Abstract
FOXM1 is implicated in genotoxic drug resistance but its role and mechanism of action remain unclear. Here, we establish that γH2AX foci, indicative of DNA double strand breaks, accumulate in a time-dependent manner in the drug sensitive MCF-7 cells but not in the resistant counterparts in response to epirubicin. We find that FOXM1 expression is associated with epirubicin sensitivity and double strand break (DSB) repair. Ectopic expression of FOXM1 can increase cell viability and abrogate DSBs sustained by MCF-7 cells following epirubicin, owing to an enhancement in repair efficiency. Conversely, alkaline comet and γH2AX foci formation assays show that Foxm1-null cells are hypersensitive to DNA damage, epirubicin and γ-irradiation. Furthermore, we find that FOXM1 is required for DNA repair by homologous recombination (HR) but not non-homologous end joining (NHEJ), using HeLa cell lines habouring an integrated direct repeat green fluorescent protein (DR-GFP) reporter for DSB repair. We also identify BRIP1 as a direct transcription target of FOXM1 by promoter analysis and chromatin-immunoprecipitation assay. In agreement, depletion of FOXM1 expression by siRNA down-regulates BRIP1 expression at the protein and mRNA levels in MCF-7 and the epirubicin resistant MCF-7 EpiR cells. Remarkably, the requirement for FOXM1 for DSB repair can be circumvented by reintroduction of BRIP1, suggesting that BRIP1 is an important target of FOXM1 in DSB repair. Indeed, like FOXM1, BRIP1 is needed for HR. These data suggest that FOXM1 regulates BRIP1 expression to modulate epirubicin-induced DNA damage repair and drug resistance.
Keywords: FOXM1, BRIP1, DNA damage, epirubicin, resistance, breast cancer
Introduction
Breast cancer is the most common malignancy in women worldwide, with 1 in 9 of all western hemisphere women developing this disease in their lifetimes (1). Anthracyclines, including epirubicin and doxorubicin, are some of the most widely used chemotherapeutic drugs for the treatment of breast cancers in the adjuvant setting (2, 3). It is also the preferred option for the management of advanced or metastatic breast cancers (2). Anthracyclines function by intercalating DNA strands, releasing free oxygen radicals, and inhibiting topoisomerase II activity (4). Acting as topoisomerase II poisons, anthracyclines form complexes with DNA and topoisomerase, preventing DNA resealing, and thus promoting double strand breaks. The resulting unrepaired breaks lead to damages to the genome and a wide range of cytotoxic effects and ultimately cell death (5, 6). Despite anthracyclines being some of the most effective chemotherapeutic drugs, patients will eventually develop resistance and relapse (3, 7). The exact molecular mechanism underlying response and resistance to epirubicin is still not fully understood.
FOXM1 is a member of the Forkhead box (FOX) superfamily of transcription factors that share a conserved winged-helix DNA-binding domain (8). It is ubiquitously expressed in actively proliferating tissues (9, 10) and plays a crucial role in a wide range of biological processes, including cell cycle progression (11, 12), angiogenesis (13-15), metastasis (14, 16, 17), apoptosis (18), tissue regeneration (19) and drug resistance (15, 20). FOXM1 has been reported to be a cellular modulator of drug sensitivity and resistance in various types of cancers. In breast cancer, FOXM1 has been shown to be involved in endocrine (20, 21), cisplatin (15), trastuzumab and paclitaxel (22), gefitinib (23) and, most recently in epirubicin resistance (24). In response to DNA damage, Chk2 has been proposed to phosphorylate and stabilise FOXM1, which in turn induces the expression of breast cancer gene 2 (BRCA2) and X-ray cross complementing group 1 (XRCC1), genes involved in DNA damage repair (25). However, recent evidence challenges the direct regulation of BRCA2 and XRCC1 by FOXM1 upon DNA damage (15). In essence, despite having a role in DNA damage repair, the mechanism of action of FOXM1 in DNA damage repair is still poorly understood.
In this study we found a link between FOXM1 and the BRCA1-interacting protein (BRIP1). BRIP1, also known as BACH1 or FANCJ, is a DNA helicase that interacts with the BRCA1 C-terminal (BRCT) domain, an important motif involved in cellular responses to DNA damage (26, 27). This BRCA1-BRIP1 interaction contributes to the DNA repair activity of BRCA1 and, on the basis of this interaction, BRIP1 has been proposed to be a potential breast cancer susceptibility gene (26, 27). Consistently, BRIP1 mutations are associated with intermediate risk of breast cancer predisposition (26, 28). BRIP1 (BRCA1 interacting protein C-terminal helicase 1) has a complex role in double stranded-break (DSB) repair. Homologous recombination (HR) is compromised in human cells after depletion of BRIP1 (29). In fact, for the high fidelity homologous recombination (HR), BRIP1 requires the integrity of the helicase domain and BRCA1 binding (30, 31), but when uncoupled from BRCA1, the error-prone microhomology-mediated non-homologous end-joining (MMEJ) or polη-dependent translesion DNA synthesis (TLS) take precedence (30, 31). Moreover, the BRCA1-BRIP1 complex formation is also essential for DNA damage-induced G2/M checkpoint control (32). In the present study, we investigated the role of FOXM1 in BRIP1 expression, DNA damage repair and epirubicin resistance in breast cancer cells and mouse embryo fibroblasts (MEFs).
Results
Increased DNA damage upon depletion of FOXM1 levels
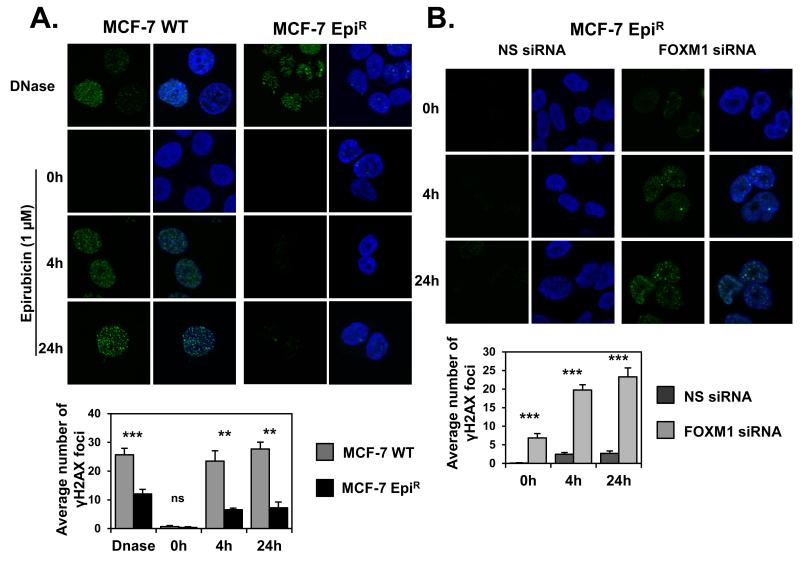
The involvement of FOXM1 in genotoxic drug resistance has led us to investigate if FOXM1 confers epirubicin resistance through enhancing DNA damage repair (24). To investigate this, we examined the formation of γH2AX-foci in MCF-7 and the previously described MCF-7 derived epirubicin resistant (MCF-7 EpiR) breast cancer cell lines following epirubicin treatment (Figure 1A) (20). We found that MCF-7 cells had significantly higher number of γH2AX foci after epirubicin treatment compared with MCF-7-EpiR cells (Figure 1A), suggesting that the parental MCF-7 cells sustained higher levels of DNA double-strand breaks (DSB) compared with the resistant cells in response to epirubicin. To test the role of FOXM1 in DNA damage, we depleted FOXM1 with siRNA in the resistant cells and found that FOXM1 knockdown significantly increased the number of γH2AX foci in comparison with those transfected with the non-specific (NS) siRNA (Figure 1B). The accumulation of γH2AX foci was observed 4 and 24 h after epirubicin treatment, suggesting that FOXM1 depletion renders resistant cells more susceptible to DSBs induced by epirubicin. Similarly, FOXM1 knockdown also sensitised MCF-7 EpiR cells to epirubicin, further supporting a role of FOXM1 in epirubicin resistance (Supplementary Figure S5B and S5C).
Figure 1. FOXM1 depletion leads to increased levels of DNA damage.
A. MCF-7 WT and MCF-7 EpiR cells were cultured on coverslips and treated with 1 μg/mL DNAse I for 60 min or with 1 μM of epirubicin for 0, 4 and 24 h, as indicated, and stained for γH2AX and 4′,6-diamidino-2-phenylindole (DAPI). Images were acquired with Leica TCS SP5 microscope. B. MCF-7 EpiR cells were transfected with control siRNA (NS siRNA) or with FOXM1 siRNA for 48 h. Cells were cultured on coverslips and treated with 1 μM of epirubicin for 0, 4 and 24 hours and stained for γH2AX and DAPI. Images were acquired with Leica TCS SP5. The panels below A. and B. show quantification of γH2AX foci number. Bars represent average of three independent experiments ± SEM. Statistical analyses were conducted using Student’s t tests against the correspondent time point. **, p ≤ 0.001; ***, p ≤ 0.0001, significant; ns, non-significant.
