Second messengers such as Ca2+, cGMP, and cAMP are known to regulate diverse cellular functions including excitability, contraction, movement, proliferation, and gene expression. Our understanding of how Ca2+ signals orchestrate such diverse cellular functions has increased dramatically over the last forty years, due in large part to the development of single-cell methods for measuring intracellular Ca2+ (Tsien, 1992). Our understanding of the subcellular localization, kinetics, and frequency of cyclic nucleotide signals has lagged far behind. Thus, we are only beginning to understand how information is encoded within cyclic nucleotide signals.
Until recently, investigators were limited by a lack of real-time, single-cell sensors for cAMP and cGMP. However, in the last decade several groups have developed various cAMP and cGMP sensors based on the binding domains of PKA, PKG, CNG channels, phosphodiesterases (PDEs), and exchange factors activated by cAMP (Epacs). Each of these sensors has inherent advantages and disadvantages. In this Perspective we first outline the strengths and limitations of several single-cell cyclic nucleotide sensors. We then consider how information may be encoded within cyclic nucleotide signals and how current cyclic nucleotide sensors may be used to decipher the mechanisms that underlie signaling specificity. We believe that a better understanding of the strengths and limitations of these biosensors will promote a quantitative understanding of cyclic nucleotide signaling and help to direct the design of the next generation of probes.
Single-cell sensors for cAMP and cGMP
PKA-based sensors.
More than twenty years ago Tsien and colleagues published the first report describing a novel, Förster resonance energy transfer (FRET)-based approach for measuring cAMP signals (Adams et al., 1991). They labeled the catalytic and regulatory subunits of PKA type I with a fluorescent donor (fluorescein) and acceptor (rhodamine). When cAMP concentrations were low, the subunits were in the holoenzyme complex, and FRET occurred between fluorescein and rhodamine. However, when cAMP levels were high, cAMP bound to the regulatory subunits, the catalytic subunits dissociated, and FRET diminished. This ingenious method has been described as a real-time cAMP sensor (Adams et al., 1991; Goaillard et al., 2001; Gorbunova and Spitzer, 2002). However, there are limitations to its use:
(1) The reassociation of PKA subunits may be slow (Rich and Karpen, 2002, and references therein).
(2) PKA is regulated by (high) physiological concentrations of cGMP (Francis and Corbin, 1999).
(3) Fluorescently labeled PKA is catalytically active (Adams et al., 1991; Goaillard et al., 2001).
(4) High concentrations of labeled PKA are required to overwhelm endogenous PKA (otherwise, binding of fluorescently labeled subunits to endogenous subunits will distort FRET signals). PKA has a high affinity for cAMP. High concentrations of high-affinity buffers will severely blunt cAMP signals (Rich and Karpen, 2002).
These limitations hinder the utility of labeled PKA as a cAMP sensor. However, this work sparked researchers from several groups to develop novel cAMP and cGMP probes, each with advantages and disadvantages.
CNG channel-based cyclic nucleotide sensors.
Two groups developed genetically modified CNG channels, which are directly opened by binding of cyclic nucleotides, as cyclic nucleotide sensors (Trivedi and Kramer, 1998; Rich et al., 2000, 2001b). Unlike many other ion channels, CNG channels do not desensitize in response to prolonged cyclic nucleotide exposure (Dhallan et al., 1990; Rich et al., 2000, 2001a), making them suitable for monitoring cyclic nucleotide levels. Open channels allow cations (Na+, K+, Ca2+) to pass through the surface membrane; thus, activation of CNG channels is readily detected with electrophysiological or Ca2+ imaging techniques. CNG channels have several other characteristics that both lend themselves to specific experimental designs and preclude them from others:
(1) CNG channels have fast kinetics (90% rise time in <0.2 s), allowing measurement of rapid changes in cyclic nucleotide levels near the plasma membrane (Rich et al., 2000).
(2) CNG channels are targeted to the plasma membrane, allowing for membrane-localized cAMP measurements. However, they cannot be readily used in other regions of the cell.
(3) CNG channel activity is readily detected at low expression levels. Thus, buffering of cyclic nucleotides is low and in most cases will not substantively alter cyclic nucleotide levels (Rich and Karpen, 2002).
(4) CNG channels are regulated by other intracellular signals including PIP3 and Ca2+ (Brady et al., 2006). Thus, careful controls are required to ensure that measured responses are indeed caused by changes in cyclic nucleotide levels.
(5) High kinetic resolution measurements require electrophysiology; electrophysiological experiments are considered technically more difficult than imaging experiments. The Ca2+ permeability of CNG channels has been used to detect changes in cAMP by monitoring intracellular Ca2+ levels (Rich et al., 2001b).
(6) CNG channels conduct Ca2+. Thus, experiments need to be conducted in the absence of extracellular Ca2+ to avoid CNG channel–mediated Ca2+ regulation of cyclic nucleotide synthesis and degradation.
This combination of strengths and limitations makes CNG channels well-suited for measuring the kinetics of near-membrane signals (Rich and Karpen, 2002). However, CNG channels are ill-suited for the study of feedback interactions between Ca2+, phosphoinositide, and cyclic nucleotide signaling pathways.
Epac-based cyclic nucleotide sensors.
Several groups developed FRET-based cAMP sensors using the cAMP binding protein Epac to measure cAMP signals in different cellular domains (Nikolaev et al., 2004; Ponsioen et al., 2004). These sensors are comprised of catalytically inactive Epac sandwiched between the fluorescent donor (CFP) and fluorescent acceptor (YFP; Ponsioen et al., 2004). In the basal state (low cAMP), efficient FRET occurs between CFP and YFP. When cAMP binds to Epac, there is a conformational change such that FRET efficiency is reduced. There are several versions of the Epac probe available (van der Krogt et al., 2008; Klarenbeek et al., 2011), as well as FRET-based probes for the measurement of cGMP (Nikolaev et al., 2006). These FRET-based probes have similar advantages and disadvantages.
(1) The probes can be readily targeted to different intracellular locations (Terrin et al., 2006).
(2) Although the kinetics of Epac activation have not been carefully measured, experimental measurements suggest that Epac-based sensors are fast enough to faithfully reproduce most cAMP signals.
(3) In general, fluorescence and FRET measurements are technically simple to perform (Börner et al., 2011). However, calibration of these sensors in intact cells is difficult because one can seldom demonstrate that both minimum and maximum fluorescence or FRET levels have been reached.
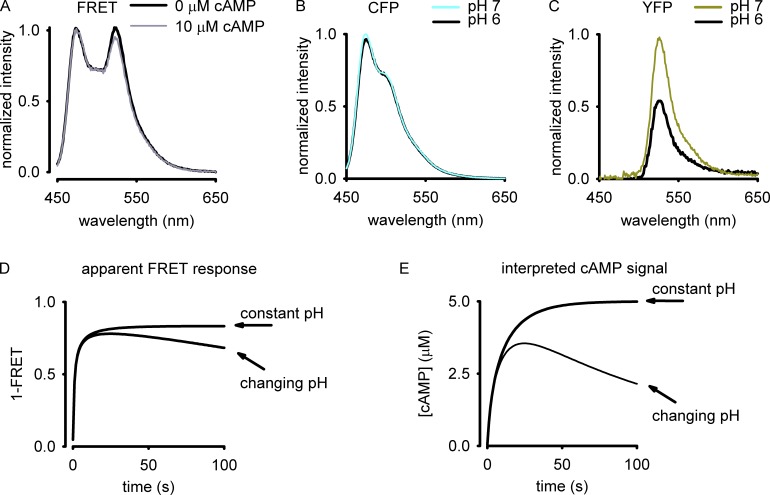
(4) Epac-based FRET probes have a limited range and high background (Fig. 1 A), leading to a low signal-to-noise ratio. This is further complicated by the lack of an appropriate comparison of commonly used FRET imaging and analysis approaches. Only recently have investigators quantitatively assessed the signal-to-noise ratio of different analysis approaches (Leavesley et al., 2013). And although substantive progress has been made in the development of FRET probes with increased range (Klarenbeek et al., 2011), the actual dynamic range of FRET probes has not been adequately assessed. Thus, it is unclear what increments of cAMP or cGMP can actually be discriminated.
