Abstract
Diet-induced obesity predisposes individuals to insulin resistance, and adipose tissue has a major role in the disease. Insulin resistance can be induced in cultured adipocytes by a variety of treatments, but it is unknown what aspects of the in vivo responses are captured by these models. We use global RNA-sequencing (RNA-Seq) to investigate changes induced by TNFα, hypoxia, dexamethasone, high insulin, and a combination of TNFα and hypoxia, comparing the results to the changes in white adipose tissue from diet-induced obese (DIO) mice. We found that different in vitro models capture distinct features of DIO adipose insulin resistance, and a combined treatment of TNFα and hypoxia is most able to mimic the in vivo changes. Using genome-wide DNase I hypersensitivity followed by sequencing (DNase-Seq), we further examined the transcriptional regulation of TNFα-induced insulin resistance, and we found that C/EPBβ is a potential key regulator of adipose insulin resistance.
Introduction
Obesity has become a global epidemic and predisposes individuals to insulin resistance, which in turn is a risk factor of many metabolic diseases (e.g., type 2 diabetes, hypertension, atherosclerosis, and cardiovascular diseases) and cancer (Reaven 2005). The 3T3-L1 cell line (Green and Meuth 1974) has been widely used to study insulin resistance in adipocytes (Knutson and Balba 1997). Many agents are used to induce insulin resistance in differentiated 3T3-L1; these include TNFα (Ruan, Hacohen et al. 2002), IL-1(Jager, Grémeaux et al. 2007), IL-6 (Rotter, Nagaev et al. 2003), free fatty acids (Nguyen, Satoh et al. 2005), dexamethasone (Sakoda, Ogihara et al. 2000), high insulin (Thomson, Williams et al. 1997), glucosamine (Nelson, Robinson et al. 2000), growth hormone (Smith, Elmendorf et al. 1997), and hypoxia (Regazzetti, Peraldi et al. 2009), among others. It is unclear what features of in vivo adipose insulin resistance are captured by each of the different in vitro models, and whether a combination of treatments would be able to capture the in vivo changes better than a single treatment.
In order to address these issues, we have examined the changes in transcription and transcriptional regulation induced by TNFα, hypoxia, dexamethasone, high insulin and a combination of TNFα and hypoxia in differentiated 3T3-L1 adipocytes. TNFα is a proinflammatory cytokine, which is secreted by adipocytes and macrophages in adipose tissue. Since the discovery of its role in obesity-linked insulin resistance (Hotamisligil, Shargill et al. 1993), it has been widely used to induce insulin resistance in cultured cells. A more recently- discovered way to induce insulin resistance is hypoxia treatment. Obese adipose tissue is hypoxic, which can lead to dysregulation of adipokine production (Hosogai, Fukuhara et al. 2007) and insulin signaling (Regazzetti, Peraldi et al. 2009). Both TNFα and hypoxia have been linked to inflammatory responses. Interestingly, dexamethasone, a synthetic glucocorticoid frequently prescribed as an anti-inflammatory agent and immunosuppressant, can also induce insulin resistance. Excessive use of dexamethasone results in Cushing’s syndrome, characterized by central obesity, insulin resistance and other metabolic abnormalities (Andrews and Walker 1999). Elevated endogenous glucocorticoid (e.g., the hormone cortisol in humans and corticosterone in rodents) can also lead to visceral obesity and aggravate high-fat-diet-induced insulin resistance (Masuzaki, Paterson et al. 2001; Wang 2005). Lastly, high levels of insulin can induce insulin resistance, and hyperinsulinemia is postulated to be both the result and the driver of insulin resistance (Shanik, Xu et al. 2008).
To understand the relationship of these models to each other and to the in vivo setting, we have made use of high-throughput RNA-sequencing (RNA-Seq) technology (Trapnell, Williams et al. 2010) and analyzed the in vitro data in parallel with adipose tissue transcriptome data from three independent diet-induced obesity (DIO) mouse models. We find that the different in vitro models show diverse transcriptional responses, each of which captures a different aspect of the in vivo data. The TNFα and hypoxia models capture the downregulation of many glucose, lipid and amino acid metabolic pathways observed in DIO mouse adipose tissue that are not detected in the high insulin and dexamethasone models. Conversely, the upregulation of the inflammatory responses in DIO adipose tissue is mainly captured by the TNFα model. Interestingly, the combination of hypoxia and TNFα treatments resembles the actual in vivo condition more than any individual treatment.
We further explored the differences in transcriptional regulation among the in vitro models using DNase I hypersensitivity followed by massively parallel sequencing (DNase-Seq), identifying many condition-specific regulatory sites. Analysis of DNase-Seq data from TNFα-induced insulin resistance revealed that in addition to NF-κB, C/EBPβ is a potential regulator of genes induced by TNFα, and loss of PPARγ binding is likely to mediate many of the gene repression changes upon TNFα treatment.
Results
Setting up diverse in vitro insulin resistance models in the 3T3-L1 cell line
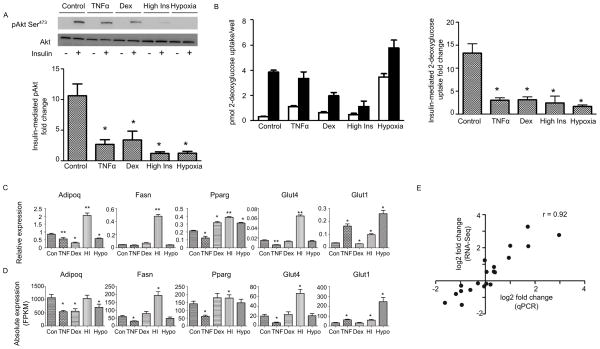
We induced insulin resistance models in mature 3T3-L1 cells using TNFα, hypoxia, dexamethasone, and high insulin following established protocols (see Methods, Figure S1). All four models exhibited compromised insulin responses as determined by phosphorylation of Akt at serine 473 (Figure 1A) and 2-deoxyglucose uptake (Figure 1B). Nevertheless, the expression of five adipocyte marker genes (Figure 1C) varied dramatically among these models. For example, the insulin-sensitizing adipokine adiponectin (Adipoq) decreases in all models except the high insulin model, and the insulin-sensitive glucose transporter Glut4 decreases only in the TNFα model. The variation in these marker genes suggests that the transcriptome shifts of the four insulin resistance models are likely to be diverse and distinct.
Figure 1. Setting up four diverse in vitro insulin resistance models in 3T3-L1, See also Figure S1 and S2 and Table S1.
(A) Western blot analysis of pAkt Ser473 before and after 10min of 20nM insulin stimulation in 3T3-L1 for the control (untreated) model and the four insulin resistance models. Bottom: Average fold-change of Akt phosphorylation level before and after insulin stimulation for each condition, each normalized to total Akt protein level (n=3). An asterisk (*) indicates p < 0.05 when compared with the control model by t-test. Error bars represent ± SEM.
(B) 2-deoxyglucose uptake in the basal state (open bars) and after 30min of 20nM insulin stimulation (solid bars) for the control (untreated) model and the four insulin resistance models. Right: Average fold change of 2-deoxyglucose uptake by dividing insulin-mediated over basal uptake for each model (n=5). An asterisk (*) indicates p < 0.05 when compared with the control model by t-test. Error bars represent ± SEM.