FOXM1 enhances DNA damage repair and confers epirubicin resistance
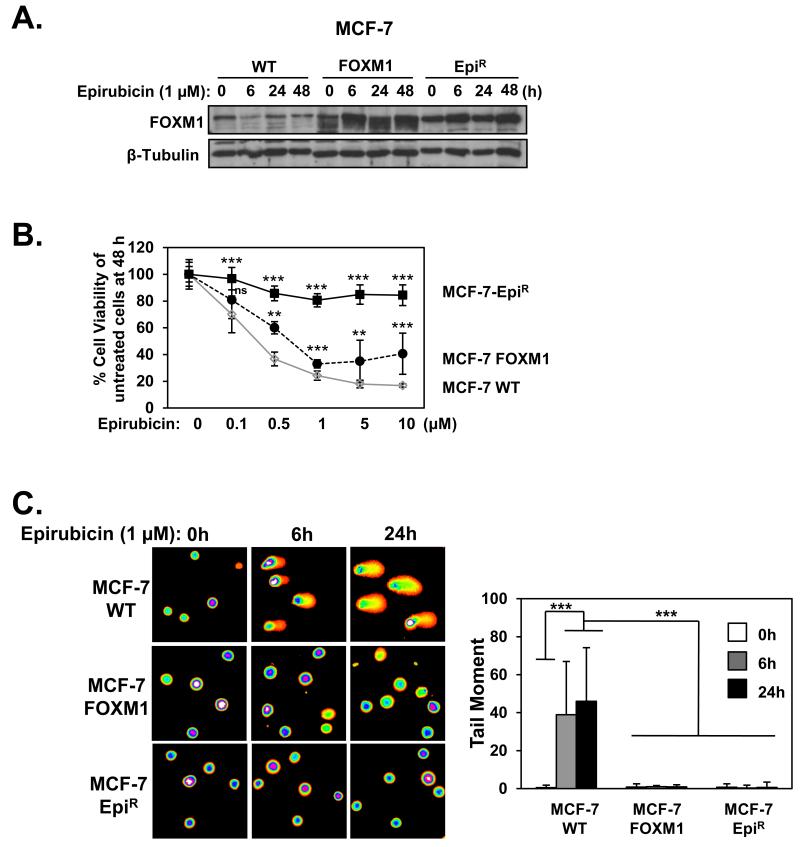
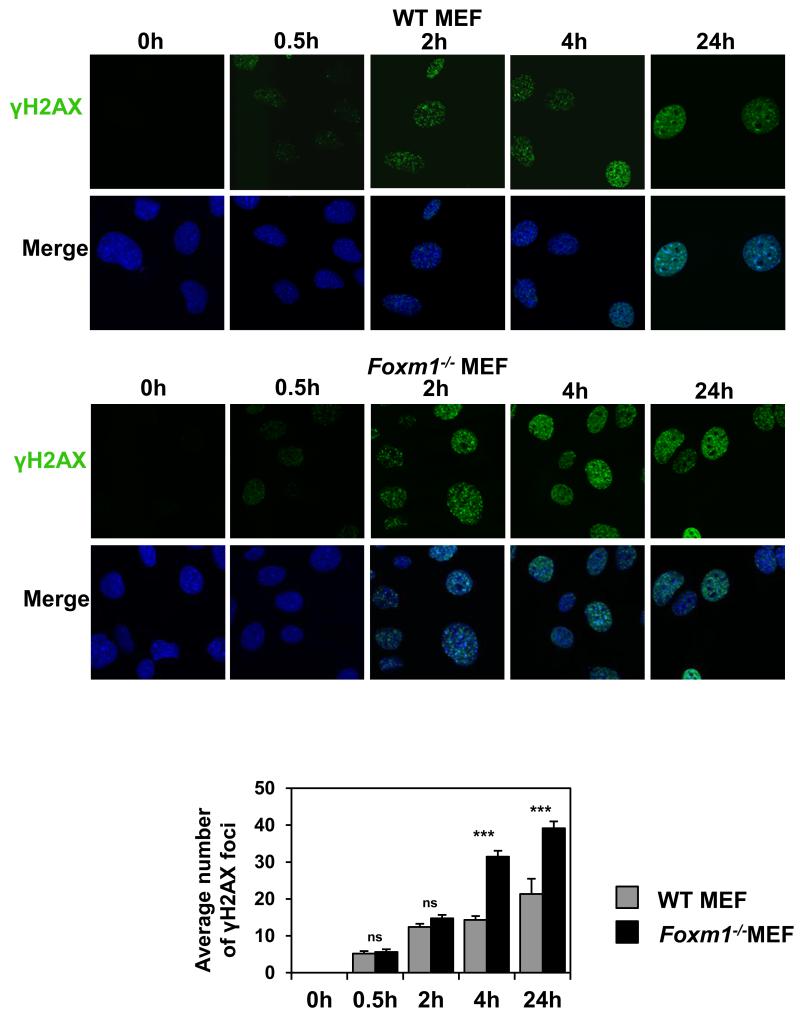
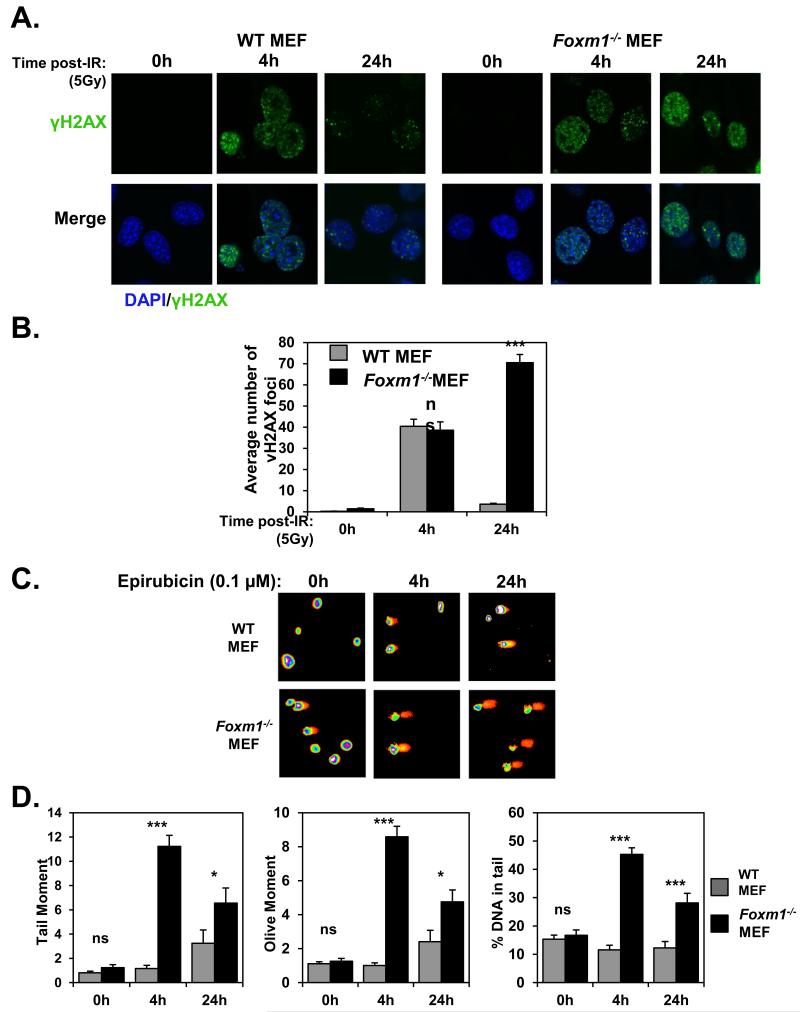
To examine the role of FOXM1 in epirubicin-resistance and DNA damage repair, we compared the proliferative rates of MCF-7, MCF-7 EpiR and MCF-7 cells stably expressing FOXM1 (MCF-7 FOXM1) in response to epirubicin (Figure 2A). SRB assay revealed that the overexpression of FOXM1 was sufficient to increase epirubicin resistance to the parental MCF-7 cells (Figure 2B). These data suggest that overexpression of FOXM1 reduces sensitivity to epirubicin and protects breast cancer cells against epirubicin-induced DNA damage. We next used alkaline comet assays to analyse the levels of damaged DNA in MCF-7, MCF-7 FOXM1 and MCF-7 EpiR cells in response to 1 μM epirubicin. As expected, overexpression of FOXM1 led to significantly lower tail moments when compared to MCF-7 WT, mimicking the lower rates of DNA damage sustained in MCF-7 EpiR cells (Figure 2C). To directly assess the influence of FOXM1 on DNA damage, we assayed the accumulation of DNA damage associated γH2AX foci on wild-type (WT) and Foxm1−/− MEFs (Figure 3) treated with 0.1 μM epirubicin, a dose which produced significant differential cytotoxic effects on WT and Foxm1−/− MEFs (Supplementary Figure S1). Consistently, when assessing DNA damage by γH2AX foci quantification a greater number of γH2AX foci was also observed at the longer times of 4 and 24 h after epirubicin treatment in the Foxm1−/− compared to WT MEFs (Figure 3). However, it is also notable that the accumulation of γH2AX foci in both WT and Foxm1−/− MEFs was at comparable rates at the earlier time points of 0.5 and 2 h, indicating that the lower levels of γH2AX foci observed in the WT is not due to the inability of epirubicin to access the genome DNA or to cause DNA damage in the FOXM1 expressing cells. The accumulation of γH2AX foci in the Foxm1−/− MEFs also suggests that the cells are less effective in repairing DSBs. To demonstrate further that the accumulation of γH2AX foci in the FOXM1-deficient cells is due to impaired DNA damage repair, WT and Foxm1−/− MEFs were γ-irradiated (5Gy) and assayed for γH2AX foci formation at 0 (mock irradiated), 4 and 24 h following irradiation (Figure 4A and 4B). The results showed that the accumulation of γH2AX foci in both WT and Foxm1−/− MEFs was at comparable rates at the earlier time points of 0 and 4 h, indicating similar levels of DNA damage induced. However, a greater number of γH2AX foci was also observed at the longer time point of 24 h following γ-irradiation in the Foxm1−/− when compared to WT MEFs, suggesting that MEFs-deficient of FOXM1 are less effective in DNA damage repair rather than more susceptible to DNA damage. To directly assess the influence of FOXM1 on DNA damage, we performed alkaline comet assay on wild-type (WT) and Foxm1−/− MEFs (Figure 4C) treated with 0.1 μM epirubicin. The result showed epirubicin induced higher levels of DNA damage in Foxm1−/− MEFs compared to the WT control after 4 and 24 h treatment as revealed by the longer comet tails (Figure 4C). Measurement of the tail moment, olive moment and percentage of DNA in tail (Figure 4D) showed that the epirubicin-induced DNA damage was significantly higher for Foxm1−/− MEFs compared to WT MEFs after epirubicin treatment.
