Introduction
The cAMP signaling pathway is one of the best-characterized transduction systems because of its presence in all tissues and systems. Since the discovery of the second messenger cAMP in the 1950s, it is widely appreciated that numerous hormones and neurotransmitters exert their physiological functions through activation of the cAMP pathway and its downstream effectors. Intracellular targets for cAMP include the canonical serine/threonine PKA, as well as the guanine nucleotide exchange factor Epac and cyclic nucleotide–gated ion channels. Despite being activated by a small diffusible second messenger, the functional consequences of PKA activation depend upon the cellular context and stimulus. This was first documented in the 1970s by Keely, Hayes, Brunton, and others, who recognized that although activation of both β-adrenergic receptors and prostaglandin E1 receptors increased cAMP concentration in the cardiac myocyte, only β-adrenergic stimulation coupled cAMP to increased contractility and regulation of glycogen metabolism (Steinberg and Brunton, 2001). This and many similar observations have led to an understanding that PKA is compartmentalized in cells, allowing for spatial-temporal control over phosphorylation events (Dessauer, 2009; Welch et al., 2010).
The molecular mechanism for localized PKA signaling involves the association of PKA with a family of scaffolding proteins called A kinase–anchoring proteins (AKAPs) (Dodge-Kafka et al., 2006). Although originally named based on their ability to bind PKA, it has become evident that AKAPs participate in compartmentation of cAMP signaling through additional mechanisms beyond conferring specific PKA substrate phosphorylation (Welch et al., 2010). Hormone binding to a seven-transmembrane domain G protein–coupled receptor and the subsequent activation of the Gαs subunit stimulates the catalytic activity of adenylyl cyclase (AC), increasing cAMP production. In turn, cAMP is hydrolyzed to 5′-adenosine monophosphate via the action of phosphodiesterases (PDEs). It is a finely tuned balance of cAMP synthesis and degradation that ultimately regulates specific cellular responses. AKAP complexes not only contain PKA but also ACs and PDEs, coupling the synthesis, function, and degradation of cAMP in a defined space surrounding the scaffold and thus providing the molecular architecture for cAMP compartmentation. This Perspective will focus on recent evidence that provides insight into the molecular mechanisms underlying AKAP-mediated control of local cAMP gradients.
AKAPs
The canonical cAMP effector is PKA, a broad specificity serine/threonine kinase that when inactive is a tetrameric holoenzyme consisting of a regulatory (R) subunit dimer bound to two catalytic (C) subunits. When two molecules of cAMP bind to each R subunit, a conformational change occurs, releasing the now active C subunit (Francis and Corbin, 1994). This action results in the phosphorylation of substrate proteins that contain a consensus sequence, typically represented as R-R-X-S/T (Kemp et al., 1977). There are three known isoforms of the C subunit (Scott, 1991). Cα and Cβ are ubiquitously expressed, whereas Cγ is found primarily in the testis. The four R subunit genes are functionally divided into two categories: RI (RIα and RIβ) and RII (RIIα and RIIβ) (Scott, 1991). Although RI and RII contain significant sequence homology in their cAMP-binding domain, they display unique characteristics in their mechanisms of activation, subcellular localization, and substrate profiles (Francis and Corbin, 1994; Cummings et al., 1996).
AKAPs are a diverse family of scaffolding proteins that are defined solely by their ability to tether PKA. The first AKAPs were considered protein contaminants that co-purified with the regulatory subunits on cAMP-agarose affinity columns, but now their significance for directing PKA action is widely appreciated (Theurkauf and Vallee, 1982; Scott, 1991). Currently, 43 genes encode the known AKAP family of proteins (Welch et al., 2010). Many of the AKAP genes encode mRNAs subject to alternative splicing, such that >70 functionally distinct AKAP proteins have been identified. Table 1 details the current list of known AKAPs and their binding partners. The defining feature of AKAPs is their ability to bind the R subunit dimer via an amphipathic helix consisting of 14–18 amino acids that binds through hydrophobic interactions to the N-terminal dimerization/docking domain contained in the RII dimer (Carr et al., 1991; Newlon et al., 1997, 1999). Although almost all AKAP PKA-binding sites may be modeled as an amphipathic helix motif, they share little primary sequence similarity, making identification of new AKAPs via BLAST or genomic searches unfeasible. Originally, AKAPs were thought to associate only with RII. However, several dual-specific AKAPs have been identified that bind both RI and RII, although RI typically displays binding affinities severalfold less than that of RII (Herberg et al., 2000; Alto et al., 2003).
Table 1.