(C) Expression of five adipocyte marker genes in the different models measured by qPCR. Relative expression is calculated by normalizing with the house keeping gene ribosomal protein S27 (Rps27). Data is presented as mean ± SEM (n=3). Statistical significance is indicated (*p < 0.05, **p < 0.01).
(D) Same as (C), but measured by RNA-Seq. Shown is the upper and lower bound expression values calculated by Cuffdiff in FPKM. Statistical significance is indicated (*q < 0.05).
(E) Pearson correlation coefficient between the gene expression fold changes (log 2) from qPCR (C) and RNA-Seq (D).
Diverse transcriptional changes associated with models of insulin resistance
In order to obtain a genome-wide picture of the transcriptional outcomes, we carried out RNA-Seq. These data generally agreed well with the qPCR-based results for the five adipocyte marker genes (Figure 1D and 1E). The diverse effects of each method of inducing insulin resistance can be seen by analyzing glycolysis and triglyceride synthesis and degradation, key pathways of adipose metabolism. The enzymes that catalyze the irreversible steps of glycolysis, including hexokinase (Hk1and Hk2), phosphofructokinase (Pfkl and Pfkp), and pyruvate kinase (Pkm2), are upregulated after the hypoxia, high insulin and TNFα treatments but not the dexamethasone treatment (Figure S2A). Regarding the triglyceride synthesis and degradation pathway (Figure S2B), diacylglycerol O-aceyltransferases (Dgat1 and Dgat2), which catalyze the reaction in which diacylglycerol is covalently joined to form long chain fatty acyl-CoAs, are repressed in the hypoxia and TNFα models but not the high insulin model. Hormone sensitive lipase (Lipe), which hydrolyses stored triglyceride to free fatty acid, is downregulated after TNFα treatment as previously reported (Ruan, Hacohen et al. 2002); it is also repressed in the hypoxia model but not in the high insulin and dexamethasone models. Gene ontology (GO) analysis (Table S1) confirms the diversity of the transcriptional responses in each condition
Anti-adipogenesis transcriptome shift of DIO mouse adipose tissue captured mainly by treatment with TNFα, hypoxia and a combination of TNFα and hypoxia
To understand how the in vitro expression changes relate to mouse insulin resistance models, we analyzed three independent microarray data sets comparing the gene expression of adipose tissue from DIO mice and normal chow diet-fed mice (Qi, Saberi et al. 2009; Fitzgibbons, Kogan et al. 2011; Fujisaka, Usui et al. 2011). Although the DIO mouse expression studies used diverse conditions (Table S2), the expression changes of DIO versus control were highly correlated.
As the in vivo data are likely to contain contributions from multiple cell types, we chose to focus our analysis on a set of genes that are most relevant to adipocytes. To this end, we identified adipogenesis-induced and adipogenesis-repressed genes that shows consistent expression changes between preadipocytes and adipocytes from three independent data sets (Schupp, Cristancho et al. 2009; Mikkelsen, Xu et al. 2010; Sun, Goff et al. 2013) (Table S3).
The in vivo and in vitro insulin resistance models demonstrate a striking expression pattern that is the opposite of that induced by adipogenesis (Figure 2A): many adipogenesis-induced genes are downregulated in the DIO mouse models, and conversely, many adipogenesis-repressed genes are upregulated. This anti-adipogenesis transcriptome shift is strongest in TNFα and hypoxia 3T3-L1 models, but it can also be detected clearly in the high insulin and dexamethasone models (Figures 2A).
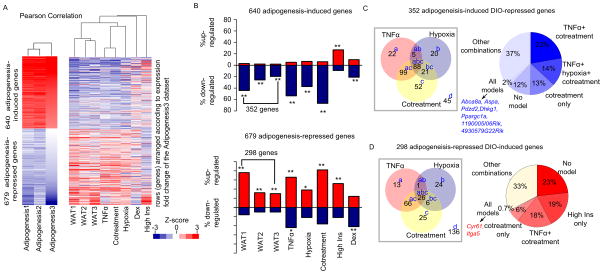
Figure 2. Anti-adipogenesis transcriptome shift of DIO mouse adipocytes captured mainly by the TNFα, hypoxia and cotreatment models, See also Figure S3 and Tables S2 and S3.
(A) Heatmaps showing the 1,319 genes that are concordantly differentially expressed between preadipocytes and differentiated adipocytes in three publications: Adipogenesis1 (Mikkelsen, Xu et al. 2010), Adipogenesis2 (Schupp, Cristancho et al. 2009), and Adipogenesis3 (Sun, Goff et al. 2013). The 1,319 genes consist of 640 that are upregulated (red) and 679 that are downregulated (blue) by greater than two-fold. Left: hierarchical clustering of the expression fold changes during adipogenesis among the three adipogenesis data sets. Right: hierarchical clustering of expression changes of the same genes in three independent mouse models of diet-induced obesity (WAT1, WAT2, and WAT3) and five 3T3-L1 insulin resistance models (TNFα, hypoxia, cotreatment, high insulin (High Ins), and dexamethasone (Dex)). The expression fold changes were calculated as follows: for the mouse models: adipose tissue from DIO vs. normal chow diet-fed mice; for the 3T3-L1 models: treated 3T3-L1 vs. untreated 3T3-L1. For both clustering analyses, expression ratios were converted to z-scores. Rows (genes) were pre-ranked according to the fold change of the Adipogenesis3 data set, while columns were clustered using the Pearson correlation similarity metric. The height of each arm of the dendrogram represents the distance between the different data sets.
(B) The percentage of the concordantly differentially expressed genes from panel A that are altered in each data set. Red bars represent the percentage of upregulated genes and blue bars represent the percentage of downregulated genes. Top panel showing the expression changes of the 640 adipogenesis-induced genes, among which 352 are downregulated in at least one of the mouse data sets; bottom panel showing the 679 adipogenesis-repressed genes, among which 298 genes are upregulated in at least one of the mouse data sets. Up- or downregulation was defined as having a log2 fold change of > 0.58 or < −0.58 when compared to the control data set. P-values (indicating whether the percentage is significantly larger than expected) were calculated using 1-sample proportion test (p < 1E-5: *; p < 1E-10: **).
(C) Left: Venn diagrams showing the overlap of genes that are downregulated in the TNFα, hypoxia or cotreatment model among the 352-adipogenesis-induced-DIO-repressed genes. The number of genes that fall into each category is indicated, and the letters next to the numbers refer to the particular gene ontology categories in Table 1. Right: The percentage breakdown of the 352-adipogenesis-induced-DIO-repressed genes according to their gene expression changes in the different combinations of models. Highlighted in blue are the 7 genes that are repressed in all the 5 in vitro models.
(D) Same as Figure 2 (C), except that the analysis was done on the 298-adipogenesis-repressed-DIO-induced genes, showing the overlap of genes that are upregulated in the TNFα, hypoxia or cotreatment model. Highlighted in red are the 2 genes that are induced in all the 5 in vitro models.
