, 21%, 18% and 32%, respectively
Table 1.
Patient Demographics
| Clinical Variable | |
|---|---|
| Total Cohort N | 1115 |
| Gender: Males N (%) | 597 (53) |
| Median disease duration at last follow up (months [range]) | 38.1 [0–541.7] |
| Median age at diagnosis (years [range]) | 15 [1–77] |
| Diagnosis ≤ 16 years of age N (%) | 632 (57) |
| Caucasian N (%) | 1062 (95) |
| Ashkenazi Jewish N (%) | 532 (48) |
| Small bowel involvement N (%) | 843 (76) |
| Stricturing and/or Internal Penetrating disease N (%) | 454 (51) |
| Quartile Sum Score groups N (%) 1 2 3 4 |
89(8) 231(21) 205(18) 362(32) |
| NOD2 positive N (%) | 321 (33) |
| Intestinal resection surgery any time N (%) | 444 (40) |
| Intestinal resection surgery within 60 months N (%) | 239 (21) |
Need for early surgery
Single SNP Genome Wide Associations
A total of 239 cases and 375 controls were included in this analysis. Controls included patients who were followed for at least 60 months and never had surgery. From the targeted analysis of the 71 known CD susceptibility loci,5were found to be associated (p ≤ 0.05) with need for surgery (Table 2). Table, Supplemental Digital Content 2, http://links.lww.com/IBD/A155, illustrates the allele frequency of the known CD loci in our cohort. Table 3 lists the results of the single SNP associations (with a p value < 5×10−5 identified in the genome-wide approach. The association with the need for surgery at the IL12B locus remains significant when corrected for multiple tests in the known susceptibility loci (pc = 0.001). No association was seen between any of the NOD2 SNPs and need for surgery.
Table 2.
Known CD susceptibility loci: SNPs associated with need for surgery (P < 0.05)
| SNP | Locus | Risk Allele* |
Mean allele frequency |
Positional Candidate Gene (s) of Interest |
OR | 95% CI | P value |
|---|---|---|---|---|---|---|---|
| rs6556412 | 5q33 | A | 0.40 | IL12B | 1.6 | 1.3 – 2.0 | 1.5×10−4 |
| rs3764147 | 13q14 | G | 0.32 | C13orf31 | 1.4 | 1.1 – 1.8 | 0.01 |
| rs12037853 | 1p31 | G | 0.29 | IL23R | 1.4 | 1.0–1.7 | 0.02 |
| rs7130588 | 11q13 | A | 0.41 | C11orf30 | 0.8 | 0.6–1.0 | 0.03 |
Risk allele as defined as risk for CD
Table 3.
Genome-wide analysis:: SNPs associated with need for surgery (p < 5.0×10−5)
| SNP | Loci | Risk Allele |
Mean Allele Frequency |
Positional candidate Gene(s) of Interest |
OR | 95% CI | P value |
|---|---|---|---|---|---|---|---|
| rs11103429 | 9q34 | G | 0.05 | RXRA, COL5A1 | 3.6 | 2.1–6.3 | 5.8 ×10-6 |
| rs11235508 | 11q13 | G | 0.13 | PDE2A, INPPL1, CLPB, ARAP1 | 2.2 | 1.5–3.0 | 1.5×10-5 |
| rs7159570 | 14q24 | G | 0.2 | JDP2,BATF,FLVCR2,TTLL5 | 0.5 | 0.4–0.9 | 1.8×10 -5 |
| rs11978472 | 7q21 | A | 0.3 | CACNA2D1 | 0.5 | 0.4–0.7 | 5.5×10-6 |
| rs7780687 | 7p21 | G | 0.2 | AHR | 2.0 | 1.4–2.8 | 2.7×10-5 |
| rs10272892 | 7p22 | G | 0.5 | SDK1 | 0.6 | 0.5–0.8 | 3.5×10-5 |
| rs2295655 | 14q32 | A | 0.2 | miRNA cluster, MEG9 | 1.8 | 1.3–2.3 | 3.6×10-5 |
| rs7029403 | 9q21 | A | 0.21 | KIF27, C9orf64, HNRNPK, RMI1, SLC28A3 | 0.5 | 0.4–0.7 | 3.3×10-5 |
| rs1722052 | 7q31 | A | 0.5 | - | 0.6 | 0.5–0.8 | 4.1×10-5 |
| rs13403289 | 2q31 | C | 0.4 | SESTD1 | 0.6 | 0.5–0.8 | 4.6 ×10-5 |
Multivariate analysis
Using logistic regression, we examined the significance of the associations of known CD susceptibility loci (p ≤ 0.05) (Table 2) together with the genome-wide identified loci (p < 10−5) (Table 3). IL12B remained independently associated with need for surgery (p = 0.0017).