Figure 2. Overexpression of FOXM1 confers epirubicin resistance and impairs DNA damage.
A. The MCF-7 WT, MCF-7-FOXM1 and MCF-7 EpiR cell lines were treated with 1 μM of epirubicin at indicated times and analysed for FOXM1 and β-Tubulin. B. MCF-7 WT, FOXM1 and EpiR cells were treated with increasing concentrations of epirubicin for 48 h and their cell viability measured by SRB assay. Representative data from 3 independent experiments are shown. Statistical analyses were performed using Student’s t tests and compared to the MCF-7 WT for the correspondent epirubicin concentration. *, p ≤ 0.05; ***, p ≤ 0.0001. C. MCF-7 WT, FOXM1 and EpiR cells were exposed to 1 μM of epirubicin for 0, 6 and 24 h and assayed for comet assay. The DNA damage was quantified using the tail moment (right panel). Represented data is average of two independent experiments (100 comets were measured per experiment). Error bars denote standard deviation. Statistical significance was performed using Student’s t tests (***, p ≤ 0.0001, significant). Results were tested for significance against the correspondent time point in the MCF-7 WT cell line.
Figure 3. Foxm1−/− MEFs cells accumulate higher levels of γH2AX foci than WT MEFs in response to epirubicin treatment.
WT and Foxm1−/− MEFs cultured in chamber culture slides were treated with 0.1 μM of epirubicin for 0, 0.5, 2, 4 and 24 h and stained for γH2AX (green). Nuclei were counterstained with DAPI (blue). Images were acquired with Leica TCS SP5. γH2AX foci quantification is shown in the lower panel. Bars represent average of three independent experiments ± SEM. Statistical analyses were conducted using Student’s t tests. ***, p ≤ 0.0001, significant; ns, non-significant.
Figure 4. Foxm1−/− MEFs accumulate sustained higher levels of DNA damage in reponse to γ-irradiation and epirubicin treatment.
WT and Foxm1−/− MEF seeded on culture chamber slides were either non-radiated (control; 0h) or exposed to 5 Gy of γ-irradiation for 4 and 24 h. Cells were then fixed and immunostained for anti-γH2AX with Alexa Fluor 488 anti-rabbit sera (green). Nuclei were counterstained with DAPI (blue). Images were acquired with Leica TCS SP5 (63× magnification). B. For each time point, images of at least 100 cells were captured and used for quantification of γH2AX foci number. The average number of foci per each image was quantified using the CellProfiler software. Results represent average of 3 independent experiments ± SEM. Statistical analyses were conducted using Student’s t tests against the correspondent time point. ***, p ≤ 0.0001 significant; ns, non-significant. C. WT and Foxm1−/− MEFs were exposed to 0.1 μM of epirubicin for 0, 4 and 24 h and subjected to comet assay. Representative images of three independent assays are shown. The DNA migration pattern with the comet head represents intact DNA and the comet tail broken DNA strands that migrate towards the anode. D. DNA damage was measured with Comet Assay IV and analysed for the following parameters: Tail moment (left panel), Olive moment (middle panel), % of DNA in tail (right panel). The represented data is average of two independent experiments (100 comets were measured per experiment). Bars represent average ± SEM. Statistical analyses were conducted using Student’s t tests against the correspondent time point. **, p ≤ 0.001; ***, p ≤ 0.0001, significant; ns, non-significant.
FOXM1 reconstitution in Foxm1−/− MEFs abrogates the accumulation of γH2AX foci
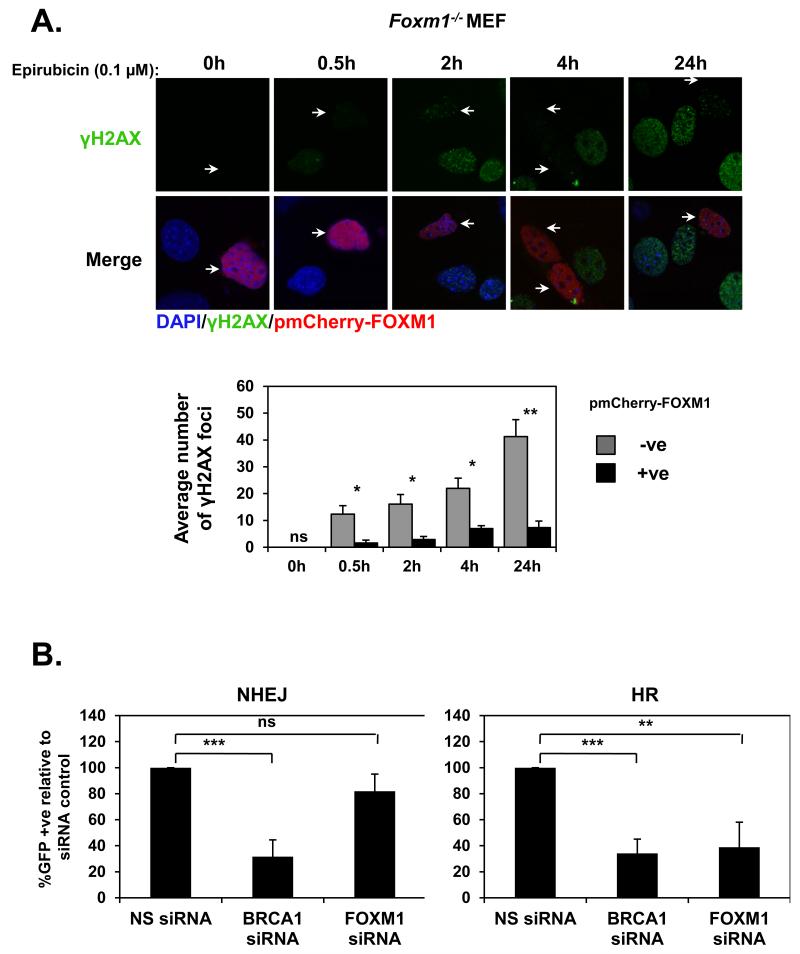
To demonstrate definitely that the lack of FOXM1 in Foxm1−/− MEFs is responsible for the progressive accumulation of γH2AX foci upon epirubicin treatment, we next sought to determine whether reintroduction of FOXM1 to Foxm1−/− MEFs was able to abolish the accumulation of γH2AX foci. As seen in Figure 5A, cells that were transfected with pmCherry-FOXM1 (red) displayed significantly fewer foci compared with the neighbouring non-transfected cells. However, Foxm1−/− MEFs transfected with the empty-pmCherry control have similar kinetics for γH2AX foci accumulation as the non-transfected cells (see Figure 7). All together these findings suggest that FOXM1 has a pivotal role in the accumulation DSB-DNA damage upon epirubicin.
Figure 5. FOXM1 decreases γH2AX foci accumulation in Foxm1−/− MEFs and is involved in homologous recombination repair.