Having observed that the TNFα and hypoxia models appear to recapitulate the anti-adipogenesis transcriptome shift as seen in the DIO mouse, we investigated if a combination of TNFα and hypoxia treatments (hereafter known as cotreatment) would be better able to capture the in vivo changes. Hierarchical clustering analysis shows that the cotreatment model, like the TNFα and hypoxia model, also exhibits the anti-adipogenesis transcriptome shift (Figure 2A). Of the 640 adipogenesis-induced genes, 352 (55%) are repressed in at least one of the three DIO mouse models (Figure 2B). Conversely, 298 (44%) of the 679 adipogenesis-repressed genes are induced in at least one of the three DIO mouse models. Cotreatment with TNFα and hypoxia recapitulates the anti-adipogenesis transcriptome shift more than either the TNFα or the hypoxia model (Figure 2B).
We went on to explore the special features of the different in vitro models. The 352 adipogenesis-induced DIO-repressed genes are highly enriched in ones encoding proteins involved in oxidation reduction (p: 6.3E-17), fat cell differentiation (p: 1.2E-8), and various metabolic processes (p < 1E-4); 88% of these genes are repressed by the TNFα, hypoxia or cotreatment model (Figure 2C): those that are repressed by TNFα are most enriched in fat cell differentiation processes, while those repressed by hypoxia and cotreatment are most enriched in oxidation reduction reactions (Table 1). Of these 352 genes, 13% are captured only by the cotreatment model (Figure 2C). These are most enriched in the cellular component mitochondria, suggesting that the cotreatment model recapitulates the mitochondrial dysfunction during insulin resistance.
Table 1.
Gene ontology analysis of genes that undergo anti-adipogenesis transcriptome shift in the different in vitro models
| Of the 352 adipogenesis-induced DIO-repressed genes | ||
|---|---|---|
| Gene repression is captured by | % of Total | Enriched GO |
| TNFα (a+ab+ac+abc) | 61% | Mitochondrion (3.6E-15) fat cell differentiation (8.4E-06) fatty acid metabolic process (3.1E-5) |
| Hypoxia (b+ab+bc+abc) | 38% | Mitochondrion (1.7E-12) oxidation reduction(2.0E-4) |
| Cotreatment (c+ac+bc+abc) | 74% | Mitochondrion (4.3E-30) oxidation reduction(5.7E-11) |
| TNFα+cotreatment (ac) | 28% | Mitochondrion (4.1E-8) fat cell differentiation (6.1E-03) |
| Hypoxia+cotreatment (bc) | 6% | Mitochondrion (4.3E-4) |
| TNFα+hypoxia+ Cotreatment (abc) | 25% | Mitochondrion (4.1E-5) Propanoate metabolism (4.4E-4) |
| Only cotreatment (c) | 15% | Mitochondrion (1.8E-10) oxidation reduction(1.2E-5) |
| None of TNFα, hypoxia, or Cotreatment (d) | 13% | Mitochondrion (9.9E-6) oxidation reduction(7.1E-3) |
| Of the 298 adipogenesis-repressed DIO-induced genes | ||
|---|---|---|
| Gene induction is captured by | % of Total | Enriched GO |
| TNFα (a+ab+ac+abc) | 36% | Immune response (6.8E-3) Chemotaxis (3.2E-2) |
| Hypoxia (b+ab+bc+abc) | 19% | M phase (2.7E-5) Cell cycle (4.0E-5) |
| Cotreatment (c+ac+bc+abc) | 41% | Chemotaxis (1.4E-3) Cell cycle (1.1E-3) |
| TNFα+cotreatment but not hypoxia (ac) | 31% | Chemotaxis (5.0E-3) |
| Hypoxia+cotreatment (bc) | 2% | No enrichment |
| TNFα+hypoxia+ cotreatment (abc) | 9% | No enrichment |
| Only cotreatment (c) | 8% | No enrichment |
| None of TNFα, hypoxia, orcotreatment | 46% | Cell cycle (6.8E-21) M phase (3.3E-20) |
The different categories are indicated in Figure 2C & 2D.
P-values were calculated by Fisher-Exact tests assessing the significance of over-representation. Representative top GOs for each category are shown. Multiple hypothesis testings were corrected by Benjamini-Hochberg correction. Some combinations were not shown as there was no significant enrichment.
The 298 adipogenesis-repressed DIO-induced genes are enriched in cell-cycle related categories such as M-phase (p: 1.9E-28) and DNA replication (p: 5.3E-13), and inflammation-related processes such as cellular response to stress (p: 5.8E-3) and chemotaxis (p: 0.03). Of these genes, 54% are recapitulated in the TNFα, hypoxia or cotreatment models (Figure 2D). The TNFα model captures the upregulation of immune response and chemotaxis genes, while the hypoxia model captures the expression of genes related to cell cycle processes. Importantly, the cotreatment model captures the main feature of both the TNFα and the hypoxia models (Table 1).
Systemic transcriptome changes in adipose insulin resistance revealed by global pathway analysis
To better visualize the data and to identify groups of genes that set the different models apart, we carried out principal component analysis (PCA) of the adipogenesis-related genes across the 8 different models. PCA is a standard technique for reducing the dimensionality of data sets involving a large number of measurements while retaining as much variability as possible. The first principal component (PC1) explains 35% of the variance of the expression changes among the models. Projecting each data set along this axis reveals that the mouse models are well separated from the in vitro models (Figure 3A). Of the in vitro models, cotreatment is the closest to the mouse models. Genes making the most contribution to define PC1 are enriched in M phase, chemokine activity, fat cell differentiation, and various lipid metabolic processes (Table 2). The second principal component (PC2) captures 18.5% of the data set variance, with the TNFα, dexamethasone and high insulin models being closest to the mouse models; however, genes that contribute most to define PC2 are not enriched in any particular categories.
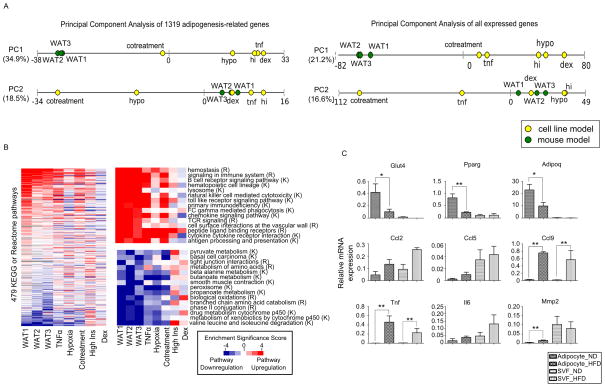
Figure 3. Systemic transcriptome changes in adipose insulin resistance revealed by global pathway analysis, See also Figure S4 and Tables S2, S3 and S4.
(A) Principal components analysis (PCA) of the eight models. Each principal component (PC1 and PC2) represents a direction of maximal variation in the matrix of expression data. The models are projected onto the first two principal components, with the cell line models shown as yellow dots and the mouse models shown as green dots. The distance between dots on each line corresponds to the distance between models along this projection. The numbers in parentheses represent the fraction of the variance in the expression fold change matrix that is explained by that particular principal component. Left: PCA using the 1,319 adipogenesis-related genes. Right: PCA using 13,043 genes with FPKM > 0.1.