Known AKAPs and their binding partners
| Gene name | Protein names and Isoforms | Binding partners | Targeting motifs/localization |
| AKAP1 | D-AKAP, s-AKAP84, AKAP121, AKAP149 | PKA I and II, protein tyrosine phosphatase PTPD1, Src, PKCα, Lfc, PDE7A, RSK1, PP1, PP2A, CaN, mRNA, AMY-1, lamin-B, HIV-1 RT | Mitochondrion, endoplasmic reticulum, nuclear envelope |
| AKAP2 | AKAP-KL | aquaporin-0, PKAII | Lens, apical surface of epithelial cells |
| AKAP5 | AKAP79, AKAP75, AKAP150 | PKAII, PKC, CaN, KCNQ2, CaV1, β1-AR, AC, SAP-97, PP1, GluR1, mGluR1/5, PSD-95, AC5/6, PP2B, Kir2.1, ASIC1a/2a, TRPV1, NMDAR, IQGAP1 | Plasmalemma and T-tubules, post |
| AKAP6 | mAKAPβ | PKAII, AC5, PDE4D3, PP2A B56δ, RyR2, CaNAβ, NFATc, HIF-1a, VHL, Siah2, Epac1, Rap1, ERK5, MEK5, RSK3, PDK1, NCX1, nesprin-1α, myopodin | Nuclear envelope |
| AKAP7 | AKAP15, AKAP18 (α, β, γ, δ) | PKAII, CaV1, NaV1.2a channel, phospholamban, inhibitor-1, PP1, PKC, AQP2, 5′-AMP | Plasmalemma and sarcoplasmic reticulum, nuclear (δ NLS), plasma membrane (α and β myristolyation, dual palmitoylation) |
| AKAP8 | AKAP95 | PKAII, PDE7A, MCM2, p68 RNA helicase, HDAC3, AMY-1, cyclin D/E, condensin | Nucleus |
| AKAP9 | Yotiao, AKAP450, AKAP350, CG-NAP, Hyperion | PKAII, AC, PP1, PP2A, PDE4D3, KCNQ1, IP3R, PKCε, PKN, casein kinase 1, chloride intracellular channel (CLIC), NMDAR, GCP2/3 | Plasmalemma, centrosomes, Golgi |
| AKAP10 | D-AKAP2 | PKA I and II, Rab11, Rab4, PDZK1, PP1, RSK1 | Outer mitochodrial membrane |
| AKAP11 | AKAP220 | PKAII, PP1, GSK3β, GABACR, AQP2 | Peroxisomes |
| AKAP12 | Gravin, SSeCKS | PKAII, PDE4D3, PKC Src, CaN, β2-AR, calmodulin, cyclin D | Actin cytoskeleton, plasma membrane (myristolyaltion) |
| AKAP13 | AKAP-Lbc, Ht31, BRX-1 | PKAII, Gα12, Rho, PKNα, MLTK, MKK3, p38α, KSR1, Raf, MEK1/2, ERK1/2, 14-3-3, PKCη, PKD, SHp2, HSP20, α-catulin, LC3 | Cytoskeleton |
| ARGEF2 | BIG2 | PKA I and II, formin-binding protein 3, PDE3A, TNFR1 | Cytoplasm, internal membranes including Golgi |
| EZR | Ezrin | PKAI and II, CFTR, EBP50/NHERF, NHE3, calmodulin, Rho kinase, actin, α1AR, E-cadherin, β-catenin, EGFR, Fas-R, PKCα, S100, FAK, SAP-97, moesin, radixin, FAK, merlin | Cytoskeleton |
| MAP2 | MAP2B | PKAII, tubulin, CaV1, myosin VIIa | Microtubules |
| CMYA5 | Myospryn | PKAII, dysbindin, titin, calpain-3, desmin, dystrophin | Sarcomere |
| SPHKAP | SKIP | PKAI, sphingosine kinase type 1 | Cytosol |
| SYNM | Synemin | PKAII, desmin, vimentin, dystrobrevin, desmuslin, zyxin, talin, vinculin | Intercalated discs, sarcolemma, Z-lines, intermediate filaments |
| TNNT2 | Troponin T | Troponin I, troponin C, actin | Sarcomere thin filaments |
| LDB3 | Cypher, ZASP | PKAII, CaN, L-type calcium channel | Sacomere Z-lines |
| C2orf88 | smAKAP | PKAI | Cell–cell junctions, filipodia |
| PCNT | Pericentrin | PKAII, calmodulin, y-tubulin | Centrosome |
| WASF-1 | Wave-1 | PKAII, actin nucleation core Arp2/3 complex, BAIAP2, profilin 1 | Actin cytoskeleton |
| ACBD3 | PAP7 | giantin, PPM1L, PKAI | Mitochondria |
| NBEA | Neurobechin | SAP102, PKA | Golgi |
| AKAP14 | Akap28 | PKAII | Ciliary axonemes |
| CBFA2T3 | Myeloid translocation gene (MTG) 8 and 16b | plexin, PKAI and PKAII | Golgi |
| RAB32 | Rab32 | PKAII | Mitochondria |
| MYRIP | Myosin-VIIa- and Rab-interacting protein | myosin VA | Perinuclear |
| NF2 | Merlin | PKAI, PKAII, HGS, ezrin, cullin-4A, syntenin-1, VPRBP, RIT1, SPTBN1, MED28, DDB1 | Soma, dendrites |
| AKAP17A | SFRS17A | PKAI, PKAII | Nucleus |
The functional significance of AKAPs has been established through manipulation of the PKA-binding domain within each AKAP (Carr et al., 1992). Small peptides have been developed that mimic the AKAP α-helix motif and globally disrupt PKA–AKAP interactions when delivered into cells. When delivered by perfusion into hippocampal neurons, these peptides functionally uncoupled PKA from the α-amino-3-5-methy-4-isoxazole propionic acid (AMPA)-type glutamate receptors and attenuated postsynaptic AMPA currents (Rosenmund et al., 1994). Others studies using similar peptides found that PKA anchoring influenced sperm motility, insulin secretion, and cardiac contraction (Lester et al., 1997; Vijayaraghavan et al., 1999; McConnell et al., 2009). New peptides have been developed that distinguish RI from RII interactions, allowing for identification of individual functions for each subunit (Alto et al., 2003; Burns-Hamuro et al., 2003; Carlson et al., 2006; Stokka et al., 2006).