Figure 1.
Measurements of apparent FRET. (A–C) Representative responses from cell lysates of HEK-293 cells expressing the Epac-based FRET probe (A), CFP (B) or YFP (C). Measurements were made as described previously (Leavesley et al., 2013; Rich et al., 2013). In brief, cell lysates were suspended in a stirred cuvette (4 × 106 cells in 4 ml) and excited at 430 nm, then emission intensity was measured from 450 to 650 nm using a spectrofluorimeter. Data were normalized to the peak intensity. (A) The change in apparent FRET response measured at 0 and 10 µM cAMP (black and gray lines, respectively). Note the modest change in FRET associated with a saturating change in cAMP. (B) Changing pH from 7 (blue line) to 6 (black line) had little effect on the measured fluorescence intensity of CFP. (C) Changing pH had a marked effect on the measured fluorescence intensity of YFP. (D and E) Simulations demonstrating the effects of changing pH on apparent FRET measurements and interpretations of the underlying cAMP signals.
(5) The (over)expression levels of Epac have not been measured; thus, the effects of cyclic nucleotide buffering cannot be adequately assessed (discussed later).
(6) Only recently have systematic, automated approaches for analysis of FRET measurements been implemented (Jalink and van Rheenen, 2009; Leavesley et al., 2013). Such approaches allow unbiased estimates of FRET efficiency for all cells within a field of view, as well as clear criteria for the exclusion of certain cells from analysis.
Artifacts associated with fluorescence- and FRET-based probes.
Fluorescent proteins, and fluorophores in general, can be modified by a variety of biochemical processes. Some of the factors that can influence fluorescence emission are considered below.
(1) Heterologously expressed proteins, especially targeted constructs, can accumulate at distinct subcellular localizations. Thus, both intermolecular and intramolecular FRET may occur.
(2) Commonly used fluorescent proteins are differentially sensitive to environmental variables including temperature, pH, reactive oxygen species (ROS), and viscosity (Bernas et al., 2005; Ha and Tinnefeld, 2012). Both spectral and lifetime measurements are subject to these artifacts (Suhling et al., 2002). To illustrate the potential problems associated with changes in the environment, we measured the effects of pH on the fluorescence emission from CFP and YFP. We observed that changing pH from 7 to 5 had little effect on the fluorescence emission from CFP (Fig. 1 B), but had a marked effect on the fluorescence emission from YFP (Fig. 1 C). To illustrate the potential problems associated with pH-mediated changes on apparent FRET measurements, we modeled the effects of constant pH 7.0 or a pH change from 7 to 6 on the apparent FRET response. In this scenario, the input cAMP signal rose from baseline to a steady plateau in ∼30 s. We assumed that the pKa of CFP was 5 and the pKa of YFP was 7. The change in pH triggered an overall reduction in FRET (Fig. 1 D). If the effects of pH were not appropriately accounted for, then the interpreted cAMP signal would be transient rather than sustained (Fig. 1 E). Although this was a worst-case scenario for pH, other environmental factors may differentially alter emission from fluorescent proteins. This may become particularly problematic in localized regions of the cell such as the mitochondria.
Certain FRET pairs have increased photostability and reduced sensitivity to pH (van der Krogt et al., 2008), and if possible these FRET pairs should be used. However, the effects of other environmental variables on these fluorophores have not been evaluated. Thus, systematic approaches for detecting and compensating for environmental changes in fluorescence and apparent FRET need to be developed.
Other real-time, single-cell cyclic nucleotide sensors.
FRET-based probes for cGMP have also been developed. These probes use cGMP binding sites from PKG, PDE2, PDE5, and CNG channels sandwiched between fluorescent donors and acceptors (Honda et al., 2005; Nikolaev et al., 2006). These probes have similar advantages and disadvantages to the FRET-based cAMP probes described earlier. However, FRET-based cGMP probes have not been as widely used, in part because Dostmann and colleagues have developed probes in which the cGMP binding site of PKG was fused with circularly permutated enhanced green fluorescent protein (Nausch et al., 2008). Although these probes have high sensitivity, high range, and likely a high dynamic range, the experimental application of these probes is not always straightforward, and fluorescence emission is subject to environmental changes as well as changes in cGMP.
Although there are limitations to the overall utility of FRET-based cyclic nucleotide probes and data interpretation is not always straightforward, these FRET probes are the most widely used cyclic nucleotide sensors to date. The relative ease of use and the ability to be targeted to different regions of a cell allow FRET-based probes to readily detect changes in localized cyclic nucleotide levels.
We next consider the strengths and limitations of the different cyclic nucleotide sensors, with an emphasis on the FRET-based sensors, in deciphering the information content of cyclic nucleotide signals.
How might information be encoded within second messenger signals?
Cyclic nucleotide signals regulate dozens of cellular processes over timescales ranging from seconds to hours (Francis and Corbin, 1999, and references therein). However, it remains unclear how the information required to differentially regulate multiple cellular functions is encoded. Several concepts have been proposed, including encoding information in the spatial distribution of signals (compartmentalization), in the frequency content of signals (frequency encoding), or in the combination of signals that are turned on or off, above or below thresholds, at a given time (digital encoding; Brooker, 1973; Hanson et al., 1994; Dolmetsch et al., 1998; Rich and Karpen, 2002; Ruf et al., 2006; Feinstein et al., 2012). These mechanisms are outlined in the following sections.
Spatial encoding of information in cyclic nucleotide signals.
Spatial encoding of cyclic nucleotide signals, or signal compartmentalization, requires that cAMP and cGMP levels be high enough to activate PKA, PKG, Epac, or CNG channels in one region of a cell, but not in others. The evidence for this phenomenon is straightforward: cAMP rises triggered by different hormones regulate distinct cellular processes. For example, in cardiac myocytes two agents that trigger similar rises in total cellular cAMP levels—isoproterenol and prostaglandin E1 (PGE1)—have markedly different downstream effects (see Saucerman et al. in this issue). Treatment of cardiac myocytes with isoproterenol, a β-adrenergic agonist, triggers cAMP-dependent activation of PKA and subsequent phosphorylation of proteins associated with cardiac excitability; however, treatment with PGE1 triggers the cAMP-dependent activation of PKA, but not phosphorylation of these proteins. This has been partially explained by the observation that these agents activate different pools of PKA: particulate and soluble (Corbin et al., 1977). The idea of localized pools of PKA activity was further supported by the discovery of A kinase–anchoring proteins (AKAPs), the scaffolds that tether PKA to cellular targets (see Kapiloff et al. in this issue). However, spatial proximity of the proteins associated with this signaling pathway, e.g., β2-adrenergic receptors, adenylyl cyclase (AC), PKA, and L-type Ca2+ channels is not enough to ensure localized responses (Feinstein et al., 2012; Saucerman et al., 2014, and references therein). The signal itself must be spatially restricted.