(B) Heatmap showing the results from gene set enrichment analysis (GSEA) of genome-wide expression data. We computed the p-value for the significance of enrichment for either upregulation or downregulation of each pathway in the KEGG and Reactome databases. The log transformed p-values were taken as the Enrichment Significance Scores: upregulated pathways are represented as red (positive Enrichment Significance Score) and downregulated pathways as blue (negative Enrichment Significance Score), with the intensity of the color representing the significance of upregulation/downregulation; white represents no upregulation/downregulation; grey indicates that the number of genes was less than 15. Left: Rows (pathways) were ranked according to the sum of the Enrichment Significance Scores of the 3 mouse models. The scale bar is indicated at the right bottom corner of the figure and is the same for all panels. Top right: Illustration of the top 15 pathways upregulated in vivo (ranked as in the left panel). Pathways from Reactome are labeled (R) and those from KEGG are labeled (K). Bottom right: Illustration of the top 15 pathways downregulated in vivo.
(C) Expression of selected adipocyte and inflammatory response-related genes in isolated adipocytes or stromal vascular fraction (SVF) from normal chow-fed mice (ND) or high fat diet-fed mice (HFD) measured by qPCR. Relative expression is calculated by normalizing with the house keeping gene ribosomal protein S27 (Rps27). Data is presented as mean ± SEM (n=3). Statistical significance is indicated (*p < 0.05, **p < 0.01).
Table 2.
Gene ontology analysis of genes that define each principal component
| Representative GO associated with genes that contribute the most to define the adipogenesis-related PCA | |||
|---|---|---|---|
| Principal Component | Loading | GO | P-val |
| 1 | Most negative | Fat cell differentiation | 9.0E-7 |
| Acylglycerol metabolic process | 2.6E-6 | ||
| Neutral lipid metabolic process | 2.6E-6 | ||
| Most positive | M phase | 2.5E-5 | |
| Chemokine activity | 2.0E-3 | ||
| Representative GO associated with genes that contribute the most to define the genome-wide PCA | |||
|---|---|---|---|
| Principal Component | Loading | GO | P-val |
| 1 | Most negative | Lysosome | 4.9E-7 |
| Immune response | 3.4E-3 | ||
| Chemotaxis | 8.1E-3 | ||
| Most positive | Oxidation reduction | 6.0E-7 | |
| Glucose metabolic process | 3.1E-5 | ||
| Fat cell differentiation | 1.6E-4 | ||
| Valine, leucine and isoleucine degradation | 2.2E-4 | ||
| Lipid metabolic process | 3.2E-3 | ||
We repeated the principal component analysis at a genome-wide level by using all 13,043 genes with FPKM > 0.1. PC1 from the genome-wide PCA explains 21% of the data set variance. It separates the mouse models from the cell line models, and once again the cotreatment model is closest to the mouse models (Figure 3A). Interestingly, although we include 10 times more genes in the genome-wide PCA, the genes that contribute most to define the genome-wide PC1 are enriched in similar gene ontology categories as those that define the adipogenesis-related PC1 (Table 2). This suggests that the set of 1,319 adipogenesis-related genes is able to capture many of the genome-wide differences of the different models.
To systematically analyze pathway changes that occurred during adipose insulin resistance, we searched for differences in expression of pathways defined in the Reactome and KEGG databases using gene set enrichment analysis (GSEA) of the genome-wide expression data (Subramanian, Tamayo et al. 2005). For a similar analysis based on the set of adipogenesis-related genes, see Table S4. Plotting the enrichment scores for each condition in a heatmap reveals pathways that are upregulated or downregulated in the different insulin resistance models (Figure 3B). Consistently downregulated pathways in vivo include various glucose, lipid, and amino acid metabolic pathways as well as several cytochrome detoxification-related pathways. The in vivo downregulation of metabolic pathways is largely captured by the TNFα, hypoxia and the cotreatment models but not the other two models (Figure 3B). It is noticeable that while these models capture the direction of change (i.e. downregulation) of these pathways, the extent of downregulation in the in vitro models is not as significant as that in the DIO mouse models, an example of which is illustrated in the KEGG valine, leucine, and isoleucine degradation pathway (Figure S4).
The heatmap also reveals some consistently upregulated pathways in vivo, many of which relate to inflammatory responses, which are mainly captured by the TNFα and cotreatment models, and to a lesser extent, by the hypoxia and high insulin models (Figure 3B), while dexamethasone treatment downregulates many of these immune-related pathways. These analyses suggest that the systematic pathway changes occurring during insulin resistance vary in the different in vitro models, with the TNFα and cotreatment models capturing many of these key changes.
One of the most salient features of the TNFα and cotreatment models is that they appear to mimic the downregulation of key metabolic pathways and the upregulation of immune-related responses in vivo. Adipose tissue is a heterogeneous tissue comprising multiple cell types. Upon high-fat feeding, there is massive infiltration of activated macrophages into white adipose tissue (Weisberg, McCann et al. 2003). As our comparison was made between multiple in vitro adipocyte models and in vivo whole adipose tissue, it is uncertain if our in vitro models were capturing the upregulation of the various inflammatory processes in the adipocytes or the associated macrophage-enriched stromal vascular fraction (SVF). In order to tease out the contribution of the different cell types in adipose tissue, we isolated adipocytes and SVF from epipdidymal fat pads of age-matched normal chow-fed and DIO mice. While we found, as expected, that the downregulation of Pparg, Glut4 and Adipoq mainly occurs in adipocytes, the upregulation of chemokines (Ccl9), metalloproteinases (Mmp2) and inflammatory cytokines (Tnf) occur in both adipocytes and SVF (Figure 3C).
Identifying keys regulators of insulin resistance
The diverse transcriptional patterns of the in vitro models suggest that different transcriptional regulators are active under these conditions. In order to identify these transcription factors, we used an unbiased strategy based on DNase I hypersensitivity followed by high-throughput sequencing (DNase-Seq) (Hesselberth, Chen et al. 2009; Siersbaek, Nielsen et al. 2011) and computational analysis of the sequences of hypersensitive regions (Eguchi, Yan et al. 2008; Ling, Sugathan et al. 2010). Our DNase-Seq data are in good agreement with the literature (Birney, Stamatoyannopoulos et al. 2007; Mikkelsen, Xu et al. 2010; Siersbaek, Nielsen et al. 2011) at well-studied loci (Figure 4A).
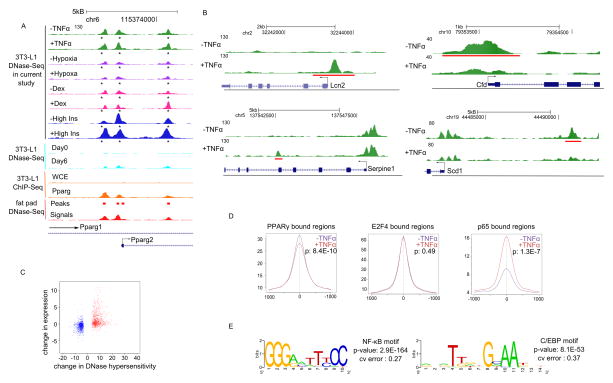
Figure 4. Identifying keys regulators of TNFα-induced insulin resistance, See also Figure S5 and Tables S5, S6 and S7.
(A) UCSC genome browser tracks showing the Pparg locus. Control track represents untreated mature 3T3-L1 cells. +TNFα, +Hypoxia, +Dex, and +High Ins represents 3T3-L1 cells treated with 24h of TNFα, hypoxia, dexamethasone, and high insulin, respectively. An asterisk (*) indicates DNase hypersensitive regions identified by the current study overlap with those in Day 6 differentiated 3T3-L1 (Siersbaek, Nielsen et al. 2011) and mouse fat pad (Birney, Stamatoyannopoulos et al. 2007), as well as PPARγ bound sites identified from PPARγ ChIP-Seq in 3T3-L1 (Mikkelsen, Xu et al. 2010). Arrows indicate the direction of transcription. Y-axis represents the height of the mapped sequenced reads and is the same for all tracks.