A. Foxm1−/− MEFs transfected with the pmCherry-FOXM1 (red) and treated with 0.1 μM of epirubicin for 0, 0.5, 2, 4 and 24 h were stained for γH2AX followed by addition of Alexa488 (green)-labelled anti-rabbit. Nuclei were counterstained with DAPI (blue). Images were acquired with Leica TCS SP5. γH2AX foci quantification is shown in the lower panel. Bars represent average of three independent experiments ± SEM. Statistical analyses were conducted using Student’s t tests. *, p ≤ 0.05; **; p ≤ 0.001, significant; ns, non-significant. −ve, negative pmCherry-FOXM1 cells; +ve, positive pmCherry-FOXM1 cells. B. Non-homologous end joining (NHEJ) and homologous recombination (HR) were assayed in HeLa cells using DR-GFP reporter system bearing integrated end-joining and gene conversion substrates. Cells were transfected with nonspecific (NS) siRNA, si smart pool against BRCA1 or si smart pool against FOXM1. Forty-eight hours after transfection, cells were transfected again with I-SceI and left for 72 h before analysis. GFP-positive cells are indicated for each siRNA, with standard deviation based on three independent experiments (a total of 50,000 events were analysed for each experiment, and experiments were performed in duplicate). Statistical analyses were conducted using Student’s t tests against the NS siRNA **, p ≤ 0.001; ***, p ≤ 0.0001, significant; ns, non-significant.
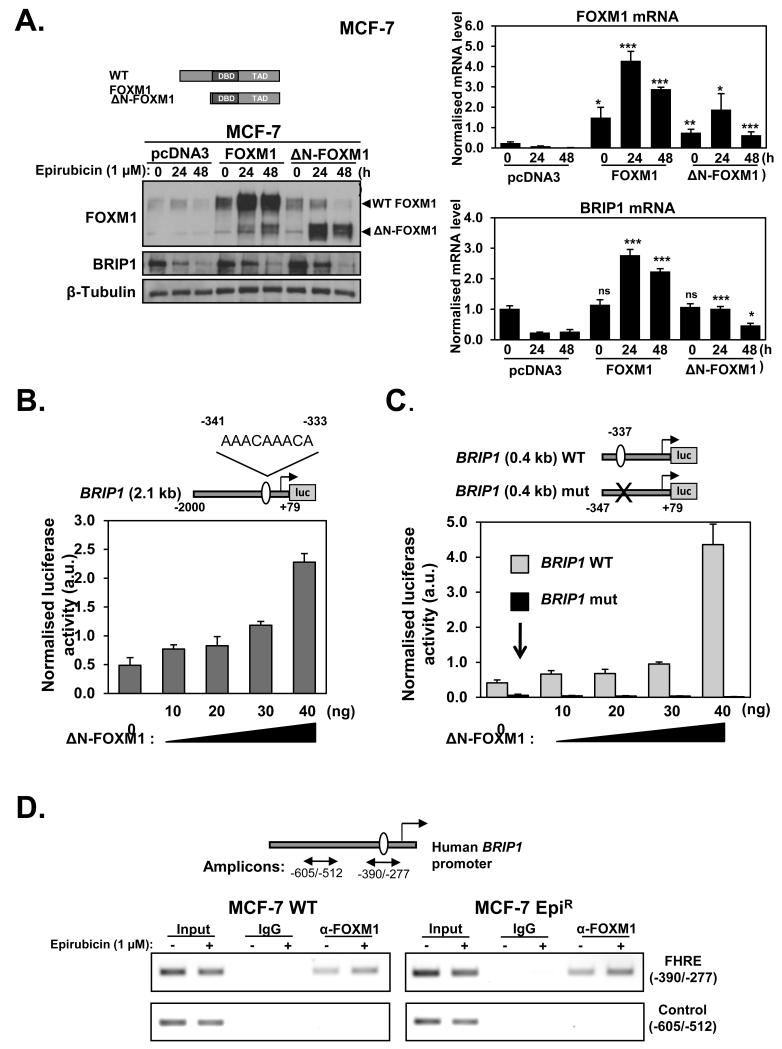
Figure 7. FOXM1 regulates BRIP1 expression through a forkhead responsive element (FHRE) consensus on its promoter.
A. MCF-7 cells were transiently transfected with 3 μg of pcDNA3, (control) pcDNA3-FOXM1 or pcDNA3-ΔN-FOXM1 and 24 h after transfection treated with 1 μM of epirubicin for 0, 24 and 48 h. The protein expression levels were determined by western blot using specific antibodies as indicated (left panel). Gene transcripts of these cells were analysed by qRT-PCR (right panels) for FOXM1 and BRIP1 normalised to L19 mRNA expression. Columns are the mean of three independent experiments in triplicate; bars, ± SD. Statistical significance was determined using Student’s t tests and compared to the correspondent time point in the control pcDNA3 transfected cells. *, p ≤ 0.05; **, p ≤ 0.001; ***, p ≤ 0.0001, significant; ns, non-significant. B. MCF-7 cells were transiently transfected with 20 ng of the pGL3-BRIP1 promoter, C. pGL3-BRIP1 (0.4kb) WT or pGL3-BRIP1 (0.4kb) mut together with increasing amounts (0, 10, 20, 30 and 40 ng) of ΔN -FOXM1 expression vector. Cells were harvested after 24 h of transfection and assayed for luciferase activity. All relative luciferase activity values are corrected for cotransfected Renilla activity. Each column represents the mean ± SD from 3 independent experiments. D. MCF-7 and MCF-7 EpiR cells untreated or treated with 1 μM of epirubicin for 16 h were used for ChIP assays by using IgG negative control and anti-FOXM1 antibody, as indicated. After reversal of cross-linking, the coimmunoprecipitated DNA was amplified by PCR, using primers amplifying the FOXM1 FHRE-binding site containing region (−390/−277) and a control region (−605/−512), and resolved in 2% agarose gel. Inverted ethidium bromide stained images are shown.
FOXM1 is required for homologous recombination repair
We next analysed a possible role for FOXM1 in DSB repair using HeLa cell lines habouring an integrated direct repeat green fluorescent protein (DR-GFP) reporter for homologous recombination (HR) or non-homologous end-joining (NHEJ) (33, 34). The I-SceI expression plasmid was transfected into DR-GFP HeLa cells with the non-specific (NS), FOXM1, or BRCA1 siRNA and the percentage of GFP-positive cells measured by flow cytometric analysis (Supplementary Figure S2). The knockdown of BRCA1 significantly decreased the percentage of GFP-positive cells in comparison with non-specific (NS) control siRNA in both DR-GFP reporter systems for HR (34.2%) and NHEJ (31.6%) (Figure 5B). Likewise, FOXM1 depletion using siRNA reduced the HR DSB repair (38.9%) by 61.1%, but had no significant effects on NHEJ repair (Figure 5B). These observations suggest that similar to BRCA1, FOXM1 contributes to HR-directed DSB repair. Consistent with the fact that homologous recombination (HR)-deficient cells are more sensitive to blockade of the base excision repair pathway through inhibition of the poly (ADP-ribose) polymerase (PARP) activity (35), the Foxm1−/− MEFs were more sensitive to PARP inhibitors, olaparib and veliparib, compared to WT MEFs (Supplementary Figure S3), further suggesting that FOXM1 has a crucial role in HR.
FOXM1 modulates the expression of BRIP1 at protein and mRNA levels
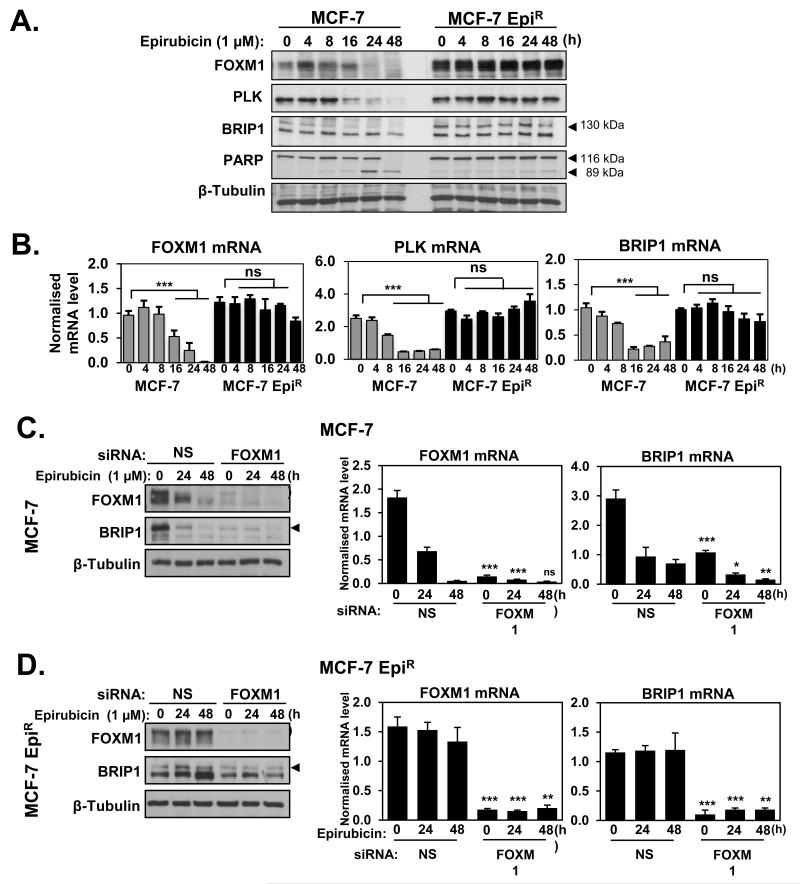
The BRIP1 protein interacts with BRCA1 and is required for the HR DSB repair activity of BRCA1 (29, 31). To explore the possibility that FOXM1 regulates BRIP1 expression, we investigated the expression patterns of FOXM1 and BRIP1 in MCF-7 and MCF-7 EpiR cells in response to epirubicin treatment. Western blot analyses showed that the expression levels of BRIP1 protein decreased in response to epirubicin in MCF-7 cells, but remained at high levels in the resistant MCF-7 EpiR cells, displaying similar kinetics as FOXM1 and its target PLK upon epirubicin treatment (Figure 6A). Cleaved PARP was also included as a marker for apoptosis. qRT-PCR analyses revealed that the BRIP1 mRNA also followed similar kinetics as the FOXM1 and PLK mRNA, suggesting FOXM1 may regulate BRIP1 expression (Figure 6B). To test this notion, both MCF-7 and MCF-7 EpiR cells were transfected with non-targeting control (NS) or FOXM1 siRNA pools for 48 h and then treated with epirubicin for 0, 24 and 48 h (Figure 6C and D). Silencing the expression of FOXM1 caused a reduction in both BRIP1 protein and mRNA levels before and after epirubicin treatment in MCF-7 and MCF-7 EpiR cells. These data clearly suggest that FOXM1 regulates BRIP1 expression at the transcription and translation levels in MCF-7 and MCF-7 EpiR breast cancer cell lines. Consistent with this idea, Foxm1−/− MEFs also express lower levels of BRIP1 compared with WT MEFs (Supplementary Figure S4).