Time to resective surgery
Single SNP Genome Wide Associations
The total number of patients included in the time to surgery analyses was 983 after principal component analysis and after exclusion of patients who underwent surgery at or within 3 months of diagnosis. Patients who went to surgery within 3 months of diagnosis likely needed surgery even at diagnosis and were excluded so to not bias the model. The known CD susceptibility loci and novel SNPs from the genome-wide analyses associated with time to surgery are shown in Tables 5 and 6.
Table 5.
Known CD susceptibility loci: SNPs associated with time to surgery (P < 0.05)
| SNP | Loci | Risk Allele |
Gene (s) of interest |
Hazard Ratio |
95% CI | P value |
|---|---|---|---|---|---|---|
| rs6556412 | 5q33 | A | IL12B | 1.5 | 1.2 – 1.9 | 0.0006 |
| rs12521868 | 5q31 | A | SLC22A4,SLC22A5,IRF1,IL3 | 1.3 | 1.0 – 1.6 | 0.0442 |
| rs1736148 | 21q21 | G | - | 1.3 | 1.0 – 1.6 | 0.0495 |
| rs3764147 | 13q14 | G | C13orf31 | 1.3 | 1.0 – 1.6 | 0.0503 |
Table 6.
Genome-wide analysis: SNPs associated with time to surgery (p < 5.0×10−5)
| SNP | Loci | Risk Allele |
Gene (s) of Interest |
Hazard Ratio | 95% CI | P value |
|---|---|---|---|---|---|---|
| rs7195303 | 16q23 | G | ZNRF1, LDHD, ZFP1, BCAR1, CTRB1/2 | 2.1 | 1.5 – 2.8 | 9.1×10−6 |
| rs471073 | 15q25 | A | AKAP13, KLHL25, | 1.8 | 1.4 – 2.3 | 1.7×10−5 |
| rs9346964 | 6q26 | A | OK1 | 1.7 | 1.3 – 2.1 | 1.8×10−5 |
| rs6469010 | 8q23 | G | ZFPM2 | 1.8 | 1.4 – 2.4 | 1.9×10−5 |
| rs4901140 | 14q21 | G | C14orf25, TTC6 | 1.7 | 1.4 – 2.3 | 1.9×10−5 |
| rs11583755 | 1p36 | A | THAP3, TAS1R1, PLEKHG5 | 0.6 | 0.4 – 0.7 | 2.3×10−5 |
| rs11235508 | 11q13 | G | CLPB, PDE2A | 1.9 | 1.4 – 2.5 | 2.5×10−5 |
| rs1998828 | 13q21 | A | - | 1.6 | 1.3 – 2.0 | 2.7×10−5 |
| rs2894207 | 6p21 | G | HLA-B/C, MICA** | 1.6 | 1.3 – 2.1 | 2.9×10−5 |
| rs9548517 | 13q13 | A | FREM2, STOML3 | 1.8 | 1.4 – 2.4 | 3.0×10−5 |
| rs2765122 | 13q12 | G | PCOTH, SPATA13, C1QTNF9B, MIPEP | 0.5 | 0.3 – 0.7 | 3.8×10−5 |
| rs13403289 | 2q31 | A | ZNF385B, SESTD1 | 0.6 | 0.5 – 0.8 | 4.1×10−5 |
| rs12531232 | 7q21 | A | ZNF804B | 1.6 | 1.3 – 2.0 | 4.3×10−5 |
| rs823559 | 12q23 | A | SLC5A8, ANO4 | 2.0 | 1.4 – 2.8 | 4.8×10−5 |
| rs6957717 | 7p15 | A | SKAP2, C7orf71 | 0.6 | 0.4 – 0.7 | 4.91×10−5 |
Locus with previous association to UC.