The unique subcellular localization of each AKAP confers specificity to PKA phosphorylation events. This is accomplished primarily via protein–protein interactions. In the heart, AKAP7δ is localized to the sarcoplasmic reticulum via a direct binding to phosopholamban (Lygren et al., 2007). Peptide-based disruption of this interaction has been shown to prevent the striated staining pattern of AKAP7δ, redistributing the AKAP to a soluble, cytosolic compartment. Although less common, protein–lipid and lipid–lipid interactions have also been documented. For example, AKAP7α contains three N-terminal amino acids that are lipid modified, allowing for insertion into the cytoplasmic face of the plasma membrane (Fraser et al., 1998). Mutation of these amino acids disrupts targeting, effectively blocking the regulation of the L-type calcium channel in HEK293 cells, although the physiological significance of this interaction in cardiac myocytes has since been questioned (Jones et al., 2012). In each of these cases, redistribution of AKAP via manipulation of its targeting domain results in the failure to recruit PKA to the subcellular location of its substrate, often with physiological consequence.
To focus the catalytic activity of PKA toward specific targets, AKAPs must simultaneously associate with both the kinase and substrate. This has been clearly demonstrated using a fluorescence resonance energy transfer–based PKA activity reporter that was modified to contain an AKAP PKA-anchoring domain (Dodge-Kafka et al., 2005; Zhang et al., 2005). Tethering PKA to the reporter increased the speed and magnitude of PKA activity reporter activation, showing that phosphorylation efficiency is augmented upon kinase compartmentation. A physiological example of this property is illustrated by using a competitive peptide inhibitor mimicking the AKAP7δ-binding domain found on its substrate phospholamban (Lygren et al., 2007). β-adrenergic stimulation of the cardiac myocyte induces PKA-mediated phospholamban phosphorylation, and this was attenuated up to 50% upon inhibition of AKAP7δ-phospholamban binding by the competitive peptide inhibitor.
Although AKAPs are most often considered in terms of cAMP compartmentation, like other scaffold proteins, they assemble multimolecular signaling complexes that can include almost any type of regulatory enzyme (Welch et al., 2010). This property was first discovered when a yeast-2-hybrid screen for novel AKAP5-binding partners identified both PKC and the phosphatase calcineurin (Coghlan et al., 1995; Klauck et al., 1996). Since this original finding, multiple enzyme–AKAP interactions have been identified, including those involving mitogen-activated protein kinases and ubiquitination regulatory enzymes, allowing for the coordination of diverse signaling pathways (Dodge-Kafka et al., 2005; Wong et al., 2008).
AKAPs orchestrate formation of cAMP microdomains
It is increasingly evident that cAMP is restricted to discrete signaling domains or compartments, allowing for locally regulated spatial-temporal control of individual phosphorylation events. The molecular architecture of these individual complexes is an active area of current research. We propose that to nucleate a cAMP microdomain, AKAPs must simultaneously bring together an AC, a PDE, PKA, and a specific PKA substrate into the relevant intracellular compartment. If any of these connections are disrupted, the efficiency and/or fidelity of signaling are lost. This concept is illustrated in the examples below.
mAKAPβ.