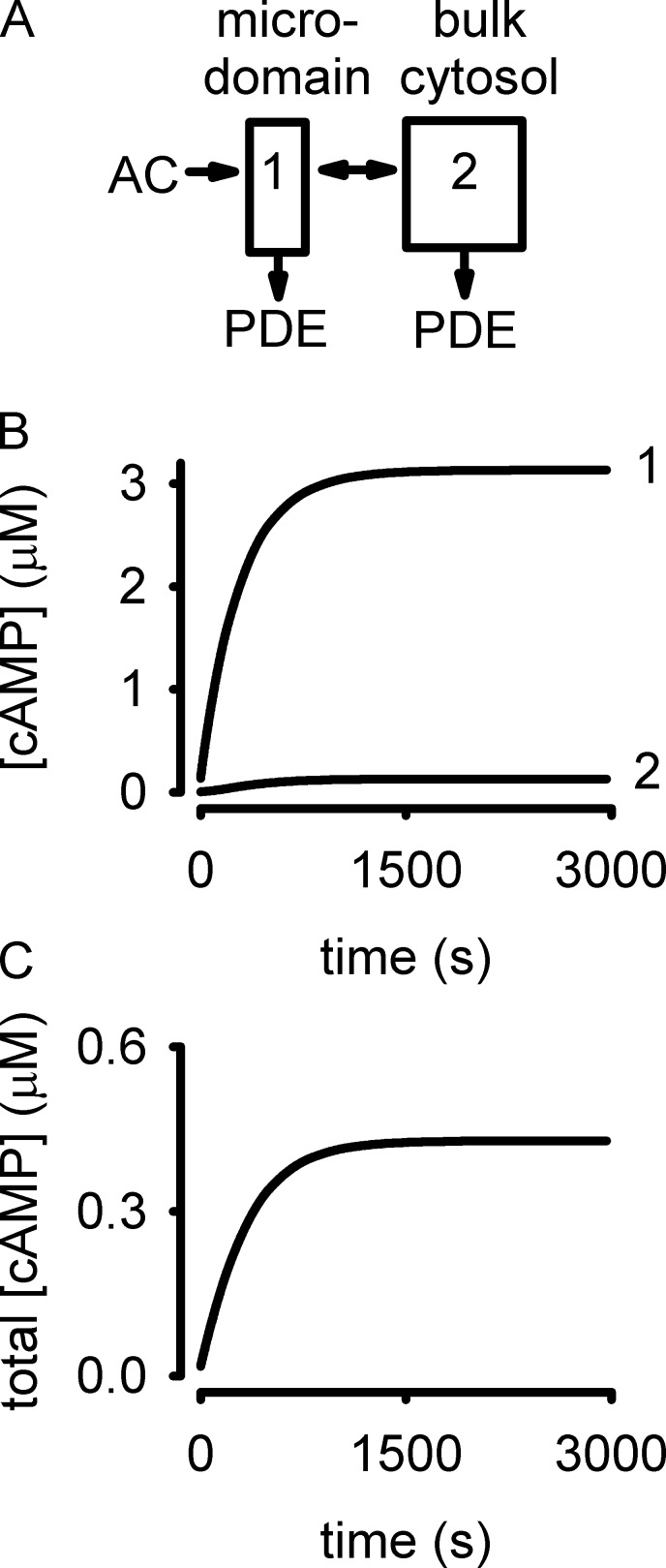
We and others have used compartmental models to describe the localization of cyclic nucleotide signals (Saucerman et al., 2014, and references therein). Here, we present an updated compartmental model based upon our recent estimates of near-membrane PDE activity (Fig. 2). In this model, cAMP is produced by AC in compartment 1. The overall PDE activity is markedly lower in compartment 1 than in compartment 2 (equations and parameter values are provided in the supplemental text). Upon stepwise activation of AC, cAMP concentrations within compartment 1 reach a plateau of >3 µM within 10 min. Diffusional restrictions that slow the flux of cAMP between compartments and the greater PDE activity in compartment 2 blunt cytosolic cAMP accumulation to <0.5 µM.
Figure 2.
Schematic model of compartmentalized cAMP signals. (A) A two-compartment model with diffusional restrictions between a membrane-localized microdomain (compartment 1) and the bulk cytosol (compartment 2). See text for details. (B) Cyclic AMP signals triggered by activation of AC. Cyclic AMP levels in compartment 1 are markedly higher than those in compartment 2. (C) Total cellular cAMP accumulation over the same time course.
The mechanisms by which this rapidly diffusible messenger is localized to different regions of a cell are not well understood. Potential mechanisms of signal localization have been discussed in detail elsewhere (Feinstein et al., 2012; Conti et al. in this issue; Saucerman et al., 2014). Here we outline four mechanisms likely to contribute to signal localization, and thus, specificity within cAMP and cGMP signaling pathways:
(1) Colocalization of signaling proteins (e.g., receptors, G-proteins, AC, PDE, PKA, phosphatases, and downstream targets) into signaling complexes is an essential component in ensuring signaling specificity. However, the localization of signaling proteins by itself is unlikely to account for spatial segregation of cAMP signals because without restrictions on the spatial spread, cAMP and cGMP would rapidly diffuse throughout the cell (Piggott et al., 2006; Feinstein et al., 2012; Saucerman et al., 2014, and references therein). Rather, these assemblies ensure that signaling proteins experience the same signals and that PKA and PKG are poised to phosphorylate specific downstream targets.
(2) Physical barriers may slow the rate of cAMP diffusion from one region of the cell to another. There is strong evidence that ER or SR come into close apposition to the plasma membrane and limit the spatial spread of Ca2+ and Na+ in specialized cells such as hair cells, neurons, and cardiac myocytes. Such barriers may partially restrict the spatial spread of cyclic nucleotides. In addition, the F-actin cortical rim may slow the movement of cAMP due to steric hindrance and charge effects. In addition, F-actin networks may trigger gelation of cytosol, dramatically lowering the local effective diffusion coefficient in the near-membrane space. For a more complete discussion see Feinstein et al. (2012) and Saucerman et al. (2014) and references therein.
(3) Buffering may contribute to the slow spatial spread of cAMP and cGMP signals. This may be particularly important when high concentrations of PKA are localized to discrete regions of the cell by AKAPs.
(4) PDE activity limits the spatial spread of cyclic nucleotide signals. Mathematical models indicate that PDE activity is particularly effective in limiting the spatial spread of cAMP and cGMP when signals are partially segregated by diffusional restrictions. The roles of PDE activity in regulating the kinetics and spatial spread of cyclic nucleotide signals have been discussed in detail elsewhere (Conti et al., 2014; Saucerman et al., 2014).
Evidence presented from several groups suggests that cyclic nucleotide compartmentalization is a critical component of signaling specificity, as discussed in Conti et al. (2014), Kapiloff et al. (2014), and Saucerman et al. (2014). The model presented here illustrates how diffusional barriers and variable PDE activity may allow distinct cyclic nucleotide signals in different subcellular locations. Multiple compartments containing scaffold-localized signaling complexes likely contribute to signaling specificity in several intracellular signaling pathways. Localized signaling complexes may also facilitate crosstalk between signaling pathways in a controlled manner, facilitating both frequency-dependent and digital signals.