Predictive Models of Time to Surgery
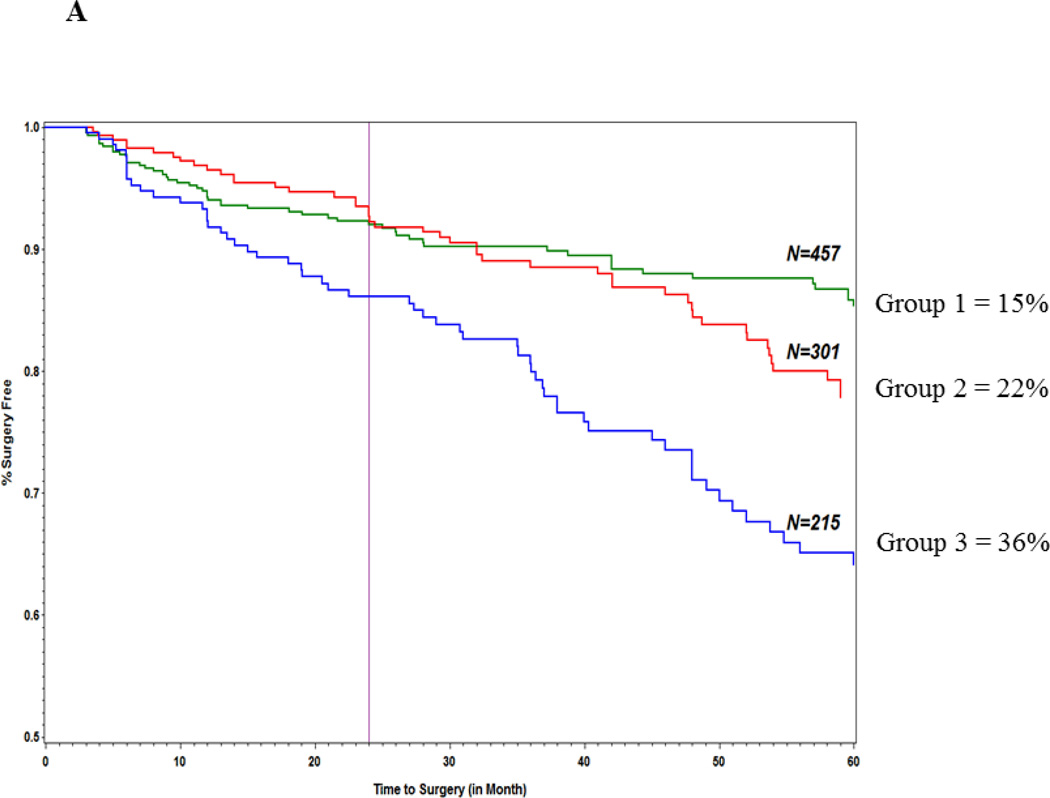
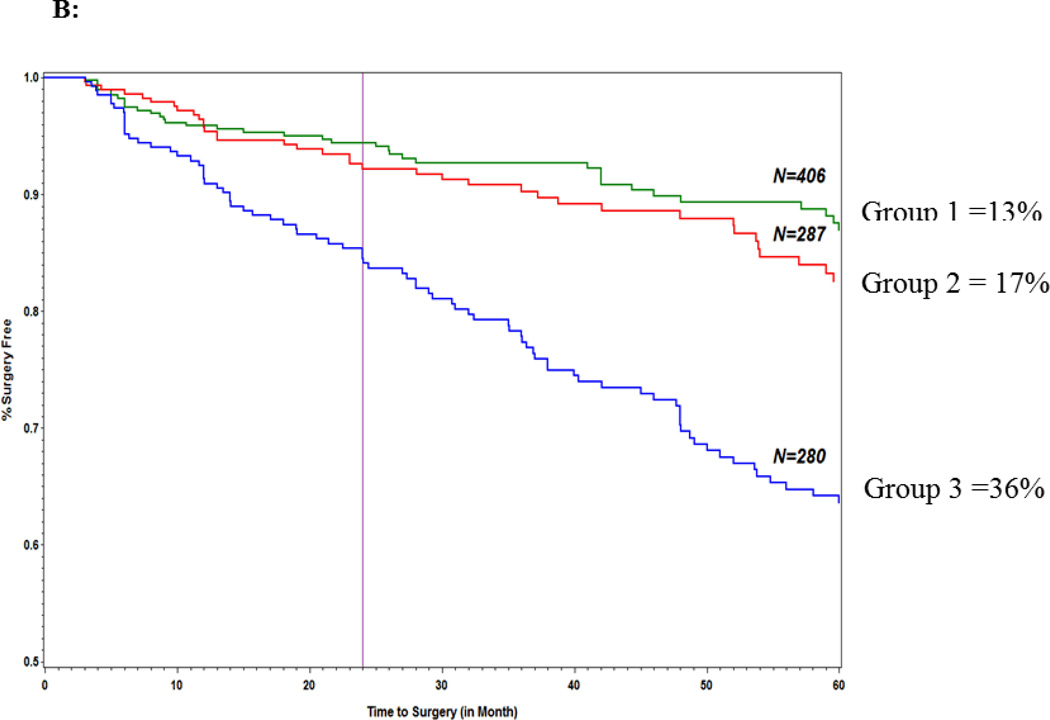
The goal of these analyses were to first determine using Cox regression modeling the variables that were independently associated with progression to surgery and then to use log rank testing to compare the survival curves for different risk strata within each model in order to determine which patients progressed to surgery faster. As previously noted in the methods section the 3 risk strata were derived based on the sum of the variables in the final predictive models: strata 1 = ≤ the 25% quartile of the sum of variables, strata 2 = between 25% and 75 % quartiles of the sum of variables and strata 3 = ≥ 75% quartile of the sum of variables. Each clinical and serology variable gets a score of 1 or 0 and an additive genetic model was adopted.Table 7 shows the hazard ratios for the genetics only model (Model I) and Figure 1A illustrates the survival curves for the 3 risk strata in this model. The proportion of patients in the lowest strata score that progressed to surgery within 60 months was significantly less (15%) than the medium(22%) and highest strata (36%) (log rank test: p=3.2×10−6). In the clinical variables only model(Model II) only small bowel predicted time to surgery (HR [95% CI] 3.3 [1.9–5.6] p=1.28×10−5) and neither age at diagnosis or gender showed statistically significant association with time to surgery. For model IV (genetics + clinical) all 4 SNPs from