The cardiac myocyte represents one of the best-characterized cell types with regard to AKAP function (Diviani et al., 2011). mAKAPβ is a relatively large scaffold predominately localized to the nuclear envelope of cardiac myocytes, where mAKAPβ plays an important role in the regulation of cardiac gene expression (Kapiloff et al., 1999; Zhang et al., 2013). Although mAKAPβ has been detected at other locations, including the sarcoplasmic reticulum and intercalated disks, the significance of mAKAPβ in these compartments is unclear, and this Perspective will focus on its nuclear functions (Marx et al., 2000; Schulze et al., 2003). Localization of mAKAPβ to the nuclear envelope is conferred by a protein–protein interaction with the integral membrane protein nesprin-1α and can be disrupted via competing peptides containing the mAKAPβ or nesprin-1α spectrin-like repeat domains, resulting in mAKAPβ redistribution throughout the cytosol (Pare et al., 2005b). mAKAPβ is only one of several perinuclear AKAPs, and loss of mAKAPβ does not significantly reduce the amount of PKA that is around the nucleus (Pare et al., 2005a; Li et al., 2010). However, the normal perinuclear localization of mAKAPβ is functionally significant, as delocalization of mAKAPβ attenuates agonist-stimulated cardiac myocyte hypertrophy. Additionally, PKA anchoring by mAKAPβ is required for adrenergic-induced myocyte hypertrophy. The hypertrophic response can be prevented via RNAi knockdown of mAKAPβ expression and is not rescued by a mAKAPβ lacking a PKA-binding site (Pare et al., 2005a). Thus, mAKAPβ complexes respond to cAMP signaling at the nucleus, focusing the actions of the kinase onto targets that regulate myocyte growth.
To understand the regulation of mAKAP-bound PKA, mAKAP immunoprecipitates isolated from rat heart extract were examined for the presence of PDE activity (Dodge et al., 2001). The use of pharmacological inhibitors found that a PDE of the type 4 family complexes with the AKAP, allowing for coupled regulation of PKA activity. Biochemical and cellular experiments demonstrated that mAKAP directly binds to the unique N terminus of PDE4D3 (Dodge et al., 2001; Carlisle Michel et al., 2004). PDE4D3 is also a substrate of PKA, with phosphorylation sites at serine residues 13 and 54 (Sette and Conti, 1996). Phosphorylation at serine 13 enhances the binding between mAKAP and PDE4D3, whereas phosphorylation at serine 54 allosterically activates the enzyme, increasing PDE activity two- to threefold (Dodge et al., 2001; Carlisle Michel et al., 2004). Together, the enhanced binding and catalytic activity of PDE4D3 constitutes a negative feedback loop for cAMP signaling. Basal PDE activity associated with mAKAP keeps PKA activity in the complex low, limiting substrate phosphorylation. Stimulation of the cardiac myocyte and the resulting increase in cAMP concentration exceeds the basal activity of mAKAP-bound PDE4D3, allowing for activation of PKA and phosphorylation of substrates including PDE4D3 at serines 13 and 54. This increases both the association and activity of mAKAP-bound PDE4D3, attenuating PKA activity and functionally resetting the system. Importantly, human coding nonsynonymous polymorphisms identified in mAKAP were found to display a significant reduction in PDE4D3 binding when the proteins were coexpressed in HEK293 cells, suggesting that these mutations may modulate mAKAP-mediated cAMP signaling (Rababa’h et al., 2013).
In 2009, an additional cAMP-related enzyme was discovered in the mAKAPβ complex when AC activity co-purified with mAKAPβ immunoprecipitates isolated from rat heart extract (Kapiloff et al., 2009). Although heterologous cell expression systems suggest that mAKAPβ can associate with multiple AC family members, AC5 contributes most of the mAKAPβ-associated cyclase activity in the heart, as no AC activity was detectable in mAKAPβ immunopreciptates isolated from AC5 knockout mice. Importantly, mAKAPβ orchestrates another negative feedback loop to limit cAMP production via phosphorylation of the cyclase, attenuating AC activity. Functionally, this confines mAKAP-bound PKA activity, as competing peptide disruption of mAKAP-AC binding increases cAMP levels, resulting in increased myocyte hypertrophy in the absence of agonist stimulation (Kapiloff et al., 2009).
Yotiao.
Yotiao is the smallest splice variant of the AKAP9 gene family and directs PKA to several substrates in the brain and heart, including the NR1 subunit of the NMDA receptor, IP3 receptors, and the KCNQ1 channel in the heart (Westphal et al., 1999; Marx et al., 2002; Tu et al., 2004). It is the interaction between yotiao and KCNQ1 that provided the first genetic evidence supporting the importance of AKAPs in compartmentalizing PKA with its substrates (Marx et al., 2002; Chen et al., 2007). KCNQ1 is responsible for the slow delayed rectifying potassium current that contributes to repolarization of the cardiac myocyte during termination of the action potential. Mutations of this channel lead to cardiac arrhythmias and the development of long Q-T syndrome. Importantly, an hKCNQ1-G589D long Q-T syndrome mutation identified in human patients prevents yotiao association and PKA phosphorylation of the channel (Marx et al., 2002). A different mutation presenting in 2% of patients with clinically robust symptoms is found within the KCNQ1-binding domain on yotiao, blocking the association of the scaffold and the channel (Chen et al., 2007). These findings exemplify the importance of AKAP–substrate interactions for the creation of cAMP microdomains.