Two basic approaches have been used to elucidate the mechanisms underlying the spatial segregation of cyclic nucleotide signals. The first approach relies on sensors localized to a discrete region of the cell, such as a CNG channel localized to the plasma membrane, and on changing the location of the cyclic nucleotide source. For example, Piggott et al. (2006) examined the ability of cGMP produced by particulate GC (pGC) and soluble GC (sGC) to activate CNG channels in both HEK-293 cells and vascular smooth muscle cells. They observed that in a 10-min timeframe, cGMP produced by pGC readily activated CNG channels, whereas cGMP produced by sGC did not, even in the presence of PDE inhibitors. In conceptually similar experiments, investigators have dialyzed known concentrations of cyclic nucleotides into cells and measured the activation of CNG channels to estimate the effective diffusion coefficient (Koutalos et al., 1995; Chen et al., 1999; Rich et al., 2000). We have recently used a similar approach to provide evidence that PDE4 is lower in the near-membrane compartment than in the bulk cytosol (unpublished data). Although these basic approaches allow a quantitative estimate of the averaged effective diffusion coefficient throughout the cell, they cannot be used to further dissect the spatial spread of cyclic nucleotide signals because measurements of CNG channel activity are localized to the plasma membrane.
A second approach compares responses measured in different subcellular locations. Different studies have compared CNG channels with changes in total cellular cyclic nucleotide levels (Rich et al., 2000, 2001a; Piggott et al., 2006), CNG channels and soluble FRET probes (Rochais et al., 2006; Willoughby et al., 2006), FRET probes based upon different cyclic nucleotide binding sites (Warrier et al., 2007), and FRET probes localized to different subcellular compartments (Saucerman et al., 2006; Terrin et al., 2006; Blackman et al., 2011). As indicated earlier, the ability of fluorescence- and FRET-based probes to be targeted to discrete subcellular domains makes them well-suited for such studies. However, data interpretation is not necessarily straightforward. For example, changes in viscosity and ROS in the near-membrane space may differentially alter fluorescence emission of the donor and acceptor (e.g., CFP and YFP). Similarly, dynamic changes in ROS and pH in the mitochondria may confound interpretation of FRET measurements. Also, as noted earlier, localized FRET probes may reach concentrations in which both intramolecular and intermolecular FRET occur. Thus, great care must be taken when comparing the magnitude or kinetics of fluorescence and FRET measurements from different subcellular locations. We believe that until more systematic approaches for monitoring the effects of changing cellular environments are developed, the best approach is to use multiple probes for the same measurement. For example, Blackman et al. (2011) used targeted FRET probes, CNG channels, radioimmunoassays, and measurements of downstream effector activity to elucidate the specific roles of PDE4 isoforms localized within discrete subcellular compartments.
Frequency encoding of information in cyclic nucleotide signals.
Although compartmentalization appears to make a major contribution to signaling specificity, other mechanisms, such as frequency-dependent signaling, are likely to contribute to the information content in second messenger systems. Frequency-dependent signaling implies that downstream effectors respond to particular frequencies in signals. In other words, effectors may respond differently to fast oscillations in cyclic nucleotide levels than to slow oscillations. Oscillations in cAMP levels were proposed almost forty years ago (Brooker, 1973; Wollenberger et al., 1973). Mathematical simulations demonstrated that feedback interactions between Ca2+-inhibitable ACs and Ca2+-permeable ion channels could lead to stable oscillations in cAMP levels (Cooper et al., 1995). Similarly, simulations indicated that Ca2+-mediated stimulation of PDE activity could also trigger stable oscillations in cAMP levels (Rich and Karpen, 2002). Experimental evidence that cAMP levels may oscillate was provided by Reisert and Matthews (2001), who observed oscillations in currents through CNG channels of olfactory cilia in response to sustained odorants. Later studies in β cells demonstrated cAMP oscillations and the importance of pulsatile cAMP production in maintaining these oscillations (Dyachok et al., 2006; Tian et al., 2012). Clever use of a Ca2+-stimulated AC8 overexpression system elucidated the potential for crosstalk between cAMP and Ca2+ pathways and coordinated oscillations due to feedback interactions between pathways (Willoughby and Cooper, 2006). Zhang and colleagues provided convincing experimental evidence that slow cAMP oscillations trigger PKA oscillations (Ni et al., 2011). Although there is substantial evidence that cyclic nucleotide oscillations occur, the number of studies describing these oscillations is small, especially compared with the abundant description of Ca2+ oscillations. Thus, we are left with the question: are experimental difficulties limiting our ability to detect cyclic nucleotide oscillations or are such oscillations rare epiphenomena?
We believe experimental limitations have impeded our ability to detect cyclic nucleotide oscillations. The PKA-based cAMP probes developed more than twenty years ago lacked the fast kinetics to detect rapid changes in cAMP levels (Rich and Karpen, 2002). Buffering of cAMP signals was also problematic due to cellular microinjection or overexpression of high probe concentrations. This was further complicated by the fact that the fluorescently labeled PKA is an active kinase. In contrast, the rapid response time of CNG channels and low buffering capacity allow detection of high-frequency cAMP oscillations (Rich et al., 2000). However, CNG channels are active proteins that allow Na+ and Ca2+ influx. Thus, kinetic experiments need to be conducted in Ca2+-free buffer to prevent CNG channel-mediated Ca2+ regulation of enzymes in the signaling system, preventing the study of Ca2+-cAMP interactions. The response time of FRET-based probes for cAMP and cGMP is more than fast enough for the measurement of oscillations with a period (T) of 30 s, and they are not active enzymes. Yet few investigators have been able to measure cyclic nucleotide oscillations with this probe. To better understand why, we have developed a realistic mathematical description of the cGMP signaling pathway (equations as well as parameter definitions and values are provided in the supplemental text).