the genetics only model along with small bowel disease location and age at diagnosis(HR [95% CI] = 3.2 [1.9 – 5.4] p=2.2×10−5 and 1.5 [1.1 – 2.0] p = 0.02, respectively)were associated with time to surgery. Figure 1B illustrates the survival curves for the 3 strata for model IV. In our final model (Model V) which included NOD2 and quartile sum score groups in addition to genetic and clinical variables, NOD2(HR [95% CI]; 1.7 [1.2–2.5]; p = 0.006), 3 of the 4 SNPS from the genetics only model (IL12B, ZNRF1 and 21q21) in addition to small bowel disease location and age at diagnosis were all associated with time to surgery. Quartile sum scores were not predictive of surgery. As seen in Model I, patients in the highest risk strata progressed to surgery faster for models IV and V(p= 1.7×10−10 and p = 1.0×10−6, respectively) Moreover, there was a greater than 20% difference in frequency of progression to surgery between the lowest risk and highest risk strata in all 3 models. The AUC for the 5 models are shown in Table 8. The AUC of all 5 models was validated by 5-fold cross validation (80/20) and no difference was seen between any of the discovery and validation cohorts. Model IV and V had the highest AUC amongst the 5 models which were also very similar AUCs (0.65 [0.62–0.68] and 0.67 [0.65–0.69], respectively). The confidence intervals of these 2 models did not overlap with the genetics only model (AUC = 0.59 [0.57–.60]. The AUC for the clinical only model was the lowest (0.24 [0.22–0.27].