The cardiac potassium current conducted by the KCNQ1 complex is also regulated by a rolipram-sensitive PDE (Terrenoire et al., 2009). To investigate the molecular mechanisms for this effect, immunoprecipitations of KCNQ1 isolated from mouse heart extract were examined for PDE association. These experiments identified PDE4D3 as a binding partner for the channel. Importantly, yotiao was responsible for mediating the interaction, and expression of the anchoring protein was required for PDE regulation of the channel. Thus, by linking the PDE to the same signaling complex as PKA, yotiao orchestrates a cAMP compartment that constrains PKA activity to regulate KCNQ1 channel properties.
As might be expected, AC activity is also associated with yotiao isolated from the brain and heart (Piggott et al., 2008; Li et al., 2012). Although in vitro biochemical experiments suggest that yotiao can bind to ACs 1, 2, 3, and 9, cellular analysis suggests that yotiao tethers AC9 to KCNQ1, as peptides mimicking the N terminus of AC9 attenuated AC activity in the complex (Piggott et al., 2008; Li et al., 2012). Importantly, formation of a KCNQ1–yotiao–AC9 complex sensitized KCNQ1 to β-adrenergic receptor stimulation, allowing the channel to respond to significantly lower concentrations of agonist (Li et al., 2012). Hence, yotiao dictates spatial-temporal control of KCNQ1 channel activity by creating a cAMP microdomain consisting of the channel, PKA, PDE4D3, and AC9.
Conclusions
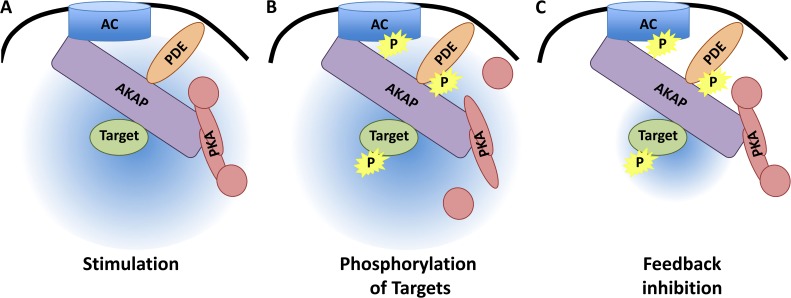
cAMP microdomains are formed via AKAP complexes by integrating specific AC, PDE, and PKA isoforms as well as individual PKA substrates into a single signaling unit or “signalosome” (Fig. 1 A). AC catalyzes the synthesis of cAMP to activate the bound kinase while PDE limits cAMP signaling through degradation of the second messenger. Depending on the isoforms, two types of negative feedback loops act to constrain AKAP-bound PKA activity: phosphorylation of the AC to attenuate cAMP production, and enhanced degradation of cAMP mediated by PKA phosphorylation of the PDE (Fig. 1, B and C). As shown in the examples above, disruption of any component of the complex upsets the finely tuned balance of PKA activity. Importantly, each of these associations could be manipulated for therapeutic treatment, as focused disruption of individual AKAP complexes would affect localized cAMP levels and ultimately downstream effectors. Such an approach makes compelling a detailed structural analysis of each AKAP complex to provide the basis for developing this translational strategy.
Figure 1.
AKAP-orchestrated cAMP microdomains. (A) Formation of the cAMP microdomain occurs when the AKAP simultaneously associates with an AC, a PDE, PKA, and a substrate. Stimulation of AC increases cAMP (depicted in blue) surrounding the AKAP complex. (B) The increase in cAMP overcomes the basal activity of the PDE, allowing for PKA to become active and phosphorylate substrates in the complex, including the AC, PDE, and target protein. (C) Phosphorylation of the AC attenuates cAMP production, whereas PDE phosphorylation increases cAMP hydrolysis. These two acts decrease cAMP concentration, allowing for feedback inhibition of PKA.
This Perspectives series includes articles by Karpen, Rich et al., Conti et al., and Saucerman et al.
Acknowledgments
This work was supported by National Institutes of Health (grants HL82705 to K. Dodge-Kafka and HL075398 to M.S. Kapiloff).
Olaf S. Andersen served as editor.