Figure 6. FOXM1 is upregulated in epirubicin resistant cell line (MCF-7 EpiR) and modulates the expression of BRIP1 at protein and mRNA levels.
A. MCF-7 and MCF-7 EpiR cells were treated with 1 μM of epirubicin for 0, 4, 8, 16, 24 and 48 h. Cells were collected at indicated times and analysed by western blot to determine the protein expression levels of FOXM1, PLK, BRIP1, PARP and β-Tubulin (arrows indicate the specific protein band) B. and by qRT-PCR to determine FOXM1, PLK and BRIP1 mRNA transcript levels normalised to L19 mRNA expression. Columns are the mean of three independent experiments in triplicate; bars, ± SD. Statistical analyses were performed using Student’s t tests against the 0 h time point. ***, p ≤ 0.0001, significant; ns, non-significant. C. MCF-7 and D. MCF-7 EpiR cells were transfected with nonspecific (NS) siRNA or siRNA smart pool against FOXM1. Twenty-four hours after transfection, cells were treated with 1 μM of epirubicin and harvested for western blot (left panel) and qRT-PCR (right panel) analyses at 0, 24 and 48 h. Protein expression levels were determined for FOXM1, BRIP1 and β-Tubulin and mRNA levels for FOXM1 and BRIP1. Statistical analyses were performed using Student’s t tests and compared to the correspondent time point in the control NS siRNA transfected cells. Columns are the mean of three independent experiments in triplicate; bars, ± SD. *, p ≤ 0.05; **, p ≤ 0.001; ***, p ≤ 0.0001, significant; ns, non-significant.
FOXM1 activates BRIP1 through a forkhead responsive element in its promoter
We next overexpressed the wild-type (WT) FOXM1 and a constitutively active ΔN-FOXM1 (15), which lacks the FOXM1 N-terminal repressor domain, in the MCF-7 cells. The result showed that overexpression of WT or ΔN-FOXM1 caused an induction of BRIP1 mRNA and protein expression levels in MCF-7 cells (Figure 7A). It is notable that BRIP1 levels decrease regardless of FOXM1 or deltaNFOXM1 overexpression after 48h of epirubicin treatment within MCF7 cells. This is likely to be due to the fact that both FOXM1 and BRIP1 are also regulated at post-transcriptional levels in response to epirubicin as reported recently (36) and as revealed by their differential mRNA and protein expression kinetics in response to epirubcin (Figure 7A). To investigate if the regulation of BRIP1 by FOXM1 is at the promoter level, MCF-7 cells were transiently co-transfected with the constitutively active FOXM1 expression construct ΔN-FOXM1 and a luciferase reporter gene under the control of a 2.1kb human BRIP1 gene region upstream of the putative transcriptional start site (Figure 7B). We observed a dose-dependent increase in the BRIP1 (2.1kb)-luciferase activity (1.7-4.6×) with the increasing amounts of ΔN-FOXM1 expression vector transfected, indicating the putative BRIP1 promoter is responsive to FOXM1 induction. We then examined the ability of FOXM1 to transactivate a 5′-truncated wild-type (WT) BRIP1 (0.4kb) promoter as well as a mutant (mut) BRIP1 (0.4kb) promoter with a putative FHRE (forkhead response element) (−337bp) mutated. The results showed that while the (WT) BRIP1 (0.4kb) promoter increased with titrated amounts of ΔN-FOXM1 (1.5-10.1×), the mutant (mut) BRIP1 (0.4kb) promoter was not inducible by ΔN-FOXM1 and had little promoter activity (Figure 7C). Collectively, these results suggest that FOXM1 is able to transactivate BRIP1 promoter via the FHRE located at position −337 bp, further confirming that BRIP1 is a target gene of FOXM1. To confirm further that FOXM1 binds to the FHRE of the endogenous BRIP1 promoter, we studied the in vivo occupancy of FHRE region of the BRIP1 promoter by FOXM1 using Chromatin-Immunoprecipitation (ChIP) before and after 16 h epirubicin treatment in both the MCF-7 and MCF-7 EpiR cells. The ChIP analysis showed that FOXM1 is recruited to the endogenous FHRE in both the MCF-7 and MCF-7 EpiR and its binding to the FHRE increases in response to epirubicin (Figure 7D).
BRIP1 is the FOXM1 target involved in epirubicin-induced DSB repair and resistance
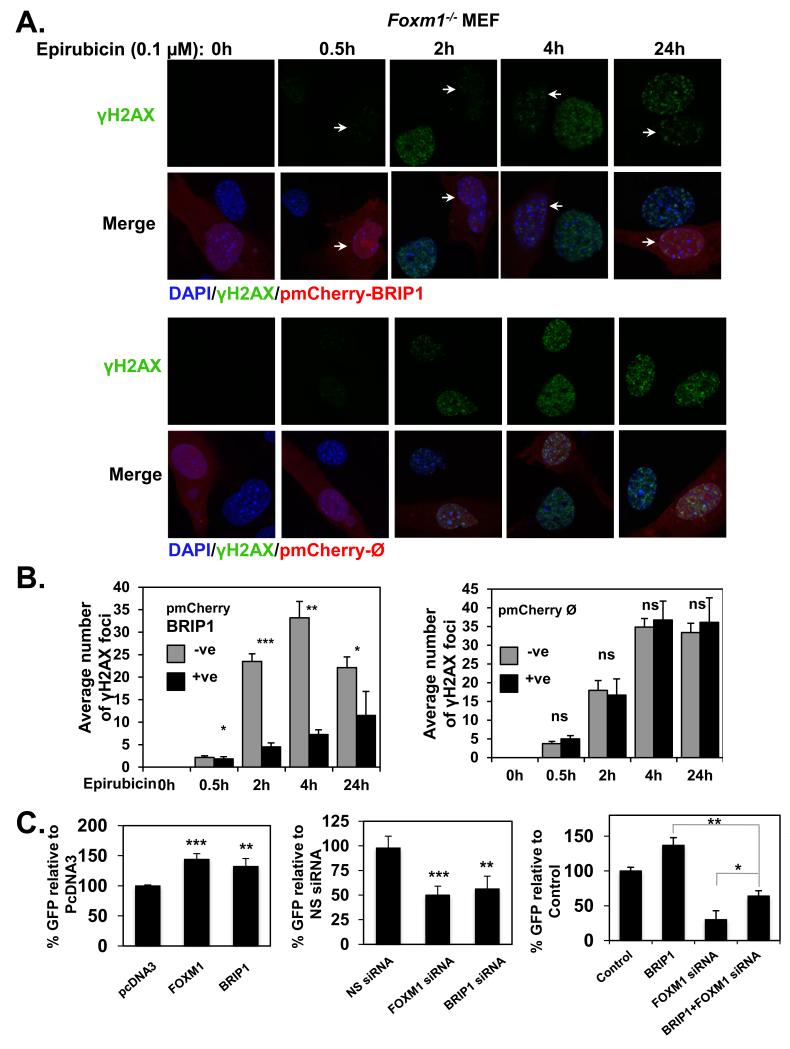
After establishing that BRIP1 is a direct transcriptional target of FOXM1, we next tested if BRIP1 could account for the DNA repair and epirubicin resistant function of FOXM1. To this end, we examined if overexpression of BRIP1 can override the defects in DSB repair, as reflected by the abnormal accumulation of γH2AX foci in Foxm1−/− MEFs. Interestingly, we found that similar to FOXM1 (Figure 5A), overexpression of BRIP1 in Foxm1-deficient cells was able to significantly reduce γH2AX foci accumulation, when compared to those neighbouring non-transfected cells and those transfected with the empty pmCherry vector (Figure 8A and 8B), confirming that BRIP1 has a central role in DSB response (26, 27) following epirubicin treatment. Consistently, the siRNA-mediated knockdown of BRIP1 was able to significantly increase γH2AX foci formation in comparison with cells transfected with the non-specific siRNA (Supplementary Figure S5A). Moreover, SRB cell viability assays revealed that knockdown of FOXM1 or BRIP1 (Supplementary Figure S5B), was able to resensitise MCF-7 EpiR cells to epirubicin treatment, further showing that FOXM1 and BRIP1 function together in response to epirubicin. To address further the role of BRIP1 on the repair of DSBs by HR, we used the HeLa cell lines integrating the DR-GFP-HR expression vector. We observed that overexpression of FOXM1 or BRIP1 increases the DSB repair via HR (Figure 8C, left panel; Supplementary Figure S6 and S7). In contrast, depletion of FOXM1 and BRIP1 by siRNA, significantly reduced the ability of cells to repair DNA via HR (Figure 8C, middle panel; Supplementary Figure S6 and S7). Importantly, BRIP1 overexpression in the presence of FOXM1 silencing enhanced HR repair over FOXM1 knockdown, but was not able to restore HR repair to levels similar to BRIP1 overexpression alone (Figure 8C, right panel; Supplementary Figure S8). Western blot analysis showed that the low HR repair activity is partly caused by the downregulation of endogenous BRIP1 through FOXM1 knockdown. Collectively, this result indicates that FOXM1 mediates HR repair in part through the regulation of BRIP1 expression, but other FOXM1 target genes are also likely to be involved in HR repair and signalling. Taken together, our present findings show that FOXM1 is a very important activator of DNA repair genes involved in the HR, such as BRIP1, which renders cells more resistant to cytotoxic drugs that cause DSBs.