Table 7.
Model I: Genetics only; Cox proportional Regression
| SNP | Top Hit SNP (TH) or GWAS SNP (GW) |
Gene (s) of Interest |
HR | 95% CI | P value |
|---|---|---|---|---|---|
| rs6556412 | TH | IL12B | 1.5 | 1.0 – 1.6 | 0.0006 |
| rs12521868 | TH | SLC22A4, SLC22A5 | 1.3 | 1.2 – 1.9 | 0.03 |
| rs1736148 | TH | 21q21 | 1.3 | 1.0 – 1.6 | 0.03 |
| rs7195303 | GW | ZNRF1, LDHD | 2.1 | 1.5 – 2.9 | 10×10−5 |
Figure 1.
Predictors of surgery: (A) genetics model; (B) combined genetic and clinical model.
Table 8.
AUC and Cross Validation for the 5 Cox Regression Models
| Model | AUC Discovery |
AUC Cross Validation |
Range |
|---|---|---|---|
| I. Known CD susceptibility loci and genome-wide identified loci | 0.587 | 0.589 | 0.574– 0.598 |
| II. Clinical only: SB, gender, Age at Diagnosis, | 0.242 | 0.242 | 0.215–0.274 |
| III. Clinical + Serologies | 0.242 | 0.242 | 0.215–0.274 |
| IV. Genetics + Clinical | 0.646 | 0.649 | 0.630–0.677 |
| V. Genetics + NOD2 status + Clinical + Serologies | 0.658 | 0.661 | 0.647–0.689 |
Discussion
The ability to identify CD patients with a more aggressive natural history would be of benefit to both clinicians and patients and a major step on the road to a more personalized or individualized approach to clinical care. This study of over 1100 well-characterized CD cases is the first to include demographic, clinical, serological and genetic parameters to predict the need for early intestinal resection surgery in CD. The most intriguing finding is the consistent association between the 1L12B locus and both the need for surgery and time to surgery, a finding that remains significant through all the model development, and remains significant after correcting for multiple-testing in our hypothesis driven analyses.IL12B (also known as natural killer cell stimulatory factor 2 or p40) associates with IL23A to form IL23, a known stimulator of the Jak-Stat signaling pathway and a pathway with proven importance in IBD (31, 32).This locus has previously been associated with complicated disease in CD and also the need for colectomy in medically refractory ulcerative colitis (19, 33). These cumulative data strongly implicate this locus not only in disease susceptibility but also in the natural history of IBD and together, with both our current understanding of the biological relevance of this pathway and also the potential role of anti-IL12 therapies in IBD, suggests the intriguing and attractive possibility of targeted anti-IL12 therapy in these high-risk groups. Other known CD loci associated in this study with the need for surgery includes the IL23R locus.
A novel aspect of this study is the whole genome-approach looking for loci associated with need and time to early surgery. However, none of the putative loci met the criteria for genome-wide level of significance (< 5×10−8). Thus, our putative disease severity associated SNPs will need to be replicated in other cohorts. Nevertheless a number of loci met genome-wide suggestive association (Tables 3 and 6), including the HLA region (tagging HLA B/C and MICA (a stress induced antigen recognized by intestinal epithelial gamma delta T cells)) associated with a number of immune related conditions including UC, Behcet’s, Type 1 Diabetes, lupus, and psoriasis. Other implicated genes of potential interest include: CACNA2D1 which has a role in apoptosis (34), PLEKHG5 (NFkB activator); THAP3 (apoptotic pathway modifier); SPATA13 (cell adhesion and associated with intestinal adenoma formation); and SKAP2 (macrophage adhesion). The major finding of this study is that the combination of different parameters provides an improved predictor of progression to surgery in CD than any single parameter. The data demonstrate that progression to surgery is more accurately predicted in patients with CD who ‘carry’ both clinical and genetic factors with clear improvements as different risk variables are added to the model. Our findings suggest that if models are to be utilized in clinical practice to facilitate the development of a more individualized approach to managing CD patients, then they should, at least, include these different parameters for maximum differentiation of risk groups and hence the greatest utility in clinical practice.