Footnotes
Abbreviations used in this paper:
- AC
- adenylyl cyclase
- AKAP
- A kinase–anchoring protein
- PDE
- phosphodiesterase
References
- Alto N.M., Soderling S.H., Hoshi N., Langeberg L.K., Fayos R., Jennings P.A., Scott J.D. 2003. Bioinformatic design of A-kinase anchoring protein-in silico: a potent and selective peptide antagonist of type II protein kinase A anchoring. Proc. Natl. Acad. Sci. USA. 100:4445–4450 10.1073/pnas.0330734100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns-Hamuro L.L., Ma Y., Kammerer S., Reineke U., Self C., Cook C., Olson G.L., Cantor C.R., Braun A., Taylor S.S. 2003. Designing isoform-specific peptide disruptors of protein kinase A localization. Proc. Natl. Acad. Sci. USA. 100:4072–4077 10.1073/pnas.2628038100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle Michel J.J., Dodge K.L., Wong W., Mayer N.C., Langeberg L.K., Scott J.D. 2004. PKA-phosphorylation of PDE4D3 facilitates recruitment of the mAKAP signalling complex. Biochem. J. 381:587–592 10.1042/BJ20040846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson C.R., Lygren B., Berge T., Hoshi N., Wong W., Taskén K., Scott J.D. 2006. Delineation of type I protein kinase A-selective signaling events using an RI anchoring disruptor. J. Biol. Chem. 281:21535–21545 10.1074/jbc.M603223200 [DOI] [PubMed] [Google Scholar]
- Carr D.W., Stofko-Hahn R.E., Fraser I.D., Bishop S.M., Acott T.S., Brennan R.G., Scott J.D. 1991. Interaction of the regulatory subunit (RII) of cAMP-dependent protein kinase with RII-anchoring proteins occurs through an amphipathic helix binding motif. J. Biol. Chem. 266:14188–14192 [PubMed] [Google Scholar]
- Carr D.W., Stofko-Hahn R.E., Fraser I.D., Cone R.D., Scott J.D. 1992. Localization of the cAMP-dependent protein kinase to the postsynaptic densities by A-kinase anchoring proteins. Characterization of AKAP 79. J. Biol. Chem. 267:16816–16823 [PubMed] [Google Scholar]
- Chen L., Marquardt M.L., Tester D.J., Sampson K.J., Ackerman M.J., Kass R.S. 2007. Mutation of an A-kinase-anchoring protein causes long-QT syndrome. Proc. Natl. Acad. Sci. USA. 104:20990–20995 10.1073/pnas.0710527105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan V.M., Perrino B.A., Howard M., Langeberg L.K., Hicks J.B., Gallatin W.M., Scott J.D. 1995. Association of protein kinase A and protein phosphatase 2B with a common anchoring protein. Science. 267:108–111 10.1126/science.7528941 [DOI] [PubMed] [Google Scholar]
- Cummings D.E., Brandon E.P., Planas J.V., Motamed K., Idzerda R.L., McKnight G.S. 1996. Genetically lean mice result from targeted disruption of the RII beta subunit of protein kinase A. Nature. 382:622–626 10.1038/382622a0 [DOI] [PubMed] [Google Scholar]
- Dessauer C.W. 2009. Adenylyl cyclase–A-kinase anchoring protein complexes: the next dimension in cAMP signaling. Mol. Pharmacol. 76:935–941 10.1124/mol.109.059345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diviani D., Dodge-Kafka K.L., Li J., Kapiloff M.S. 2011. A-kinase anchoring proteins: scaffolding proteins in the heart. Am. J. Physiol. Heart Circ. Physiol. 301:H1742–H1753 10.1152/ajpheart.00569.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge K.L., Khouangsathiene S., Kapiloff M.S., Mouton R., Hill E.V., Houslay M.D., Langeberg L.K., Scott J.D. 2001. mAKAP assembles a protein kinase A/PDE4 phosphodiesterase cAMP signaling module. EMBO J. 20:1921–1930 10.1093/emboj/20.8.1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge-Kafka K.L., Soughayer J., Pare G.C., Carlisle Michel J.J., Langeberg L.K., Kapiloff M.S., Scott J.D. 2005. The protein kinase A anchoring protein mAKAP coordinates two integrated cAMP effector pathways. Nature. 437:574–578 10.1038/nature03966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodge-Kafka K.L., Langeberg L., Scott J.D. 2006. Compartmentation of cyclic nucleotide signaling in the heart: the role of A-kinase anchoring proteins. Circ. Res. 98:993–1001 10.1161/01.RES.0000218273.91741.30 [DOI] [PubMed] [Google Scholar]
- Francis S.H., Corbin J.D. 1994. Structure and function of cyclic nucleotide-dependent protein kinases. Annu. Rev. Physiol. 56:237–272 10.1146/annurev.ph.56.030194.001321 [DOI] [PubMed] [Google Scholar]