(B) UCSC genome browser tracks showing representative examples of genomic regions with altered DNase hypersensitivity after TNFα treatment. Gain in DNase hypersensitivity: Lcn2 (top left) and Serpine1 (bottom left); loss in DNase hypersensitivity: Cfd (top right) and Scd1 (bottom right). Genomic regions with a gain or loss of DNase hypersensitivity are underlined in red. +TNFα represents DNase-Seq after 24h of TNFα treatment; −TNFα represent DNase-Seq from unstimulated cells. Arrows indicate the direction of transcription.
(C) Genes within 10kb of regions with altered DNase hypersensitivity are more likely to be differentially expressed. An increase in DNase hypersensitivity is associated with an increase in expression and vice versa.
(D) Changes of DNase hypersensitivity at the PPARγ and p65 binding sites. Plots show the average number of tag counts from DNase-Seq experiments in a 2kb window around experimentally determined binding sites for PPARγ (left), E2F4 (middle) and p65 (right). P-values were calculated based on the tag counts from the DNase hypersensitivity experiments using Wilcoxon rank sum tests.
(E) The top two DNA sequence motifs from the p65 ChIP-Seq experiment as determined by THEME. Cv error represents cross-validation errors. P-values were calculated as described in Methods.
As our DNase-Seq data for TNFα-induced insulin resistance were of particularly high quality, we examined it in the greatest detail. MACS analysis (Zhang, Liu et al. 2008) identifies regions that lose or gain DNase hypersensitivity after TNFα treatment (examples are shown in Figure 4B). Genes near regions with altered DNase hypersensitivity are more likely to be differentially expressed (p < 2.2E-16) (Figure 4C and S5). The observed correlation between changes in DNase hypersensitivity and gene expression suggests that hypersensitive sites may represent loci where there is a gain or loss of regulator binding. Motif analysis of these regions (see Methods) identified a number of potential regulators in each condition, including, as expected, NF-κB and AP-1 for TNFα treatment, glucocorticoid receptor (GR) for dexamethasone treatment, and hypoxia-inducible factor (Hif) in hypoxia (Tables S5 and S6).
Using both previously reported ChIP data and new experiments, we were able to confirm several hypotheses emerging from the motif analysis. To test our hypothesis that PPARγ regulates TNFα-repressed genes, we examined previously reported PPARγ binding data (Mikkelsen, Xu et al. 2010). Indeed, PPARγ-bound sites lose hypersensitivity in the TNFα- treated cells (p: 8.44E-10) (Figure 4D). By contrast, DNase hypersensitivity did not change at E2F4 bound regions (p = 0.49) (Figure 4D) (MacIsaac, Lo et al. 2010). Thus, it appears that PPARγ may be an important regulator of TNFα-repressed genes.
To test the hypothesis that changes in hypersensitivity can be used to predict an increase in regulator binding, we carried out a p65 ChIP-Seq experiment on the TNFα-treated cells. We found that close to 60% of the high confidence p65 bound sites (p < 1E-10, 260 out of 437) overlap with the TNFα-induced DNase hypersensitivity regions. Examples of p65 bound genes include Ccl2, Ccl7, Saa3, Hp, Lcn2, etc.; many of which are well-known targets of NF-κB. The average DNase hypersensitivity profile around p65 bound sites increases in the TNFα-treated cells (p: 1.3E-7) compared to the control (Figure 4D). It is noteworthy that the C/EBP motif is highly enriched at p65-bound sites (Figure 4E and Table S7). This observation suggests that one or more members of the C/EBP transcription factor family are not only potential regulators of TNFα-induced insulin resistance, but use some of the same regulatory sites as p65.
C/EBPβ in TNFα-induced insulin resistance
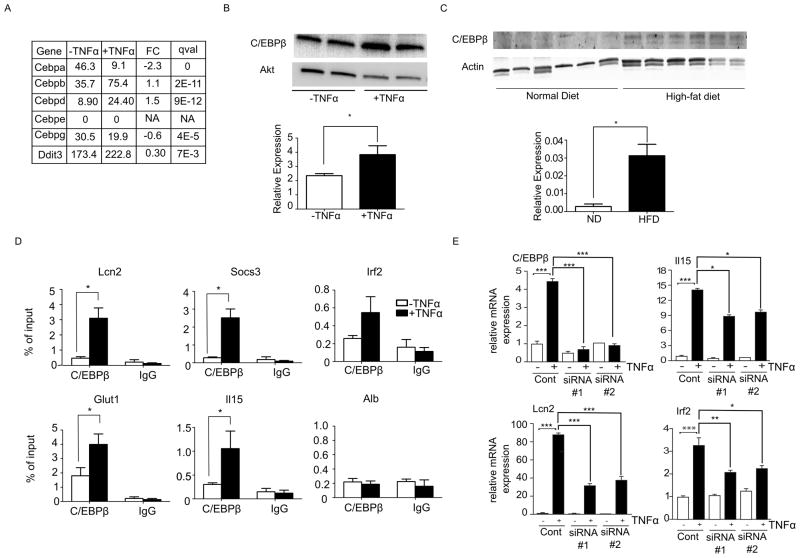
Having shown that the C/EBP motif is enriched in regions with increased DNase hypersensitivity near the TNFα-induced genes, we focused our analysis on Cebpb, which increased in expression to a relatively high level after TNFα treatment (Figure 5A). We confirmed the increased expression using qPCR (data not shown) and Western blots (Figure 5B). C/EBPβ protein expression is also higher in white adipose tissue harvested from mice fed a high-fat diet compared to mice fed a normal chow diet (Figure 5C). To assess if C/EBPβ binds to the regulatory regions of TNFα-induced genes, we carried out C/EBPβ chromatin immunoprecipitation (ChIP) experiments on selected loci with increased DNase hypersensitivity near TNFα-induced genes. We observed a significant increase in binding of C/EBPβ in the TNFα-treated cells over control at the regulatory regions of Lcn2, Socs3, Glut1, and Il15 but not at the control region (Alb) (Figure 5D). To assess whether knocking down Cebpb would affect the gene induction of the above-mentioned genes after TNFα treatment, we used two different siRNA constructs to knock down induction of this protein (Figure 5E). Upon Cebpb knock down, there was a significant reduction in TNFα-mediated induction of Lcn2, Irf2 and Il15 (Figure 5E), indicating that Cebpb indeed is required for induction of genes following TNFα treatment.
Figure 5. C/EBPβ in TNFα-induced insulin resistance.
(A) mRNA expression values in FPKM of the 6 C/EBP isoforms from untreated (−TNFα, control) and treated (+TNFα) 3T3-L1 measured by RNA-Seq. FC: log 2 fold change of expression values (+TNFα/ −TNFα), qval is determined by Cuffdiff.
(B) Western blot analysis of C/EBPβ protein expression from untreated (−TNFα, control) and treated (+TNFα) 3T3-L1. Bottom panel: Quantification of protein level normalized to total Akt loading control, scale bars are mean ± SEM (n=4). An asterisk (*) indicates p < 0.05 by t-test.