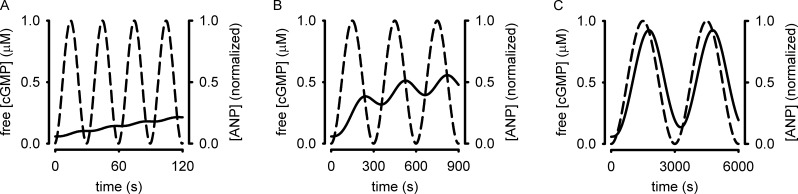
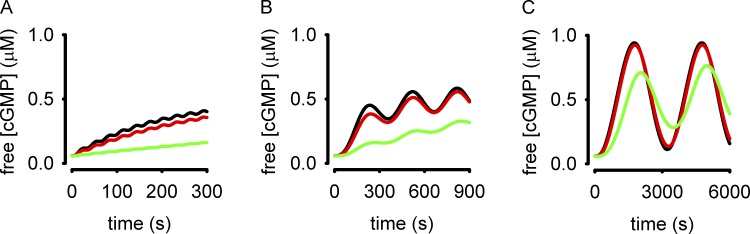
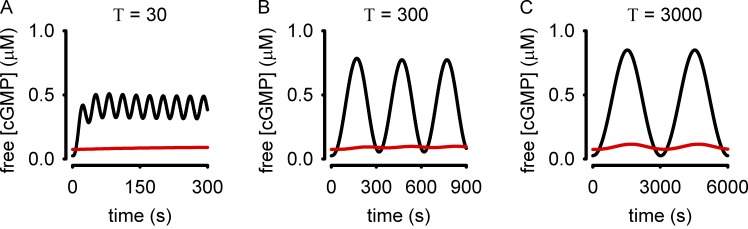
In this model, activation of pGC increases intracellular cGMP accumulation. Dephosphorylation of pGC causes receptor desensitization and a reduction in the rate of cGMP synthesis. Cyclic GMP is hydrolyzed by PDE type 5 (PDE5), which is regulated by cGMP binding to the noncatalytic site and phosphorylation. Inputs to the model were analogous to sinusoidal oscillations in atrial natriuretic peptide (ANP) with periods (T) of 30, 300, and 3,000 s. The levels of pGC activity were similar to those observed experimentally in response to 50 nM ANP. These inputs triggered sinusoidal intracellular cGMP accumulation with a lag that varied with stimulus frequency (Fig. 3). The amplitude of cGMP oscillations increased with the period of sinusoidal stimulation. A cGMP probe would require a high dynamic range (the ability to reproducibly measure small changes in cGMP) to detect the cGMP responses to ANP oscillations with periods of 30 or 300 s (Fig. 3, A and B). The cGMP FRET probes currently available lack the dynamic range to adequately sample either signal. Similarly, cAMP probes that use CFP and YFP as the fluorescent donor and acceptor lack the requisite sensitivity to measure analogous fluctuations in cAMP in intact cells. Because of their limited dynamic range, these FRET-based sensors are only able to reproducibly detect large cyclic nucleotide fluctuations, similar to those modeled in response to slow ANP oscillations (Fig. 3 C). The ability of these probes to detect small fluctuations in cyclic nucleotide levels is further limited by their buffering capacity. To illustrate this we repeated the simulations depicted in Fig. 3 with exogenous buffer concentrations of 0.05, 0.5, and 5 µM (Fig. 4, black, red, and green lines, respectively). These simulations demonstrate that buffering by the probe may alter the cyclic nucleotide signals being measured. At present, we cannot accurately estimate absolute fluorescent probe concentrations in single cells; thus, the effects of cyclic nucleotide buffering capacity of these sensors is unknown and likely varies from cell type to cell type (if not from cell to cell). Therefore the first generation cGMP and cAMP sensors using CFP and YFP lack the dynamic range to reproducibly discern small fluctuations in cyclic nucleotide levels. However, recently developed cGMP sensors by W. Dostmann and colleagues (Nausch et al., 2008) and cAMP sensors developed by K. Jalink and colleagues (Klarenbeek et al., 2011) have larger responses and likely increased dynamic range. As such, these next-generation sensors are better suited for detection of cyclic nucleotide oscillations because they are likely able to detect changes of smaller increment in cyclic nucleotide levels, and can be expressed with weaker promoters, allowing lower expression levels and reduced buffering capacity.
Figure 3.
Simulations depicting intracellular cGMP accumulation in response to an oscillatory stimulus. Broken lines represent model inputs: sinusoidal stimuli with periods (T) of 30 s (A), 300 s (B), or 3,000 s (C). Solid lines represent simulations of intracellular cGMP accumulation.
Figure 4.
Simulations depicting the effects of buffering by cyclic nucleotide probes. Simulations depict the cGMP responses in cells expressing 0.05 (black lines), 0.5 (red lines), and 5 µM (green lines) FRET probes (the parameter buf = 0.05, 0.5, and 5 µM, respectively). The K1/2 of the probe for cGMP was set to 1 µM. Input frequencies were 30, 300, and 3,000 s (as in Fig. 3). Note the different timescales for each simulation. Equations as well as parameter definitions and values are given in the supplemental text.
In addition to difficulties in detecting cyclic nucleotide oscillations inherent to the characteristics of the first generation of genetically encoded cyclic nucleotide sensors, it may be difficult to detect cyclic nucleotide oscillations localized to discrete subcellular compartments. To illustrate this, we modeled the effects of compartmentalization on oscillatory cGMP signals (Fig. 5). pGC activity was constrained to a near-membrane compartment (C1) that comprised 10% of the cell volume (depicted in Fig. 2). Thus, in C1 enzymes, concentrations were 10-fold higher than listed in Table S2. We next added both PDE5 activity (maximal PDE5 activity of 2 µM/min) and basal sGC activity (0.026 µM/min) in the cytosolic compartment (C2). No stimulation of sGC was considered. We modeled a sinusoidal stimulus to trigger oscillatory cGMP fluctuations. Oscillatory cGMP responses occurred in C1 in response to all the stimuli (T = 30, 300, and 3,000 s; Fig. 5, black lines). The amplitude of cGMP oscillations in C1 was greater at the faster input frequencies compared with the one-compartment model (compare Figs. 3 and 5). Only small cGMP responses occurred in C2 (Fig. 5, red lines). Assuming that soluble FRET probes would be uniformly distributed throughout the cell (this may not be the case, see Raymond et al., 2007), then 90% of the signal would be from C2, where cGMP levels are low. In addition, using membrane-localized cGMP probes would increase the concentration of the probe in C1, and in turn increase the cGMP buffering capacity of the probe in C1. The situation becomes more difficult if the cyclic nucleotide fluctuations occur near highly localized receptor complexes rather than in the sizeable subcellular compartment used in this simulation. Thus, the choice of probe and experimental conditions required for triggering and detecting cyclic nucleotide oscillations may be based upon trial, error, persistence, and a bit of luck. The frequency content of larger fluctuations may be accurately measured, but the amplitude of fluctuations and the effects of cyclic nucleotide buffering by the probes are difficult to assess.
Figure 5.
Simulations depicting intracellular cGMP accumulation in a two-compartment model in response to an oscillatory stimulus. In this model cGMP is produced by pGC in a near-membrane compartment (C1). cGMP levels rapidly equilibrate in C1, but the flux of cGMP into the larger cytosolic compartment (C2) is slow. PDE5 activity is present in both C1 and C2. Basal sGC activity maintains a low baseline cGMP level in C2. Black lines represent cGMP concentration in C1 in response to sinusoidal stimuli oscillations with periods (T) of 30 s (A), 300 s (B), or 3,000 s (C). Red lines represent cGMP accumulation in C2. Simulations assumed no cGMP sensors were present, thus the parameter buf was set to 0 µM. Note the different timescales for each simulation. Model details are given in the supplemental text.
A final thought on the measurement of cyclic nucleotide oscillations: investigators typically use high agonist concentrations to trigger cyclic nucleotide production. This often allows for measurement of (relatively) large responses, and the responses are also typically more reproducible because experiments are conducted well above the EC50 for cyclic nucleotide production. Although this may allow unraveling of potentially complex interacting pathways, it may not be physiological and, importantly, may trigger damped overshoot or transient responses due to feedback regulation within signaling systems. Thus, careful titration is needed to evaluate the information content within physiological signals.
Frequency-dependent downstream effectors.
While there is growing evidence that cyclic nucleotide signals oscillate, at least under certain experimental conditions, it is unclear how these oscillations are interpreted by downstream effectors. We have previously used simulations of CNG channels and PKA to examine how each effector would respond to cAMP oscillations (Rich and Karpen, 2002). We observed that CNG channels would faithfully reproduce fast cAMP oscillations (T = ∼2 s). Simulations also indicated that PKA activity would track slow cAMP oscillations (T = ∼1 min). However, as the period of oscillations decreased, PKA activation would reach a plateau, largely because of the slow reassociation of catalytic and regulatory subunits. Zhang and colleagues used mathematical models of oscillatory circuits and came to a similar conclusion (Ni et al., 2011). Simulations provided by both groups indicated that the level of PKA activation was proportional to the frequency and duty cycle of the oscillations. Thus, a particular level of PKA activity could be sustained by altering the frequency and duty cycle of cAMP fluctuations rather than maintaining steady intracellular cAMP concentrations. Similarly, it seems likely that positive feedback provided by autophosphorylation of PKG would allow the frequency and duty cycle of cGMP oscillations to dictate the level of PKG activity.