- Fraser I.D., Tavalin S.J., Lester L.B., Langeberg L.K., Westphal A.M., Dean R.A., Marrion N.V., Scott J.D. 1998. A novel lipid-anchored A-kinase anchoring protein facilitates cAMP-responsive membrane events. EMBO J. 17:2261–2272 10.1093/emboj/17.8.2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herberg F.W., Maleszka A., Eide T., Vossebein L., Tasken K. 2000. Analysis of A-kinase anchoring protein (AKAP) interaction with protein kinase A (PKA) regulatory subunits: PKA isoform specificity in AKAP binding. J. Mol. Biol. 298:329–339 10.1006/jmbi.2000.3662 [DOI] [PubMed] [Google Scholar]
- Jones B.W., Brunet S., Gilbert M.L., Nichols C.B., Su T., Westenbroek R.E., Scott J.D., Catterall W.A., McKnight G.S. 2012. Cardiomyocytes from AKAP7 knockout mice respond normally to adrenergic stimulation. Proc. Natl. Acad. Sci. USA. 109:17099–17104 10.1073/pnas.1215219109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapiloff M.S., Schillace R.V., Westphal A.M., Scott J.D. 1999. mAKAP: an A-kinase anchoring protein targeted to the nuclear membrane of differentiated myocytes. J. Cell Sci. 112:2725–2736 [DOI] [PubMed] [Google Scholar]
- Kapiloff M.S., Piggott L.A., Sadana R., Li J., Heredia L.A., Henson E., Efendiev R., Dessauer C.W. 2009. An adenylyl cyclase-mAKAPbeta signaling complex regulates cAMP levels in cardiac myocytes. J. Biol. Chem. 284:23540–23546 10.1074/jbc.M109.030072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp B.E., Graves D.J., Benjamini E., Krebs E.G. 1977. Role of multiple basic residues in determining the substrate specificity of cyclic AMP-dependent protein kinase. J. Biol. Chem. 252:4888–4894 [PubMed] [Google Scholar]
- Klauck T.M., Faux M.C., Labudda K., Langeberg L.K., Jaken S., Scott J.D. 1996. Coordination of three signaling enzymes by AKAP79, a mammalian scaffold protein. Science. 271:1589–1592 10.1126/science.271.5255.1589 [DOI] [PubMed] [Google Scholar]
- Lester L.B., Langeberg L.K., Scott J.D. 1997. Anchoring of protein kinase A facilitates hormone-mediated insulin secretion. Proc. Natl. Acad. Sci. USA. 94:14942–14947 10.1073/pnas.94.26.14942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Negro A., Lopez J., Bauman A.L., Henson E., Dodge-Kafka K., Kapiloff M.S. 2010. The mAKAPβ scaffold regulates cardiac myocyte hypertrophy via recruitment of activated calcineurin. J. Mol. Cell. Cardiol. 48:381–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Chen L., Kass R.S., Dessauer C.W. 2012. The A-kinase anchoring protein Yotiao facilitates complex formation between adenylyl cyclase type 9 and the IKs potassium channel in heart. J. Biol. Chem. 287:29815–29824 10.1074/jbc.M112.380568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lygren B., Carlson C.R., Santamaria K., Lissandron V., McSorley T., Litzenberg J., Lorenz D., Wiesner B., Rosenthal W., Zaccolo M., et al. 2007. AKAP complex regulates Ca2+ re-uptake into heart sarcoplasmic reticulum. EMBO Rep. 8:1061–1067 10.1038/sj.embor.7401081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx S.O., Reiken S., Hisamatsu Y., Jayaraman T., Burkhoff D., Rosemblit N., Marks A.R. 2000. PKA phosphorylation dissociates FKBP12.6 from the calcium release channel (ryanodine receptor): defective regulation in failing hearts. Cell. 101:365–376 10.1016/S0092-8674(00)80847-8 [DOI] [PubMed] [Google Scholar]
- Marx S.O., Kurokawa J., Reiken S., Motoike H., D’Armiento J., Marks A.R., Kass R.S. 2002. Requirement of a macromolecular signaling complex for beta adrenergic receptor modulation of the KCNQ1-KCNE1 potassium channel. Science. 295:496–499 10.1126/science.1066843 [DOI] [PubMed] [Google Scholar]
- McConnell B.K., Popovic Z., Mal N., Lee K., Bautista J., Forudi F., Schwartzman R., Jin J.P., Penn M., Bond M. 2009. Disruption of protein kinase A interaction with A-kinase-anchoring proteins in the heart in vivo: effects on cardiac contractility, protein kinase A phosphorylation, and troponin I proteolysis. J. Biol. Chem. 284:1583–1592 10.1074/jbc.M806321200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newlon M.G., Roy M., Hausken Z.E., Scott J.D., Jennings P.A. 1997. The A-kinase anchoring domain of type IIalpha cAMP-dependent protein kinase is highly helical. J. Biol. Chem. 272:23637–23644 10.1074/jbc.272.38.23637 [DOI] [PubMed] [Google Scholar]
- Newlon M.G., Roy M., Morikis D., Hausken Z.E., Coghlan V., Scott J.D., Jennings P.A. 1999. The molecular basis for protein kinase A anchoring revealed by solution NMR. Nat. Struct. Biol. 6:222–227 10.1038/6663 [DOI] [PubMed] [Google Scholar]
- Pare G.C., Bauman A.L., McHenry M., Michel J.J., Dodge-Kafka K.L., Kapiloff M.S. 2005a. The mAKAP complex participates in the induction of cardiac myocyte hypertrophy by adrenergic receptor signaling. J. Cell Sci. 118:5637–5646 10.1242/jcs.02675 [DOI] [PubMed] [Google Scholar]