(C) Western blot analysis of C/EBPβ protein expression from white adipose tissues from ND mice and HFD mice (n=6). Bottom panel: Quantification of protein level normalized to actin loading control, scale bars are mean ± SEM (n=6). An asterisk (*) indicates p < 0.01 by t-test.
(D) ChIP-PCR analysis of control (open bars) and TNFα-treated (close bars) 3T3-L1 showing C/EBPβ occupancy on selected loci. C/EBPβ binding to negative control region (Alb) is shown for comparison. % of input on selected loci from ChIP experiment with C/EBPβ antibody or non-specific IgG is indicated. Error bars indicate mean ± SEM (n=3). An asterisk (*) indicates p < 0.05 by t-test.
(E) Cebpb expression was suppressed in fully differentiated 3T3-L1 cells using siRNA and the effect of Cebpb repression on TNFα-induced transcriptional changes was analyzed. Gene expression was measured by qPCR using Taqman probes specific for Cebpb Il15, Lcn2, and Irf2. The data obtained were normalized to the amount of 18s mRNA detected in each sample. The data are presented as the mean ± standard deviation (SD, n=3). Statistically significant differences between −TNFα and +TNFα in the control knock down (non-targeting siRNA) are shown. In addition, statistically significant differences between +TNFα control and +TNFα siRNA#1 and +TNFα siRNA#2 (two separate siRNAs against Cebpb) are indicated (*, p < 0.05; **, p < 0.01; *** p < 0.001).
Discussion
We have presented a detailed global transcriptome analysis of five different in vitro insulin resistance models and compared them with three independent DIO mouse models. Our results show that different models capture distinct aspects of the in vivo changes. We find specific pathways that are altered in vivo and are captured by the individual models, and we are able to identify several transcriptional regulators that are likely to drive these changes.
It is not surprising that no single in vitro model captures all the features of DIO adipose insulin resistance, which are complicated phenotypes depending on multiple factors (e.g., mouse strain, high-fat diet formulation, and duration of high-fat feeding). Nevertheless, the TNFα and hypoxia models, and even more so the cotreatment model, are able to recapitulate a wide range of the DIO transcriptional changes associated with metabolism. The impairment of metabolic pathways is not limited to the relatively well-studied glucose and lipid metabolic pathways. For example, the cytochrome P450 metabolic pathways are downregulated in vivo and in the TNFα, hypoxia and cotreatment models. White fat was suggested to have a prominent detoxification function (Forner, Kumar et al. 2009), and our analysis suggests that this function may be impaired in DIO mouse adipose tissue and also in these in vitro models. Moreover, we observed downregulation of branched-chain amino acid catabolic pathways in vivo and in the TNFα, hypoxia, and cotreatment in vitro models, but not in the high insulin or dexamethasone models. Levels of branched-chain amino acids (valine, leucine, and isoleucine) are elevated in obese (Newgard, An et al. 2009) and diabetes-prone (Wang, Larson et al. 2011) humans. In addition, oxidation enzymes for branched-chain amino acids are downregulated in adipose tissue of obese and insulin-resistant humans (Pietilainen, Naukkarinen et al. 2008). Our study highlights the downregulation of these pathways in DIO mice and shows that the TNFα, hypoxia and cotreatment models capture downregulation of these pathways.
Furthermore, our studies identify features captured by the five in vitro models uniquely or jointly. For example, the TNFα and the cotreatment models capture the dedifferentiation-, chemotaxis- and inflammation-related features that are observed in vivo. In particular, we found that the two TNFα-related models are the only models among the five that can mimic the upregulation of genes related to chemotaxis. Indeed, three chemotaxis genes upregulated by TNFα (Ccl2, Ccl7, and Ccl9) are among the six chemotactic factors that are consistently upregulated in adipose tissue, and predominantly adipocytes, of ob/ob and DIO mice (Jiao, Chen et al. 2009). The chemotactic nature of DIO adipocytes is suggested to contribute to macrophage infiltration and the ensuing chronic inflammatory responses (Weisberg, McCann et al. 2003). As for the inflammatory responses associated with insulin resistance, the TNFα and cotreatment models are also largely able to replicate these. Conversely, hypoxia, high insulin and dexamethasone are not good models to capture the inflammation-related aspect of adipose insulin resistance. Given that dexamethasone is anti-inflammatory in nature, this is not surprising. However, it is rather unexpected that hypoxia could not model the DIO-induced inflammatory responses well because adipose hypoxia has been associated with an increase in expression of many inflammatory genes and the activation of NF-κB and TNFα (Ye, Gao et al. 2007). We are confident that our hypoxic treatment was working since various hypoxia-responsive genes (e.g., Glut1, heme oxygenase 1 (Hmox1), pyruvate dehydrogenase kinase 1(Pdk1), and vascular endothelial growth factor (Vegfa)) were markedly upregulated in the hypoxia-treated cells; however, we cannot rule out that a longer duration of hypoxic treatment (> 24h) is required to trigger the inflammatory responses in vitro.
While the TNFα and the cotreatment models capture the various immune response-related features, the high insulin model, and to a lesser extent, the hypoxia model, capture the upregulation of genes related to cell cycle processes and mitosis. This is in agreement with a recent study comparing gene expression of adipose tissue from insulin-resistant and insulin-sensitive subjects with matched BMI (Elbein, Kern et al. 2011), in which many genes related to cell cycle progression and cell adhesion were differentially expressed.
In our analysis, dexamethasone appears to be the model that is the least relevant to DIO adipose insulin resistance at the transcriptional level. However, we cannot exclude the possibility that dexamethasone induces proteomic changes that are similar to those in vivo. It is also plausible that the dexamethasone model is a better model for capturing features of insulin resistance of a different origin, such as insulin resistance associated with Cushing’s syndrome. One important in vitro model of insulin resistance that we did not investigate in detail is fatty-acid induced insulin resistance (Van Epps-Fung, Williford et al. 1997). The conditions that we tested (800μM of palmitate for 24h to 48h) induce only a minor impairment of insulin-stimulation of glucose uptake and AKT phosphorylation, expression changes in ~100 genes (Figure S3), and enrichment in a limited number of gene sets (Table S4). The subtle changes associated with palmitate treatment could represent early stages in development of insulin resistance and warrant further study. Besides analyzing the transcriptional profiles of the diverse models of adipose insulin resistance, we explored in detail the transcriptional regulation of TNFα-induced insulin resistance by combining genome-wide RNA-Seq with DNase-Seq analysis. In addition to known regulators such as PPARγ and NF-κB, we found that C/EBPβ is also a potential mediator of TNFα-induced insulin resistance. Whole-body C/EBPβ deletion protects against obesity and insulin resistance upon high-fat diet treatment (Millward, Heaney et al. 2007) and reduces adiposity and hepatic steatosis in db/db mice (Schroeder-Gloeckler, Rahman et al. 2007). C/EBPβ has been extensively studied in the context of adipogenesis (Steger, Grant et al. 2010; Siersbaek, Nielsen et al. 2011); however, its role in TNFα-induced insulin resistance has not been explored. We show that C/EBPβ protein expression increases upon TNFα treatment and high-fat diet feeding, and that it binds to the regulatory regions of several induced genes in TNFα-treated 3T3-L1 cells. Importantly, induction of several TNFα-responsive genes is diminished upon Cebpb knock down. Furthermore, DNA motif analysis suggests that AP-1 related motifs are enriched in regions with increased hypersensitivity after TNFα, dexamethasone and hypoxia treatments. Activation of the transcription factor AP-1 is downstream of the activation of JNK, giving rise to the possibility that JNK activation is a common feature of multiple forms of insulin resistance.