Digital encoding of information in cyclic nucleotide signals.
Thus far we have discussed the strengths and weaknesses of current cyclic nucleotide probes when used to measure localized and oscillating signals (spatial and frequency encoding). We briefly discuss a third mechanism by which information can be encoded in second messenger signals: digital encoding. Digital encoding is simply the combination of a set of signals that are on or off. For example, consider the case with three signals: cAMP, cGMP, and Ca2+. The information contained in a signal in which cAMP is on (above a threshold), cGMP is off, and Ca2+ is off is different from the information contained in the signal in which cAMP is on, cGMP is off, and Ca2+ is on. The information content is 2N, where N is the number of signals (in this case, N = 3).
Biology is seldom this simple. There may be multiple effectors, for example PKA and Epac. These effectors have different affinities for cAMP, and thus there would be three states: off (in which cAMP levels are not high enough to substantively activate PKA), low (in which cAMP is high enough to substantively activate PKA but not Epac), and high (in which cAMP levels are high enough to substantively activate both PKA and Epac). There may also be multiple inputs for one signaling pathway, for example epinephrine-mediated activation of β2-adrenergic receptors and PGE2-mediated activation of EP2 receptors. Both of these inputs trigger increases in cAMP; however, the information contained in each signal may be additive or just different. Thus, there may be information encoded in the combination of signals that are on or off. Digital encoding of information is also likely to work in conjunction with spatial and frequency encoding. Thus, to decipher the information encoded in cyclic nucleotide signals we must simultaneously monitor other intracellular signals such as Ca2+. State-of-the-art measurements are primarily fluorescence- and FRET-based probes. How can we quantitatively measure signals from several fluorescence- and FRET-based probes in cells and tissues?
Hyperspectral imaging and analysis of cyclic nucleotide signals.
One promising approach to measure several intracellular signals simultaneously is hyperspectral imaging and analysis. Hyperspectral imaging approaches were developed by the Department of Defense and NASA to solve remote sensing problems associated with satellite imaging. These approaches have been used to study biological systems and second messenger signaling (Leavesley et al., 2013; Rich et al., 2013; Woehler, 2013). Hyperspectral approaches allow for measurements of the fluorescence emission spectrum of a sample, allowing accurate detection and quantification of the abundance of unique spectral signatures, e.g., the emission spectra of CFP, YFP, and Hoechst 33342 (a nuclear stain), within cells and tissue. As such, hyperspectral approaches are well suited for simultaneously monitoring fluorescence emission from several probes (Leavesley et al., 2013; Rich et al., 2013). The tradeoff when using commercially available spectral implementations is reduced temporal resolution. For example, acousto-optical tunable filters (AOTFs) attenuate light by 80%, and long acquisition times are required. Recent advances in filter technology attenuate light by only 5%, and thus allow for faster image acquisition (Favreau et al., 2013). Finally, it should be also noted that even with the advances in technology, choosing the microscope system best suited for hyperspectral imaging is not always straightforward (Annamdevula et al., 2013), and spectral calibration may be required (Leavesley et al., 2012).
Another aspect of interpreting cyclic nucleotide signals is unbiased data analysis. Traditional image analysis approaches require investigators to select regions of interest (ROIs) for automated analysis of emission intensities. This has become a point of contention because manually selecting ROIs may inadvertently bias data interpretation, especially when examining signals from intact tissues rather than cell monolayers. Recent advances in automated image analysis overcome this limitation, selecting ROIs based upon event detection above noise thresholds or geometrical constraints (Francis et al., 2012; Leavesley et al., 2013). Thus, ROIs can be quantified in a defined manner, thresholds for signal response can be set based upon baseline noise levels, and signals above baseline noise can then be automatically identified and analyzed.
A bright future for decoding the information content in cyclic nucleotide signals
In the last fifteen years a great deal of progress has been made in measurement of cyclic nucleotide signals. Here we have discussed several cyclic nucleotide sensors, highlighted their strengths and weaknesses, and summarized three mechanisms for encoding information within intracellular signals. And although all protein-based fluorescent probes have limitations in terms of dynamic range and buffering of the signals, the latest generation of probes has increased range, and hyperspectral imaging approaches can be used to improve the signal-to-noise ratio of fluorescence and FRET measurements. These advances should allow detection and quantification of the localized cyclic nucleotide dynamics in both single cell and tissue preparations. Hyperspectral imaging also allows measurement of multiple probes to simultaneously track fluctuations of several intracellular signals. Automated image analysis provides a set of tools that allow investigators to analyze data in an unbiased fashion. These data can be used to validate and test mathematical descriptions of signaling systems in cells and tissues. In turn, mathematical models can be used to direct our experimental design. In the last decade, the technologies required to decode the information content of intracellular signals have developed to the point that we can begin to decipher the flux of information between cells and their environment.
This Perspectives series includes articles by Karpen, Kapiloff et al., Conti et al., and Saucerman et al.
Equations describing compartmentalized and oscillatory cyclic nucleotide signals are provided. Table S1 provides the parameter values, descriptions, initial conditions, and references from which parameter values were obtained or estimated for compartmental models describing subcellular cyclic nucleotide distribution. Table S2 shows provides parameter values, descriptions, initial conditions, and references from which the parameter values were obtained or estimated for models describing oscillatory cGMP signals. Online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.201311095/DC1.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grant P01H06629.
Olaf S. Andersen served as editor.