Figure 8. BRIP1 decreases γH2AX foci accumulation in Foxm1−/− MEFs and is involved in homologous recombination repair with FOXM1.
Foxm1−/− MEFs transfected with the pmCherry-BRIP1 or pmCherry-ᴓ (red) and treated with 0.1 μM of epirubicin for 0, 0.5, 2, 4 and 24 h were stained for γH2AX followed by addition of Alexa488 (green)-labelled anti-rabbit. Nuclei were counterstained with DAPI (blue). Images were acquired with Leica TCS SP5. B. γH2AX foci quantification is shown. Bars represent average of three independent experiments ± SEM. *, p ≤ 0.05; **, p ≤ 0.001; ***, p ≤ 0.0001, significant; ns, non-significant. −ve, negative pmCherry cells; +ve, positive pmCherry cells. C. Homologous recombination (HR) was assayed in HeLa cells using DR-GFP reporter system bearing an integrated gene conversion substrate. Cells were either transfected with control pcDNA3, pcDNA3-FOXM1 or pcDNA3-myc-his-BRIP1 (left panel) or with nonspecific (NS) siRNA, FOXM1 siRNA or BRIP1 siRNA (middle panel), or control, pcDNA3-myc-his-BRIP1, FOXM1 siRNA, or pcDNA3-myc-his-BRIP1 plus FOXM1 siRNA (right panel). Forty-eight hours after transfection, cells were transfected again with I-SceI and left for 72h before analysis. GFP-positive cells are indicated for each transfection, with standard deviation based on three independent experiments (a total of 50,000 events were analysed for each experiment, and experiments were performed in duplicate). Bars represent average of three independent experiments ± SD. Significance was determined using Student’s t tests and compared to the controls pcDNA3 or NS siRNA transfected cells, accordingly. *, p ≤ 0.05; **, p ≤ 0.001; ***, p ≤ 0.0001, significant.
Discussion
Epirubicin is widely used in the clinical management of solid cancers, yet the development of acquired resistance during the course of treatment constitutes a major limitation to the clinical use of this drug (37). It has been shown that anthracycline-based chemotherapeutics induce DNA double-strand breaks (DSBs) (5). DSBs are among the most cytotoxic of DNA lesions that exist within the cell and generally their repair is carried out by two distinct and complementary pathways: HR and NHEJ. Accumulating evidence reveals FOXM1 as a cellular mediator of chemotherapeutic drug sensitivity and resistance in various cancer types. For example, in breast cancer, FOXM1 has been shown to be involved in the development of resistance to hormonal (20, 21), cisplatin (15), trastuzumab, paclitaxel (22), gefitinib (23) and epirubicin treatment (24). FOXM1 has been proposed to have a role in modulating genotoxic drug resistance but its exact role and mechanism of action remain elusive. Analyses of epirubicin sensitive and resistant MCF-7 cells revealed that MCF-7 has higher levels of γH2AX foci compared with the drug resistant MCF-7 EpiR after prolonged epirubicin treatment (4 and 24 h), suggesting MCF-7 cells accumulate damaged DNA in response to epirubicin treatment and that MCF-7 EpiR cells may be more effective in DSB repair. This notion is supported by the alkaline comet assay results showing that MCF-7 EpiR sustained lower levels of DNA damage compared with the parental MCF-7 cells upon epirubicin treatment. The fact that MCF-7 EpiR express higher levels of FOXM1 compared with the parental MCF-7 cells in response to epirubicin treatment suggests that FOXM1 has a role in DNA damage repair and thus in genotoxic drug resistance. Consistent with this, alkaline comet assays showed that Foxm1−/− MEFs sustained higher levels of DNA damage compared with WT MEFs following epirubicin treatment. Similarly, γH2AX staining also demonstrated that Foxm1−/− MEFs sustained higher levels of DSBs compared to WT MEFs upon epirubicin treatment and γ-irradiation. Furthermore, overexpression of FOXM1 can decrease the levels of DNA damage and epirubicin sensitivity in wild-type MCF-7 cells, as confirmed by comet and cell viability assays, whereas depletion of FOXM1 expression by siRNA can resensitise MCF-7 EpiR cells to epirubicin. Crucially, ectopic expression of a pmCherry-FOXM1 fusion in Foxm1−/− MEFs could suppress the accumulation of γH2AX foci, whilst the neighbouring bystander non-transfected cells as well as cells transfected with the empty pmCherry vector displayed a time-dependent accumulation of γH2AX foci following epirubicin. Lastly, in agreement with a recently published report which shows that FOXM1 is important for HR (38), DR-GFP DNA repair reporter assays indicate that FOXM1 is essential for HR but is dispensable for NHEJ. The finding that NHEJ activity is unaffected by FOXM1 depletion probably indicates that redundant DSB-repair signalling pathways exist; however, this does not preclude FOXM1 also having a role in regulating NHEJ. Notably, the lack of γH2AX foci accumulation in the resistant cells is not because of the inability of epirubicin to access the nuclear DNA or induce DNA damage, as depletion of FOXM1 in MCF-7 EpiR cells and WT MEFs can restore the epirubicin-induced DNA damage, as revealed by comet assays. It is unlikely that it is due to the regulation of drug efflux transporters by FOXM1, as the γH2AX foci formation rates are at comparable levels at the earlier time points in both the wild-type and Foxm1−/− MEFs upon epirubicin treatment and γ-irradiation. Consistent with this conclusion is the observation that high levels of γH2AX foci accumulated in Foxm1−/− and not WT MEFs upon γ-irradiation at the longer time point of 24 h. The observations that these defects in DSB repair are correlated with a reduction in cell viability in response to epirubicin led us to consider that FOXM1 modulates epirubicin resistance through enhancing DSB-repair. In agreement, we also found that FOXM1-deficient cells are more sensitive to other DSB-inducing anticancer agents, including γ-irradiation, as well as PARP inhibitors, whereas Fluorouracil (5-FU)-resistant breast cancer cells also express higher levels of FOXM1 (Supplementary Figure S9). Recently, BRIP1 has been found to have a role in replicative checkpoint control, and its knockdown impairs Chk1 phosphorylation in response to Hydroxyurea (HU) (39). However, despite the fact that Chk1 phosphorylation is compromised in response to epirubicin in FOXM1-deficient MEFs, Chk1 phosphorylation was not affected in FOXM1-null MEFs upon HU-treatment (Supplementary Figure S10). An explanation could be that BRIP1 and FOXM1 have distinct roles in different checkpoints and DNA damage repair mechanisms.