We have shown that analysis of the mouse adipocyte DNase-Seq data is able to identify known and novel regulators of gene expression. In order to make this resource more broadly available, we have launched a web-based software AdipoSight (http://fraenkel.mit.edu/adipo_sight). Based on a list of user-supplied genes, the software will identify enriched DNA sequence motifs in the DNase hypersensitive regions in the proximity of the genes (see Methods).
In conclusion, our study highlights the particular features that the five in vitro models capture. This comprehensive and accurate description of the transcriptome changes of the five 3T3-L1 insulin resistance models will be a rich resource for future studies.
Materials and Methods
In-vitro cellular insulin resistance models
Cells were washed with PBS and changed to serum-free, low-glucose (1g/L) DMEM with 0.5% BSA. Insulin resistance was induced with one of the following: 2.5nM of TNFα (R & D Systems) for 24h; incubation in a 1% oxygen chamber (Powers, Millman et al. 2010) for 24h; treatment with both 2.5nM TNFα and 1% oxygen for 24h; 1μM dexamethasone (Sigma) for 24h;100nM insulin (Sigma) in high glucose (4.5g/L) medium for 24h; 800μM of palmitate (dissolved in 70% ethanol) for 48h in DMEM containing 1% serum and 2% BSA.
RNA-Seq library preparation, sequencing and analysis
RNA-Seq experiments were performed on biological triplicates. 10ug of total RNA was used for each RNA-Seq library preparation according to the manufacturer’s instructions (Illumina). Quality of RNA was verified using bioanalyzer (Agilent); only RNA with a RIN > 9 was used. Libraries were prepared and sequenced(Illumina GAII) in a pair-end, 36-bp format, except for the cotreatment samples which were sequenced by Hi-Seq in a single-end, 50-bp format. Reads from each sample were aligned to the mouse genome (mm9 build) using TopHat (version 1.1.0). Differential expression was quantified using Cuffdiff (Trapnell, Williams et al. 2010) (version 1.0.3). Differentially expressed genes are those that have a log2 fold change of > 0.58 or < −0.58 and a q-value < 0.05 when compared to the control condition. We also required that the differentially expressed genes used for downstream analysis have a FPKM greater than 0.1 in the control condition. Primers to verified RNA-Seq results are listed in Table S8.
DNase hypersensitivity followed by sequencing (DNase-Seq)
Intact nuclei were isolated from differentiated 3T3-L1 using a nuclei isolation kit (Sigma: NUC201), and prepared as described (Sabo, Kuehn et al. 2006). At least 30 million nuclei were used for each experiment; 50U/ml of DNase I (Promega RQ1 RNase-free DNase; lot number: 25308616) was used for digesting 10M cells at 37°C for 2 min followed by a SDS- and EDTA-based stop buffer. Digested nuclei were incubated at 55°C overnight with proteinase K, extracted using phenol chloroform, and the “2-hit” DNA fragments were isolated using a sucrose gradient. Isolated DNA fragments were purified, subjected to the standard Illumina library preparation, and sequenced using Illumina GA II. 36-bp sequenced reads were mapped to the reference genome mm9 using bowtie. Differential DNase hypersensitive regions were identified using MACS (Zhang, Liu et al. 2008) using a P-value threshold of 1E-10: treatment- induced DNase hypersensitivity regions were called with the treated cells as foreground and the untreated control as background. Conversely, treatment-repressed DNase hypersensitivity regions were called with the untreated control as foreground and the treated cells as background. For the TNFα, dexamethasone, and hypoxia treated samples, control DNase-Seq data set 1 (control 1) was used. For high insulin treatment, control DNase-Seq data set 2 (control 2) was used.
Supplementary Material
6. Highlights.
Different in vitro models capture different aspects of in vivo insulin resistance
TNFα & hypoxia cotreatment is most able to mimic changes in diet-induced obese mice
C/EBPβ is likely to mediate TNFα-induced adipose insulin resistance
AdipoSight is a tool to explore adipocyte transcription regulation
Acknowledgments
KAL was a recipient of the Singapore A*STAR National Science Scholarship. HFL is supported by NIH grant DK-068348. These studies were supported by NIH grants R24 DK-090963 (to RJD and EF) and R01GM-089903 (to EF). This work used computing resources funded by the National Science Foundation under Award No. DB1-0821391 and sequencing support from NIH (P30-ES002109). RJD is an investigator of the Howard Hughes Medical Institute. We would like to thank S. Carol Huang, A Soltis and C Ng for helpful discussion.
Footnotes
Accession number
The raw data for the RNA-Seq, ChIP-Seq and DNase-Seq experiments were deposited in Gene Expression Omnibus (GEO) with the accession number GSE35724.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andrews RC, Walker BR. Glucocorticoids and insulin resistance: old hormones, new targets. Clin Sci. 1999;96(5):513–523. doi: 10.1042/cs0960513. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eguchi J, Yan QW, et al. Interferon Regulatory Factors Are Transcriptional Regulators of Adipogenesis. Cell metabolism. 2008;7(1):86–94. doi: 10.1016/j.cmet.2007.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbein SC, Kern PA, et al. Global gene expression profiles of subcutaneous adipose and muscle from glucose-tolerant, insulin-sensitive, and insulin-resistant individuals matched for BMI. Diabetes. 2011;60(3):1019–1029. doi: 10.2337/db10-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgibbons TP, Kogan S, et al. Similarity of mouse perivascular and brown adipose tissues and their resistance to diet-induced inflammation. American Journal of Physiology - Heart and Circulatory Physiology. 2011;301(4):H1425–H1437. doi: 10.1152/ajpheart.00376.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner F, Kumar C, et al. Proteome Differences between Brown and White Fat Mitochondria Reveal Specialized Metabolic Functions. Cell metabolism. 2009;10(4):324–335. doi: 10.1016/j.cmet.2009.08.014. [DOI] [PubMed] [Google Scholar]
- Fujisaka S, Usui I, et al. Telmisartan Improves Insulin Resistance and Modulates Adipose Tissue Macrophage Polarization in High-Fat-Fed Mice. Endocrinology. 2011;152(5):1789–1799. doi: 10.1210/en.2010-1312. [DOI] [PubMed] [Google Scholar]
- Green H, Meuth M. An established pre-adipose cell line and its differentiation in culture. Cell. 1974;3(2):127–133. doi: 10.1016/0092-8674(74)90116-0. [DOI] [PubMed] [Google Scholar]