Footnotes
Abbreviations used in this paper:
- AC
- adenylyl cyclase
- ANP
- atrial natriuretic peptide
- Epac
- exchange factor activated by cAMP
- FRET
- Förster resonance energy transfer
- PDE
- phosphodiesterase
- pGC
- particulate GC
- ROS
- reactive oxygen species
- sGC
- soluble GC
References
- Adams S.R., Harootunian A.T., Buechler Y.J., Taylor S.S., Tsien R.Y. 1991. Fluorescence ratio imaging of cyclic AMP in single cells. Nature. 349:694–697 10.1038/349694a0 [DOI] [PubMed] [Google Scholar]
- Annamdevula N.S., Sweat B., Favreau P., Lindsey A.S., Alvarez D.F., Rich T.C., Leavesley S.J. 2013. An approach for characterizing and comparing hyperspectral microscopy systems. Sensors (Basel). 13:9267–9293 10.3390/s130709267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernas T., Robinson J.P., Asem E.K., Rajwa B. 2005. Loss of image quality in photobleaching during microscopic imaging of fluorescent probes bound to chromatin. J. Biomed. Opt. 10:064015 10.1117/1.2136313 [DOI] [PubMed] [Google Scholar]
- Blackman B.E., Horner K., Heidmann J., Wang D., Richter W., Rich T.C., Conti M. 2011. PDE4D and PDE4B function in distinct subcellular compartments in mouse embryonic fibroblasts. J. Biol. Chem. 286:12590–12601 10.1074/jbc.M110.203604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner S., Schwede F., Schlipp A., Berisha F., Calebiro D., Lohse M.J., Nikolaev V.O. 2011. FRET measurements of intracellular cAMP concentrations and cAMP analog permeability in intact cells. Nat. Protoc. 6:427–438 10.1038/nprot.2010.198 [DOI] [PubMed] [Google Scholar]
- Brady J.D., Rich E.D., Martens J.R., Karpen J.W., Varnum M.D., Brown R.L. 2006. Interplay between PIP3 and calmodulin regulation of olfactory cyclic nucleotide-gated channels. Proc. Natl. Acad. Sci. USA. 103:15635–15640 10.1073/pnas.0603344103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooker G. 1973. Oscillation of cyclic adenosine monophosphate concentration during the myocardial contraction cycle. Science. 182:933–934 10.1126/science.182.4115.933 [DOI] [PubMed] [Google Scholar]
- Chen C.H., Nakamura T., Koutalos Y. 1999. Cyclic AMP diffusion coefficient in frog olfactory cilia. Biophys. J. 76:2861–2867 10.1016/S0006-3495(99)77440-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti M., Mika D., Richter W. 2014. Perspectives on: Cyclic nucleotide microdomains and signaling specificity: Cyclic AMP compartments and signaling specificity: Role of cyclic nucleotide phosphodiesterases. J. Gen. Physiol. 143:29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper D.M.F., Mons N., Karpen J.W. 1995. Adenylyl cyclases and the interaction between calcium and cAMP signalling. Nature. 374:421–424 10.1038/374421a0 [DOI] [PubMed] [Google Scholar]
- Corbin J.D., Sugden P.H., Lincoln T.M., Keely S.L. 1977. Compartmentalization of adenosine 3′:5′-monophosphate and adenosine 3′:5′-monophosphate-dependent protein kinase in heart tissue. J. Biol. Chem. 252:3854–3861 [PubMed] [Google Scholar]
- Dhallan R.S., Yau K.-W., Schrader K.A., Reed R.R. 1990. Primary structure and functional expression of a cyclic nucleotide-activated channel from olfactory neurons. Nature. 347:184–187 10.1038/347184a0 [DOI] [PubMed] [Google Scholar]
- Dolmetsch R.E., Xu K., Lewis R.S. 1998. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 392:933–936 10.1038/31960 [DOI] [PubMed] [Google Scholar]
- Dyachok O., Isakov Y., Sågetorp J., Tengholm A. 2006. Oscillations of cyclic AMP in hormone-stimulated insulin-secreting beta-cells. Nature. 439:349–352 10.1038/nature04410 [DOI] [PubMed] [Google Scholar]
- Favreau P., Hernandez C., Lindsey A.S., Alvarez D.F., Rich T., Prabhat P., Leavesley S.J. 2013. Thin-film tunable optical filters for hyperspectral microscopy. J. Biomed. Opt. 19:011017 10.1117/1.JBO.19.1.011017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein W.P., Zhu B., Leavesley S.J., Sayner S.L., Rich T.C. 2012. Assessment of cellular mechanisms contributing to cAMP compartmentalization in pulmonary microvascular endothelial cells. Am. J. Physiol. Cell Physiol. 302:C839–C852 10.1152/ajpcell.00361.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis S.H., Corbin J.D. 1999. Cyclic nucleotide-dependent protein kinases: intracellular receptors for cAMP and cGMP action. Crit. Rev. Clin. Lab. Sci. 36:275–328 10.1080/10408369991239213 [DOI] [PubMed] [Google Scholar]
- Francis M., Qian X., Charbel C., Ledoux J., Parker J.C., Taylor M.S. 2012. Automated region of interest analysis of dynamic Ca²+ signals in image sequences. Am. J. Physiol. Cell Physiol. 303:C236–C243 10.1152/ajpcell.00016.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goaillard J.M., Vincent P.V., Fischmeister R. 2001. Simultaneous measurements of intracellular cAMP and L-type Ca2+ current in single frog ventricular myocytes. J. Physiol. 530:79–91 10.1111/j.1469-7793.2001.0079m.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbunova Y.V., Spitzer N.C. 2002. Dynamic interactions of cyclic AMP transients and spontaneous Ca(2+) spikes. Nature. 418:93–96 10.1038/nature00835 [DOI] [PubMed] [Google Scholar]
- Ha T., Tinnefeld P. 2012. Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annu. Rev. Phys. Chem. 63:595–617 10.1146/annurev-physchem-032210-103340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson P.I., Meyer T., Stryer L., Schulman H. 1994. Dual role of calmodulin in autophosphorylation of multifunctional CaM kinase may underlie decoding of calcium signals. Neuron. 12:943–956 10.1016/0896-6273(94)90306-9 [DOI] [PubMed] [Google Scholar]
- Honda A., Sawyer C.L., Cawley S.M., Dostmann W.R. 2005. Cygnets: in vivo characterization of novel cGMP indicators and in vivo imaging of intracellular cGMP. Methods Mol. Biol. 307:27–43 [DOI] [PubMed] [Google Scholar]
- Jalink K., van Rheenen J. 2009. Filter FRET: Quantitative imaging of sensitized emission. Laboratory Techniques in Biochemistry and Molecular Biology, Vol. 33. Gadella T.W.J., editor Elsevier, Oxford: 289–349 [Google Scholar]
- Kapiloff M.S., Rigatti M., Dodge-Kafka K.L. 2014. Perspectives on: Cyclic nucleotide microdomains and signaling specificity: Architectural and functional roles of A kinase–anchoring proteins in cAMP microdomains. J. Gen. Physiol. 143:9–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarenbeek J.B., Goedhart J., Hink M.A., Gadella T.W., Jalink K. 2011. A mTurquoise-based cAMP sensor for both FLIM and ratiometric read-out has improved dynamic range. PLoS ONE. 6:e19170 10.1371/journal.pone.0019170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutalos Y., Nakatani K., Yau K.-W. 1995. Cyclic GMP diffusion coefficient in rod photoreceptor outer segments. Biophys. J. 68:373–382 10.1016/S0006-3495(95)80198-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavesley S.J., Annamdevula N., Boni J., Stocker S., Grant K., Troyanovsky B., Rich T.C., Alvarez D.F. 2012. Hyperspectral imaging microscopy for identification and quantitative analysis of fluorescently-labeled cells in highly autofluorescent tissue. J. Biophotonics. 5:67–84 10.1002/jbio.201100066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavesley S.J., Britain A.L., Cichon L.K., Nikolaev V.O., Rich T.C. 2013. Assessing FRET using spectral techniques. Cytometry A. In press 10.1002/cyto.a.22340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nausch L.W., Ledoux J., Bonev A.D., Nelson M.T., Dostmann W.R. 2008. Differential patterning of cGMP in vascular smooth muscle cells revealed by single GFP-linked biosensors. Proc. Natl. Acad. Sci. USA. 105:365–370 10.1073/pnas.0710387105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Q., Ganesan A., Aye-Han N.N., Gao X., Allen M.D., Levchenko A., Zhang J. 2011. Signaling diversity of PKA achieved via a Ca2+-cAMP-PKA oscillatory circuit. Nat. Chem. Biol. 7:34–40 10.1038/nchembio.478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev V.O., Bünemann M., Hein L., Hannawacker A., Lohse M.J. 2004. Novel single chain cAMP sensors for receptor-induced signal propagation. J. Biol. Chem. 279:37215–37218 10.1074/jbc.C400302200 [DOI] [PubMed] [Google Scholar]