We recognize that this model will need to be validated in another cohort which is currently underway. Additionally a limitation to this model is the unknown effect of therapy on intestinal resection rates in the context of genetic and clinical risk factors and this will be the next step in determining whether therapy alters the natural history in the face of markers that predict risk of surgery. The effect of smoking on the adult subset was examined in both the logistic and survival analyses and smoking was not shown to be an independent risk factor for surgery in our cohort (data not shown). This however may be best analyzed in the context of treatment.
The approach outlined in this manuscript may also help identify novel targets for therapeutic intervention in CD and the IL12B associations demonstrated here, as well as its previous association with both severe UC and CD, provides compelling evidence that studying a phenotype of predicted severe disease is a potential fruitful area. Our data suggests that models built using these modalities may well be able to distinguish patients to such a degree that they provide clinical utility and also suggest that these studies need to be performed in other populations in order to confirm these findings and the validity of this approach. Furthermore these results also suggest that stratified trials, based on confirmed findings such as the risk of more severe disease in those carrying a risk IL12B, for early use of biologic therapy are warranted for patients at high risk of surgery.
Supplementary Material
Table 4.
Model I: Genetics Only Logistic Regression
| SNP | Top Hit SNP (TH) or GWAS SNP (GW) |
Gene of Interest |
OR | 95% CI | P value |
|---|---|---|---|---|---|
| rs6556412 | TH | IL12B | 1.5 | 1.1–2.0 | 0.001 |
| rs12037853 | TH | IL23R | 1.3 | 1.0–1.8 | 0.03 |
| rs7130588 | TH | C11orf30 | 0.8 | 0.6–1.0 | 0.04 |
| rs11103429 | GW | RXRA, COL5A1 | 4.0 | 2.2–7.0 | 2.7×10−6 |
| rs11978472 | GW | CACNA2D1 | 0.5 | 0.4–0.7 | 3.5×10−6 |
Acknowledgments
Funding
This study was supported in part by NIH/NIDDK grant P01-DK046763; Cedars-Sinai Medical Center Inflammatory Bowel/ Immunobiology Institute Research Funds; The Feintech Family Chair in IBD (SRT); The Cedars-Sinai Board of Governors’ Chair in Medical Genetics (JIR); The Abe and Claire Levine Chair in Pediatric IBD (MD), Joshua L and Lisa Z. Greer Chair in IBD Genetics (DPBM), DK062413 (DPBM), CTSI Grant UL1RR033176 and DERC grant DK063491. Additional support was received from The Leona M. and Harry B. Helmsley Charitable Trust
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing Interest: None to declare.