γH2AX foci formation on the DSB sites is one of the earliest events following the induction of DNA damage by genotoxic drugs (40). These γH2AX foci help to assemble DNA repair proteins, including BRCA1, RAD51 and 53BP1, at the sites of damaged DNA to initiate the HR repair process (41). In search for FOXM1 targets that function downstream of H2AX phosphorylation, we found that FOXM1 regulates BRIP1 at the protein and mRNA levels through an FHRE on its promoter in response to epirubicin treatment. BRIP1 is a key regulator of HR DNA repair, most prominently through its ability to directly interact with and activate BRCA1. Resembling FOXM1, BRIP1 overexpression decreases the accumulation of damaged DNA upon epirubicin treatment as assayed by γH2AX foci formation. Furthermore, like FOXM1, BRIP1 overexpression can increase the HR DNA repair activity, whereas its depletion by siRNA decreases HR activity. These results led us to conclude that BRIP1 is a crucial downstream target of FOXM1, which mediates the HR DNA repair activity of FOXM1. However, it is likely that other FOXM1 targets are also involved in mediating the DNA damage response. Indeed, a recent cDNA array has identified a number of FOXM1 targets which have a putative role in DNA damage repair, but their regulation by FOXM1 in response to genotoxic drugs and their involvement in genotoxic drug-Induced DNA damage repair are yet to be elucidated (38). Nevertheless, the ability of BRIP1 ectopic expression to repress the accumulation of damaged DNA-associated γH2AX in Foxm1−/− MEFs confirms that BRIP1 is a key FOXM1 target gene in DNA damage response. The fact that FOXM1 participates in genotoxic drug-induced DNA damage repair also suggests that targeting FOXM1 can potentially enhance the efficiency of genotoxic drug and overcome drug resistance (8, 42). In agreement, the natural compound thiostrepton has been shown to specifically target FOXM1 and repress its oncogenic activity. Furthermore, thiostrepton can synergise with genotoxic drugs to eliminate cancer cells. In fact, thiostrepton has been shown to reduce the transcriptional activity by binding directly to FOXM1 and inhibiting the association of FOXM1 with its genomic targets (43). In summary, this study identifies BRIP1 as a key FOXM1 target gene in DNA damage response and shows that FOXM1 recruitment to the BRIP1 promoter is associated with its transcriptional activation.
Methods and Materials
Cell Culture
The human breast carcinoma MCF-7 cell line was originated from the American Type Culture Collection and was acquired from the Cell Culture Service, Cancer Research UK, where it was tested and authenticated. Mouse embryonic fibroblasts (MEFs) isolated from Foxm1−/− and WT mouse have been previously described (44). The HeLa cells containing DR-GFP reporter system were stabilised as described (45) and were a gift from Dr. Maria Jasin (Memorial Sloan-Kettering Cancer Center, New York USA). All cells were cultured in DMEM supplemented with 10% FCS, 2 mM glutamine, 100units/ml penicillin/streptomycin and maintained at 37 °C in a humidified incubator with 10% CO2. The MCF-7 EpiR was maintained in 10μmol/L Epirubicin (Medac, Hamburg, Germany), as previously described (20). The MCF-7 FOXM1 cell line was established from MCF-7 cells transfection with the pcDNA-FOXM1 plasmid. Hydroxyurea (HU; H8627-5G) and Fluorouracil (5FU; F6627-10G) were both purchased from Sigma, Poole, UK and were dissolved in water and DMSO, respectively. The PARP inhibitors veliparib (ABT-888; Enzo Life Sciences, Exeter, UK) olaparib (AZD2281; Selleckchem, Houston, TX, USA) were solublised in DMSO.
Plasmids and promoter assays
For the generation of human BRIP1 promoter constructs: 2.1 kb BRIP1 promoter was cloned into the Xho I and Bgl lI sites of the pGL3-Basic vector (Promega Madison, WI, USA) using the following oligonucleotides: sense: 5′-TAAATGGCAATGCAAGGTGA-3′ and anti-sense: 5′-GCTCTGAGCTCCGATTCACT-3′; to generate the 0.4 kb WT and mut BRIP1 promoter fragments the sense: 5′-TACTTTAAACAAACACTAGGGATTTGCTGG-3′ and 5′-TACTGTAGGCAAGCACTAGGGATTTGCTGG-3′ primers were used, respectively with the anti-sense: 5′-GCTCTGAGCTCCGATTCACT-3′ and cloned into the Xho I and Bgl lI sites of the pGL3-Basic vector. The pcDNA3-FOXM1 and the constitutively active form of FOXM1, pcDNA3-ΔN-FOXM1, have been described previously (15) and the pmCherry-FOXM1 was generated by cloning the full-length FOXM1 cDNA from pcDNA3-FOXM1 into the EcoRI and BamHI sites of the pmCherry-N1 vector (Clontech). The pcDNA3-myc-his-BACH1 WT was acquired from addgene (Cambridge, MA, USA) and the pCMV-I-SceI has been described (33, 46).
Comet assay
The comet assay was performed under alkaline conditions (pH > 13) by a modified method by (47). Also see supplementary Materials and Methods.
γH2AX immunofluorescent staining and foci quantification
Cells grown on chamber culture slides were fixed in 4% paraformaldehyde (Thermo Scientific, Rockford, IL, USA) for 15 min followed by permeabilisation for 10 min with 0.2% Triton X-100 in PBS, and blocking with 5% goat serum for 30 min at RT. The slides were incubated with primary antibody anti-γH2AX Ser139 (20E3) (Cell Signaling, Danvers, MA, USA) overnight at 4°C. Following washes with PBS, the secondary antibody Alexa Fluor 488 (Invitrogen, Eugene, Oregon, USA) was added for 45 min at RT. Cells were then counterstained with DAPI for 10 min before mounting with Vectashield mounting medium (Vector Laboratories). Also see supplementary Materials and Methods.
Western blotting
Western blotting was performed on whole cell extracts by lysing cells in buffer as described (23). Antibodies FOXM1 (C-20), PLK (F-8), β-Tubulin (H-235) and I-SceI were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The BRIP1 antibody was purchased from Abcam (Cambridge, UK) and PARP from Cell Signaling (Danvers, USA). The mouse monoclonal c-myc (9E10) antibody was acquired from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Primary antibodies were detected using horseradish peroxidase–linked anti-mouse or anti-rabbit conjugates as appropriate (Dako, Glostrup, Denmark) and visualised using the ECL detection system (Amersham Biosciences, Pollards Wood, UK).
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted with the RNeasy Mini Kit (Qiagen, Hilden, Germany). Complementary DNA generated by Superscript III reverse transcriptase and oligo-dT primers (Invitrogen, USA) was analysed by quantitative real-time PCR (qRT-PCR) as described (48). Transcript levels were quantified using the standard curve method. L19, a non-regulated ribosomal housekeeping gene was used as an internal control to normalise input cDNA. The following gene-specific primers were used for human: L19-sense: 5′-GCGGAAGGGTACAGCCAAT-3′ and L19-antisense: 5′-GCAGCCGGCGCAAA-3′; FOXM1-sense: 5′-TCCTCCACCCCGAGCAA-3′ and FOXM1-antisense: 5′-CGTGAGCCTCCAGGATTCAG-3′; BRIP1-sense: 5′-GATTGATGCCACCCTTACTAGAAAA-3′ and BRIP1-antisense: 5′-ATCCAGGGCTTCTTCAGAACAG-3′; PLK-sense: 5′-TGATGGCAGCCGTGACCTA-3′ and PLK-antisense: 5′-GGCGGTATGTGCGGAAGT-3′ and for mice: L19-sense: 5′-CCCGTCAGCAGATCAGGAA-3′ and L19-antisense: 5′-GTCACAGGCTTGCGGATGA-3′; FOXM1-sense: 5′-AGAAATGTGACCATCAAAACTGAAAT-3′ and FOXM1-antisense: 5′-GAGGGAGCAGAGGCTTCATCTT-3′; BRIP1-sense: 5′-TCACAGGTTTGGGTTGGTACTG-3′ and BRIP1-antisense: 5′-TGAAAGGTAGCGCAGAGATTCC-3′; PLK-sense: 5′-TGGGTGGACTATTCGGACAAG-3′ and PLK-antisense: 5′-ACCCCCACACTGTTGTCACA-3′.
Gene Silencing with Small Interfering RNAs (siRNAs)
For gene silencing, cells were transiently transfected with siRNA SMARTpool reagents purchased from Thermo Scientific Dharmacon (Lafayette, CO, USA) using Oligofectamine (Invitrogen, USA) according to the manufacturer’s instructions. SMARTPool siRNAs used were: siRNA FOXM1 (L-009762-00), siRNA BRIP1 (L-010587-00) and the non-specific control siRNA, confirmed to have minimal targeting of known genes (D-001810-10-05).
Two-step cross-linking chromatin immunoprecipitation
Dual cross-linking chromatin immunoprecipitation (ChIP) using formaldehyde and di (N- succimidyl) glutarate was performed on MCF-7 cells, as described (49). Primers used to amplify BRIP1 genomic region 5′-3′ (−390/−277): AGACTCTATCGCCGGTTTCA and AAACAAGGGCTCAAGGTACG; CCGATGTCACAGAGCCTTCT and TACCCAGCTTTGCAGTAGCC (−605/−512). Also see supplementary Materials and Methods.
Sulforhodamine B assay and cell-cycle analysis
Sulforhodamine B (SRB) assays were performed and analysed as described (50).
Repair Assays
Homologous recombination and non-homologous end-joining cell assays were performed as previously described (33, 45, 46).
Supplementary Material
Acknowledgements
E.W-F. Lam; A.R. Gomes; R.C. Coombes were supported by grants from Cancer Research UK, E.W-F. Lam and C.-Y. Koo by grants from Breast Cancer Campaign and Imperial College NHS Turst, L.J. Monteiro grants from Fundação para a Ciência e a Tecnologia, and P. Khongkow and M. Kongsema from the Royal Thai Government Scholarships. We also thank Dr Keng Heng for help with the microscopy and flow cytometry work.