- Pare G.C., Easlick J.L., Mislow J.M., McNally E.M., Kapiloff M.S. 2005b. Nesprin-1alpha contributes to the targeting of mAKAP to the cardiac myocyte nuclear envelope. Exp. Cell Res. 303:388–399 10.1016/j.yexcr.2004.10.009 [DOI] [PubMed] [Google Scholar]
- Piggott L.A., Bauman A.L., Scott J.D., Dessauer C.W. 2008. The A-kinase anchoring protein Yotiao binds and regulates adenylyl cyclase in brain. Proc. Natl. Acad. Sci. USA. 105:13835–13840 10.1073/pnas.0712100105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rababa’h A., Craft J.W., Jr, Wijaya C.S., Atrooz F., Fan Q., Singh S., Guillory A.N., Katsonis P., Lichtarge O., McConnell B.K. 2013. Protein kinase A and phosphodiesterase-4D3 binding to coding polymorphisms of cardiac muscle anchoring protein (mAKAP). J. Mol. Biol. 425:3277–3288 10.1016/j.jmb.2013.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund C., Carr D.W., Bergeson S.E., Nilaver G., Scott J.D., Westbrook G.L. 1994. Anchoring of protein kinase A is required for modulation of AMPA/kainate receptors on hippocampal neurons. Nature. 368:853–856 10.1038/368853a0 [DOI] [PubMed] [Google Scholar]
- Schulze D.H., Muqhal M., Lederer W.J., Ruknudin A.M. 2003. Sodium/calcium exchanger (NCX1) macromolecular complex. J. Biol. Chem. 278:28849–28855 10.1074/jbc.M300754200 [DOI] [PubMed] [Google Scholar]
- Scott J.D. 1991. Cyclic nucleotide-dependent protein kinases. Pharmacol. Ther. 50:123–145 10.1016/0163-7258(91)90075-W [DOI] [PubMed] [Google Scholar]
- Sette C., Conti M. 1996. Phosphorylation and activation of a cAMP-specific phosphodiesterase by the cAMP-dependent protein kinase. Involvement of serine 54 in the enzyme activation. J. Biol. Chem. 271:16526–16534 10.1074/jbc.271.28.16526 [DOI] [PubMed] [Google Scholar]
- Steinberg S.F., Brunton L.L. 2001. Compartmentation of G protein-coupled signaling pathways in cardiac myocytes. Annu. Rev. Pharmacol. Toxicol. 41:751–773 10.1146/annurev.pharmtox.41.1.751 [DOI] [PubMed] [Google Scholar]
- Stokka A.J., Gesellchen F., Carlson C.R., Scott J.D., Herberg F.W., Taskén K. 2006. Characterization of A-kinase-anchoring disruptors using a solution-based assay. Biochem. J. 400:493–499 10.1042/BJ20060962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrenoire C., Houslay M.D., Baillie G.S., Kass R.S. 2009. The cardiac IKs potassium channel macromolecular complex includes the phosphodiesterase PDE4D3. J. Biol. Chem. 284:9140–9146 10.1074/jbc.M805366200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theurkauf W.E., Vallee R.B. 1982. Molecular characterization of the cAMP-dependent protein kinase bound to microtubule-associated protein 2. J. Biol. Chem. 257:3284–3290 [PubMed] [Google Scholar]
- Tu H., Tang T.S., Wang Z., Bezprozvanny I. 2004. Association of type 1 inositol 1,4,5-trisphosphate receptor with AKAP9 (Yotiao) and protein kinase A. J. Biol. Chem. 279:19375–19382 10.1074/jbc.M313476200 [DOI] [PubMed] [Google Scholar]
- Vijayaraghavan S., Liberty G.A., Mohan J., Winfrey V.P., Olson G.E., Carr D.W. 1999. Isolation and molecular characterization of AKAP110, a novel, sperm-specific protein kinase A-anchoring protein. Mol. Endocrinol. 13:705–717 10.1210/me.13.5.705 [DOI] [PubMed] [Google Scholar]
- Welch E.J., Jones B.W., Scott J.D. 2010. Networking with AKAPs: context-dependent regulation of anchored enzymes. Mol. Interv. 10:86–97 10.1124/mi.10.2.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal R.S., Tavalin S.J., Lin J.W., Alto N.M., Fraser I.D., Langeberg L.K., Sheng M., Scott J.D. 1999. Regulation of NMDA receptors by an associated phosphatase-kinase signaling complex. Science. 285:93–96 10.1126/science.285.5424.93 [DOI] [PubMed] [Google Scholar]
- Wong W., Goehring A.S., Kapiloff M.S., Langeberg L.K., Scott J.D. 2008. mAKAP compartmentalizes oxygen-dependent control of HIF-1alpha. Sci. Signal. 1:ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Hupfeld C.J., Taylor S.S., Olefsky J.M., Tsien R.Y. 2005. Insulin disrupts beta-adrenergic signalling to protein kinase A in adipocytes. Nature. 437:569–573 10.1038/nature04140 [DOI] [PubMed] [Google Scholar]
- Zhang L., Malik S., Pang J., Wang H., Park K.M., Yule D.I., Blaxall B.C., Smrcka A.V. 2013. Phospholipase Cε hydrolyzes perinuclear phosphatidylinositol 4-phosphate to regulate cardiac hypertrophy. Cell. 153:216–227 10.1016/j.cell.2013.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]