- Nikolaev V.O., Gambaryan S., Lohse M.J. 2006. Fluorescent sensors for rapid monitoring of intracellular cGMP. Nat. Methods. 3:23–25 10.1038/nmeth816 [DOI] [PubMed] [Google Scholar]
- Piggott L.A., Hassell K.A., Berkova Z., Morris A.P., Silberbach M., Rich T.C. 2006. Natriuretic peptides and nitric oxide stimulate cGMP synthesis in different cellular compartments. J. Gen. Physiol. 128:3–14 10.1085/jgp.200509403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponsioen B., Zhao J., Riedl J., Zwartkruis F., van der Krogt G., Zaccolo M., Moolenaar W.H., Bos J.L., Jalink K. 2004. Detecting cAMP-induced Epac activation by fluorescence resonance energy transfer: Epac as a novel cAMP indicator. EMBO Rep. 5:1176–1180 10.1038/sj.embor.7400290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond D.R., Wilson L.S., Carter R.L., Maurice D.H. 2007. Numerous distinct PKA-, or EPAC-based, signalling complexes allow selective phosphodiesterase 3 and phosphodiesterase 4 coordination of cell adhesion. Cell. Signal. 19:2507–2518 10.1016/j.cellsig.2007.08.005 [DOI] [PubMed] [Google Scholar]
- Reisert J., Matthews H.R. 2001. Responses to prolonged odour stimulation in frog olfactory receptor cells. J. Physiol. 534:179–191 10.1111/j.1469-7793.2001.t01-1-00179.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich T.C., Karpen J.W. 2002. Review article: cyclic AMP sensors in living cells: what signals can they actually measure? Ann. Biomed. Eng. 30:1088–1099 10.1114/1.1511242 [DOI] [PubMed] [Google Scholar]
- Rich T.C., Fagan K.A., Nakata H., Schaack J., Cooper D.M.F., Karpen J.W. 2000. Cyclic nucleotide-gated channels colocalize with adenylyl cyclase in regions of restricted cAMP diffusion. J. Gen. Physiol. 116:147–162 10.1085/jgp.116.2.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich T.C., Fagan K.A., Tse T.E., Schaack J., Cooper D.M.F., Karpen J.W. 2001a. A uniform extracellular stimulus triggers distinct cAMP signals in different compartments of a simple cell. Proc. Natl. Acad. Sci. USA. 98:13049–13054 10.1073/pnas.221381398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich T.C., Tse T.E., Rohan J.G., Schaack J., Karpen J.W. 2001b. In vivo assessment of local phosphodiesterase activity using tailored cyclic nucleotide-gated channels as cAMP sensors. J. Gen. Physiol. 118:63–78 10.1085/jgp.118.1.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich T.C., Britain A.L., Stedman T., Leavesley S.J. 2013. Hyperspectral imaging of FRET-based cGMP probes. Methods Mol. Biol. 1020:73–88 10.1007/978-1-62703-459-3_5 [DOI] [PubMed] [Google Scholar]
- Rochais F., Abi-Gerges A., Horner K., Lefebvre F., Cooper D.M.F., Conti M., Fischmeister R., Vandecasteele G. 2006. A specific pattern of phosphodiesterases controls the cAMP signals generated by different Gs-coupled receptors in adult rat ventricular myocytes. Circ. Res. 98:1081–1088 10.1161/01.RES.0000218493.09370.8e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruf F., Park M.J., Hayot F., Lin G., Roysam B., Ge Y., Sealfon S.C. 2006. Mixed analog/digital gonadotrope biosynthetic response to gonadotropin-releasing hormone. J. Biol. Chem. 281:30967–30978 10.1074/jbc.M606486200 [DOI] [PubMed] [Google Scholar]
- Saucerman J.J., Zhang J., Martin J.C., Peng L.X., Stenbit A.E., Tsien R.Y., McCulloch A.D. 2006. Systems analysis of PKA-mediated phosphorylation gradients in live cardiac myocytes. Proc. Natl. Acad. Sci. USA. 103:12923–12928 10.1073/pnas.0600137103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucerman J.J., Greenwald E.C., Polanska-Grabowska R. 2014. Perspectives on: Cyclic nucleotide microdomains and signaling specificity: Mechanisms of cyclic AMP compartmentalization revealed by computational models. J. Gen. Physiol. 143:39–48 [Google Scholar]
- Suhling K., Siegel J., Phillips D., French P.M., Lévêque-Fort S., Webb S.E., Davis D.M. 2002. Imaging the environment of green fluorescent protein. Biophys. J. 83:3589–3595 10.1016/S0006-3495(02)75359-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terrin A., Di Benedetto G., Pertegato V., Cheung Y.F., Baillie G., Lynch M.J., Elvassore N., Prinz A., Herberg F.W., Houslay M.D., Zaccolo M. 2006. PGE(1) stimulation of HEK293 cells generates multiple contiguous domains with different [cAMP]: role of compartmentalized phosphodiesterases. J. Cell Biol. 175:441–451 10.1083/jcb.200605050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G., Sågetorp J., Xu Y., Shuai H., Degerman E., Tengholm A. 2012. Role of phosphodiesterases in the shaping of sub-plasma-membrane cAMP oscillations and pulsatile insulin secretion. J. Cell Sci. 125:5084–5095 10.1242/jcs.107201 [DOI] [PubMed] [Google Scholar]
- Trivedi B., Kramer R.H. 1998. Real-time patch-cram detection of intracellular cGMP reveals long-term suppression of responses to NO and muscarinic agonists. Neuron. 21:895–906 10.1016/S0896-6273(00)80604-2 [DOI] [PubMed] [Google Scholar]
- Tsien R.Y. 1992. Intracellular signal transduction in four dimensions: from molecular design to physiology. Am. J. Physiol. 263:C723–C728 [DOI] [PubMed] [Google Scholar]
- van der Krogt G.N., Ogink J., Ponsioen B., Jalink K. 2008. A comparison of donor-acceptor pairs for genetically encoded FRET sensors: application to the Epac cAMP sensor as an example. PLoS ONE. 3:e1916 10.1371/journal.pone.0001916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warrier S., Ramamurthy G., Eckert R.L., Nikolaev V.O., Lohse M.J., Harvey R.D. 2007. cAMP microdomains and L-type Ca2+ channel regulation in guinea-pig ventricular myocytes. J. Physiol. 580:765–776 10.1113/jphysiol.2006.124891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willoughby D., Cooper D.M. 2006. Ca2+ stimulation of adenylyl cyclase generates dynamic oscillations in cyclic AMP. J. Cell Sci. 119:828–836 10.1242/jcs.02812 [DOI] [PubMed] [Google Scholar]
- Willoughby D., Wong W., Schaack J., Scott J.D., Cooper D.M.F. 2006. An anchored PKA and PDE4 complex regulates subplasmalemmal cAMP dynamics. EMBO J. 25:2051–2061 10.1038/sj.emboj.7601113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehler A. 2013. Simultaneous quantitative live cell imaging of multiple FRET-based biosensors. PLoS ONE. 8:e61096 10.1371/journal.pone.0061096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberger A., Babskii E.B., Krause E.G., Genz S., Blohm D., Bogdanova E.V. 1973. Cyclic changes in levels of cyclic AMP and cyclic GMP in frog myocardium during the cardiac cycle. Biochem. Biophys. Res. Commun. 55:446–452 10.1016/0006-291X(73)91107-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.