- Hesselberth JR, Chen X, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Meth. 2009;6(4):283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosogai N, Fukuhara A, et al. Adipose Tissue Hypoxia in Obesity and Its Impact on Adipocytokine Dysregulation. Diabetes. 2007;56(4):901–911. doi: 10.2337/db06-0911. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS, Shargill NS, et al. Adipose expression of tumor necrosis factor-alpha: direct role in obesity-linked insulin resistance. Science. 1993;259(5091):87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- Jager J, Grémeaux T, et al. Interleukin-1β-Induced Insulin Resistance in Adipocytes through Down-Regulation of Insulin Receptor Substrate-1 Expression. Endocrinology. 2007;148(1):241–251. doi: 10.1210/en.2006-0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao P, Chen Q, et al. Obesity-Related Upregulation of Monocyte Chemotactic Factors in Adipocytes. Diabetes. 2009;58(1):104–115. doi: 10.2337/db07-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutson VP, Balba Y. 3T3-L1 adipocytes as a cell culture model of insulin resistance. In Vitro Cell Dev Biol Anim. 1997;33(2):77–81. doi: 10.1007/s11626-997-0025-2. [DOI] [PubMed] [Google Scholar]
- Ling G, Sugathan A, et al. Unbiased, genome-wide in vivo mapping of transcriptional regulatory elements reveals sex differences in chromatin structure associated with sex-specific liver gene expression. Mol Cell Biol. 2010;30(23):5531–5544. doi: 10.1128/MCB.00601-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIsaac KD, Lo KA, et al. A quantitative model of transcriptional regulation reveals the influence of binding location on expression. PLoS Comput Biol. 2010;6(4):e1000773. doi: 10.1371/journal.pcbi.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuzaki H, Paterson J, et al. A Transgenic Model of Visceral Obesity and the Metabolic Syndrome. Science. 2001;294(5549):2166–2170. doi: 10.1126/science.1066285. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Xu Z, et al. Comparative Epigenomic Analysis of Murine and Human Adipogenesis. Cell. 2010;143(1):156–169. doi: 10.1016/j.cell.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millward CA, Heaney JD, et al. Mice with a deletion in the gene for CCAAT/enhancer-binding protein beta are protected against diet-induced obesity. Diabetes. 2007;56(1):161–167. doi: 10.2337/db06-0310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BA, Robinson KA, et al. High glucose and glucosamine induce insulin resistance via different mechanisms in 3T3-L1 adipocytes. Diabetes. 2000;49(6):981–991. doi: 10.2337/diabetes.49.6.981. [DOI] [PubMed] [Google Scholar]
- Newgard CB, An J, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab. 2009;9(4):311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MTA, Satoh H, et al. JNK and Tumor Necrosis Factor-α Mediate Free Fatty Acid-induced Insulin Resistance in 3T3-L1 Adipocytes. Journal of Biological Chemistry. 2005;280(42):35361–35371. doi: 10.1074/jbc.M504611200. [DOI] [PubMed] [Google Scholar]
- Pietilainen KH, Naukkarinen J, et al. Global transcript profiles of fat in monozygotic twins discordant for BMI: pathways behind acquired obesity. PLoS Med. 2008;5(3):e51. doi: 10.1371/journal.pmed.0050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Saberi M, et al. Adipocyte CREB Promotes Insulin Resistance in Obesity. Cell metabolism. 2009;9(3):277–286. doi: 10.1016/j.cmet.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reaven GM. The insulin resistance syndrome: definition and dietary approaches to treatment. Annu Rev Nutr. 2005;25:391–406. doi: 10.1146/annurev.nutr.24.012003.132155. [DOI] [PubMed] [Google Scholar]
- Regazzetti C, Peraldi P, et al. Hypoxia decreases insulin signaling pathways in adipocytes. Diabetes. 2009;58(1):95–103. doi: 10.2337/db08-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotter V, Nagaev I, et al. Interleukin-6 (IL-6) Induces Insulin Resistance in 3T3-L1 Adipocytes and Is, Like IL-8 and Tumor Necrosis Factor-α, Overexpressed in Human Fat Cells from Insulin-resistant Subjects. Journal of Biological Chemistry. 2003;278(46):45777–45784. doi: 10.1074/jbc.M301977200. [DOI] [PubMed] [Google Scholar]
- Ruan H, Hacohen N, et al. Tumor Necrosis Factor-α Suppresses Adipocyte-Specific Genes and Activates Expression of Preadipocyte Genes in 3T3-L1 Adipocytes. Diabetes. 2002;51(5):1319–1336. doi: 10.2337/diabetes.51.5.1319. [DOI] [PubMed] [Google Scholar]
- Sakoda H, Ogihara T, et al. Dexamethasone-induced insulin resistance in 3T3-L1 adipocytes is due to inhibition of glucose transport rather than insulin signal transduction. Diabetes. 2000;49(10):1700–1708. doi: 10.2337/diabetes.49.10.1700. [DOI] [PubMed] [Google Scholar]
- Schroeder-Gloeckler JM, Rahman SM, et al. CCAAT/enhancer-binding protein beta deletion reduces adiposity, hepatic steatosis, and diabetes in Lepr(db/db) mice. J Biol Chem. 2007;282(21):15717–15729. doi: 10.1074/jbc.M701329200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupp M, Cristancho AG, et al. Re-expression of GATA2 Cooperates with Peroxisome Proliferator-activated Receptor-γ Depletion to Revert the Adipocyte Phenotype. Journal of Biological Chemistry. 2009;284(14):9458–9464. doi: 10.1074/jbc.M809498200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanik MH, Xu Y, et al. Insulin resistance and hyperinsulinemia: is hyperinsulinemia the cart or the horse? Diabetes Care. 2008;31(Suppl 2):S262–268. doi: 10.2337/dc08-s264. [DOI] [PubMed] [Google Scholar]
- Siersbaek R, Nielsen R, et al. Extensive chromatin remodelling and establishment of transcription factor /’hotspots/’ during early adipogenesis. EMBO J. 2011;30(8):1459–1472. doi: 10.1038/emboj.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TR, Elmendorf JS, et al. Growth hormone-induced insulin resistance: role of the insulin receptor, IRS-1, GLUT-1, and GLUT-4. Am J Physiol. 1997;272(6 Pt 1):E1071–1079. doi: 10.1152/ajpendo.1997.272.6.E1071. [DOI] [PubMed] [Google Scholar]
- Steger DJ, Grant GR, et al. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 2010;24(10):1035–1044. doi: 10.1101/gad.1907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Goff LA, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A. 2013;110(9):3387–3392. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson MJ, Williams MG, et al. Development of Insulin Resistance in 3T3-L1 Adipocytes. Journal of Biological Chemistry. 1997;272(12):7759–7764. doi: 10.1074/jbc.272.12.7759. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotech. 2010;28(5):511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Epps-Fung M, Williford J, et al. Fatty acid-induced insulin resistance in adipocytes. Endocrinology. 1997;138(10):4338–4345. doi: 10.1210/endo.138.10.5458. [DOI] [PubMed] [Google Scholar]
- Wang M. The role of glucocorticoid action in the pathophysiology of the Metabolic Syndrome. Nutr Metab (Lond) 2005;2(1):3. doi: 10.1186/1743-7075-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang TJ, Larson MG, et al. Metabolite profiles and the risk of developing diabetes. Nat Med. 2011;17(4):448–453. doi: 10.1038/nm.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg SP, McCann D, et al. Obesity is associated with macrophage accumulation in adipose tissue. The Journal of Clinical Investigation. 2003;112(12):1796–1808. doi: 10.1172/JCI19246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Gao Z, et al. Hypoxia is a potential risk factor for chronic inflammation and adiponectin reduction in adipose tissue of ob/ob and dietary obese mice. American Journal of Physiology - Endocrinology And Metabolism. 2007;293(4):E1118–E1128. doi: 10.1152/ajpendo.00435.2007. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.