Contributors Statement:
conception and design, or analysis and interpretation of data: all authors
drafting the article or revising it critically for important intellectual content: all authors
final approval of the version to be published: all authors
Statistical analyses: Soonil Kwon
Disclosures:
Marla Dubinsky, consultant Prometheus Labs
References
- 1.Cosnes J, Cattan S, Blain A, et al. Long-term evolution of disease behavior of Crohn's disease. Inflammatory Bowel Diseases. 2002;8:244–250. doi: 10.1097/00054725-200207000-00002. [DOI] [PubMed] [Google Scholar]
- 2.Cosnes J, Nion-Larmurier I, Beaugerie L, et al. Impact of the increasing use of immunosuppressants in Crohn's disease on the need for intestinal surgery. Gut. 2005;54:237–241. doi: 10.1136/gut.2004.045294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaugerie L, Seksik P, Nion-Larmurier I, et al. Predictors of Crohn's disease. Gastroenterology. 2006;130:650–656. doi: 10.1053/j.gastro.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 4.Dubinsky MC, Lin YC, Dutridge D, et al. Serum immune responses predict rapid disease progression among children with Crohn's disease: immune responses predict disease progression. Am J Gastroenterol. 2006;101:360–367. doi: 10.1111/j.1572-0241.2006.00456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubinsky MC, Kugathasan, Mei L, et al. Increased immune reactivity predicts aggressive complicating Crohn's disease in children. Clin Gastro and Hep. 2008;6:1105–111. doi: 10.1016/j.cgh.2008.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papp M, Altorjay I, Dotan N, et al. New serological markers for inflammatory bowel disease are associated with earlier age at onset, complicated disease behavior, risk for surgery, and NOD2/CARD15genotype in a Hungarian IBD cohort. Am J Gastroenterol. 2008;103:665–681. doi: 10.1111/j.1572-0241.2007.01652.x. [DOI] [PubMed] [Google Scholar]
- 7.Ferrante M, Henckaerts L, Joossens M, et al. New serological markers in inflammatory bowel disease are associated with complicated disease behavior. Gut. 2007;56:1394–1403. doi: 10.1136/gut.2006.108043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sands BE, Arsenault JE, Rosen MJ, et al. Risk of early surgery for Crohn's disease: implications for early treatment strategies. Am J Gastroenterol. 2003;98:2712–2718. doi: 10.1111/j.1572-0241.2003.08674.x. [DOI] [PubMed] [Google Scholar]
- 9.Amre DK, Lu SE, Costea F, et al. Utility of serological markers in predicting the early occurrence of complications and surgery in pediatric Crohn's disease patients. Am J Gastroenterol. 2006;101:645–652. doi: 10.1111/j.1572-0241.2006.00468.x. [DOI] [PubMed] [Google Scholar]
- 10.Mow WS, Vasiliauskas EA, Lin YC, et al. Association of antibody responses to microbial antigens and complications of small bowel Crohn's disease. Gastroenterology. 2004;126:414–424. doi: 10.1053/j.gastro.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 11.Arnott ID, Landers CJ, Nimmo EJ, et al. Sero-reactivity to microbial components in Crohn's disease is associated with disease severity and progression, but not NOD2/CARD15 genotype. Am J Gastroenterol. 2004;99:2376–2384. doi: 10.1111/j.1572-0241.2004.40417.x. [DOI] [PubMed] [Google Scholar]
- 12.Siegel CA, Siegel LS, Hyams JS, et al. A Real- Time tool to display the predicted disease course and treatment response for children with Crohn's disease. Inflammatory Bowel Diseases. 2011;17:30–38. doi: 10.1002/ibd.21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lesage S, Zouali H, Cezard JP, et al. CARD15/NOD2 mutational analysis and genotype-phenotype correlation in 612 patients with inflammatory bowel disease. Am J of Hum Genet. 2002;70:845–857. doi: 10.1086/339432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abreu MT, Taylor KD, Lin YC, et al. Mutations in NOD2 are associated with fibrostenosing disease in patients with Crohn's disease. Gastroenterology. 2002;123:679–688. doi: 10.1053/gast.2002.35393. [DOI] [PubMed] [Google Scholar]
- 15.Kugathasan S, Maresso K, Collins N, et al. L1007Fsinc variant of CARD15/NOD2 is strongly associated with early onset and fibrostenosing behavior in pediatric Crohn’s disease. Clin Gastroenterol Hepatol. 2004;2:1003–1009. doi: 10.1016/s1542-3565(04)00452-5. [DOI] [PubMed] [Google Scholar]
- 16.Russell RK, Drummond HE, Nimmo EE, et al. Genotype-phenotype analysis in childhood-onset Crohn's disease: NOD2/CARD15 variants consistently predict phenotypic characteristics of severe disease. Inflammatory Bowel Diseases. 2005;11:955–964. doi: 10.1097/01.mib.0000183423.38037.f3. [DOI] [PubMed] [Google Scholar]
- 17.Alvarez-Lobos M, Arostegui JI, Sans M, et al. Crohn's disease patients carrying Nod2/CARD15 gene variants have an increased and early need for first surgery due to stricturing disease and higher rate of surgical recurrence. Ann Surg. 2005;242:693–700. doi: 10.1097/01.sla.0000186173.14696.ea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seiderer J, Brand S, Herrmann KA, et al. Predictive value of the CARD15 variant 1007fs for the diagnosis of intestinal stenoses and the need for surgery in Crohn's disease in clinical practice: results of a prospective study. Inflammatory bowel diseases. 2006;12:1114–1121. doi: 10.1097/01.mib.0000235836.32176.5e. [DOI] [PubMed] [Google Scholar]
- 19.Henckaerts L, Van Steen K, Verstreken I, et al. Genetic risk profiling and prediction of disease course in Crohn's disease patients. Clin Gastro Hep. 2009;7:972–980. doi: 10.1016/j.cgh.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 20.Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nature Genetics. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett JC, Hansoul S, Nicolae DL, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nature Genetics. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGovern DP, Jones MR, Taylor KD, et al. Fucosyltransferase 2 (FUT2) non-secretor status is associated with Crohn's disease. Human Molecular Genetics. 2010;19:3468–3476. doi: 10.1093/hmg/ddq248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schunkert H, König IR, Kathiresan S, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nature Genetics. 2011;43:333–338. doi: 10.1038/ng.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Okser S, Lehtimäki T, Elo LL, et al. Genetic variants and their interactions in the prediction of increased pre-clinical carotid atherosclerosis: the cardiovascular risk in young Finns study. PLoS Genet. 2010;30:6. doi: 10.1371/journal.pgen.1001146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Targan SR, Landers CJ, Yang H, et al. Antibodies to CBir1 flagellin define a unique response that is associated independently with complicated Crohn's disease. Gastroenterology. 2005;128:2020–2028. doi: 10.1053/j.gastro.2005.03.046. [DOI] [PubMed] [Google Scholar]
- 26.Vasiliauskas EA, Kam LY, Karp LC, et al. Marker antibody expression stratifies Crohn's disease into immunologically homogeneous subgroups with distinct clinical characteristics. Gut. 2000;47:487–496. doi: 10.1136/gut.47.4.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gunderson KL, Steemers FJ, Lee G, et al. A genome-wide scalable SNP genotyping assay using microarray technology. Nat Genet. 2005;37:549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- 28.Morin PA, Saiz R, Monjazeb A. High-throughput single nucleotide polymorphism geno-typing by fluorescent 5’ exonuclease assay. Biotechniques. 1999;27:538–552. doi: 10.2144/99273rr02. [DOI] [PubMed] [Google Scholar]
- 29.Price AL, Patterson NJ, Plenge RM, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 30.Terry Therneau and original Splus-R port by Thomas Lumley. Survival analysis, including penalized likelihood. R package version 2.36-5. 2011 http://CRAN.R-project.org/package=survival. [Google Scholar]
- 31.McGovern DP, Rotter JI, Mei L, et al. Genetic epistasis of IL23/IL17 pathway genes in Crohn's disease. Inflammatory Bowel Diseases. 2009;15:883–889. doi: 10.1002/ibd.20855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang K, Zhang H, Kugathasan S, Annese V, et al. Diverse genome-wide association studies associate the IL12/IL23 pathway with Crohn Disease. Am J Hum Genet. 2009;84:399–405. doi: 10.1016/j.ajhg.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haritunians T, Taylor KD, Targan SR, et al. Genetic predictors of medically refractory ulcerative colitis. Inflammatory Bowel Diseases. 2010;16:1830–1840. doi: 10.1002/ibd.21293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carboni GL, Gao B, Nishizaki M, et al. CACNA2D2-mediated apoptosis in NSCLC cells is associated with alterations of the intracellular calcium signaling and disruption of mitochondria membrane integrity. Oncogene. 2003;22:615–626. doi: 10.1038/sj.onc.1206134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.