Footnotes
Conflict of interest
The authors declare that they have no conflicts of interest.
References
- 1.Lin SX, Chen J, Mazumdar M, Poirier D, Wang C, Azzi A, et al. Molecular therapy of breast cancer: progress and future directions. Nat Rev Endocrinol. 2010 Sep;6(9):485–93. doi: 10.1038/nrendo.2010.92. [DOI] [PubMed] [Google Scholar]
- 2.Alvarez RH, Valero V, Hortobagyi GN. Emerging targeted therapies for breast cancer. J Clin Oncol. [Research Support, Non-U.S. Gov’t Review] 2010 Jul 10;28(20):3366–79. doi: 10.1200/JCO.2009.25.4011. [DOI] [PubMed] [Google Scholar]
- 3.Zelnak A. Overcoming taxane and anthracycline resistance. Breast J. 2010 May-Jun;16(3):309–12. doi: 10.1111/j.1524-4741.2010.00911.x. [DOI] [PubMed] [Google Scholar]
- 4.Stearns V, Davidson NE, Flockhart DA. Pharmacogenetics in the treatment of breast cancer. Pharmacogenomics J. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S. Review] 2004;4(3):143–53. doi: 10.1038/sj.tpj.6500242. [DOI] [PubMed] [Google Scholar]
- 5.Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev. [Research Support, Non-U.S. Gov’t Review] 2004 Jun;56(2):185–229. doi: 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- 6.Tewey KM, Rowe TC, Yang L, Halligan BD, Liu LF. Adriamycin-induced DNA damage mediated by mammalian DNA topoisomerase II. Science. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.] 1984 Oct 26;226(4673):466–8. doi: 10.1126/science.6093249. [DOI] [PubMed] [Google Scholar]
- 7.Raguz S, Yague E. Resistance to chemotherapy: new treatments and novel insights into an old problem. Br J Cancer. [Research Support, Non-U.S. Gov’t Review] 2008 Aug 5;99(3):387–91. doi: 10.1038/sj.bjc.6604510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Myatt SS, Lam EW. The emerging roles of forkhead box (Fox) proteins in cancer. Nat Rev Cancer. [Research Support, Non-U.S. Gov’t Review] 2007 Nov;7(11):847–59. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 9.Korver W, Roose J, Heinen K, Weghuis DO, de Bruijn D, van Kessel AG, et al. The human TRIDENT/HFH-11/FKHL16 gene: structure, localization, and promoter characterization. Genomics. [Research Support, Non-U.S. Gov’t] 1997 Dec 15;46(3):435–42. doi: 10.1006/geno.1997.5065. [DOI] [PubMed] [Google Scholar]
- 10.Yao KM, Sha M, Lu Z, Wong GG. Molecular analysis of a novel winged helix protein, WIN. Expression pattern, DNA binding property, and alternative splicing within the DNA binding domain. J Biol Chem. 1997 Aug 8;272(32):19827–36. doi: 10.1074/jbc.272.32.19827. [DOI] [PubMed] [Google Scholar]
- 11.Fu Z, Malureanu L, Huang J, Wang W, Li H, van Deursen JM, et al. Plk1-dependent phosphorylation of FoxM1 regulates a transcriptional programme required for mitotic progression. Nat Cell Biol. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, Non-P.H.S.] 2008 Sep;10(9):1076–82. doi: 10.1038/ncb1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park HJ, Costa RH, Lau LF, Tyner AL, Raychaudhuri P. Anaphase-promoting complex/cyclosome-CDH1-mediated proteolysis of the forkhead box M1 transcription factor is critical for regulated entry into S phase. Mol Cell Biol. [Research Support, N.I.H., Extramural] 2008 Sep;28(17):5162–71. doi: 10.1128/MCB.00387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karadedou CT, Gomes AR, Chen J, Petkovic M, Ho KK, Zwolinska AK, et al. FOXO3a represses VEGF expression through FOXM1-dependent and -independent mechanisms in breast cancer. Oncogene. 2011 Aug 22; doi: 10.1038/onc.2011.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Q, Zhang N, Jia Z, Le X, Dai B, Wei D, et al. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] 2009 Apr 15;69(8):3501–9. doi: 10.1158/0008-5472.CAN-08-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwok JM, Peck B, Monteiro LJ, Schwenen HD, Millour J, Coombes RC, et al. FOXM1 confers acquired cisplatin resistance in breast cancer cells. Mol Cancer Res. [Research Support, Non-U.S. Gov’t] 2010 Jan;8(1):24–34. doi: 10.1158/1541-7786.MCR-09-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ahmad A, Wang Z, Kong D, Ali S, Li Y, Banerjee S, et al. FoxM1 down-regulation leads to inhibition of proliferation, migration and invasion of breast cancer cells through the modulation of extra-cellular matrix degrading factors. Breast Cancer Res Treat. 2010 Jul;122(2):337–46. doi: 10.1007/s10549-009-0572-1. [DOI] [PubMed] [Google Scholar]
- 17.Dai B, Kang SH, Gong W, Liu M, Aldape KD, Sawaya R, et al. Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] 2007 Sep 13;26(42):6212–9. doi: 10.1038/sj.onc.1210443. [DOI] [PubMed] [Google Scholar]
- 18.Koo CY, Muir KW, Lam EW. FOXM1: From cancer initiation to progression and treatment. Biochim Biophys Acta. 2012 Jan;1819(1):28–37. doi: 10.1016/j.bbagrm.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Ackermann AM, Gusarova GA, Lowe D, Feng X, Kopsombut UG, et al. The FoxM1 transcription factor is required to maintain pancreatic beta-cell mass. Mol Endocrinol. 2006 Aug;20(8):1853–66. doi: 10.1210/me.2006-0056. [DOI] [PubMed] [Google Scholar]
- 20.Millour J, Constantinidou D, Stavropoulou AV, Wilson MS, Myatt SS, Kwok JM, et al. FOXM1 is a transcriptional target of ERalpha and has a critical role in breast cancer endocrine sensitivity and resistance. Oncogene. [Research Support, Non-U.S. Gov’t] 2010 May 20;29(20):2983–95. doi: 10.1038/onc.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madureira PA, Varshochi R, Constantinidou D, Francis RE, Coombes RC, Yao KM, et al. The Forkhead box M1 protein regulates the transcription of the estrogen receptor alpha in breast cancer cells. J Biol Chem. [Research Support, Non-U.S. Gov’t] 2006 Sep 1;281(35):25167–76. doi: 10.1074/jbc.M603906200. [DOI] [PubMed] [Google Scholar]
- 22.Carr JR, Park HJ, Wang Z, Kiefer MM, Raychaudhuri P. FoxM1 mediates resistance to herceptin and paclitaxel. Cancer Res. [Research Support, N.I.H., Extramural] 2010 Jun 15;70(12):5054–63. doi: 10.1158/0008-5472.CAN-10-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGovern UB, Francis RE, Peck B, Guest SK, Wang J, Myatt SS, et al. Gefitinib (Iressa) represses FOXM1 expression via FOXO3a in breast cancer. Mol Cancer Ther. [Research Support, Non-U.S. Gov’t] 2009 Mar;8(3):582–91. doi: 10.1158/1535-7163.MCT-08-0805. [DOI] [PubMed] [Google Scholar]
- 24.Millour J, de Olano N, Horimoto Y, Monteiro LJ, Langer JK, Aligue R, et al. ATM and p53 regulate FOXM1 expression via E2F in breast cancer epirubicin treatment and resistance. Mol Cancer Ther. [Research Support, Non-U.S. Gov’t] 2011 Jun;10(6):1046–58. doi: 10.1158/1535-7163.MCT-11-0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan Y, Raychaudhuri P, Costa RH. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol. [Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t] 2007 Feb;27(3):1007–16. doi: 10.1128/MCB.01068-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cantor S. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proceedings of the National Academy of Sciences. 2004;101(8):2357–62. doi: 10.1073/pnas.0308717101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, P.H.S.] 2001 Apr 6;105(1):149–60. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 28.Turnbull C, Rahman N. Genetic predisposition to breast cancer: past, present, and future. Annu Rev Genomics Hum Genet. [Review] 2008;9:321–45. doi: 10.1146/annurev.genom.9.081307.164339. [DOI] [PubMed] [Google Scholar]
- 29.Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, et al. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. [Research Support, Non-U.S. Gov’t] 2005 Sep;8(3):255–65. doi: 10.1016/j.ccr.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Xie J, Litman R, Wang S, Peng M, Guillemette S, Rooney T, et al. Targeting the FANCJ-BRCA1 interaction promotes a switch from recombination to poleta-dependent bypass. Oncogene. [Research Support, N.I.H., Extramural] 2010 Apr 29;29(17):2499–508. doi: 10.1038/onc.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dohrn L, Salles D, Siehler SY, Kaufmann J, Wiesmuller L. BRCA1-mediated repression of mutagenic end-joining of DNA double-strand breaks requires complex formation with BACH1. Biochem J. [Research Support, Non-U.S. Gov’t] 2012 Feb 1;441(3):919–26. doi: 10.1042/BJ20110314. [DOI] [PubMed] [Google Scholar]
- 32.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. [Research Support, Non-U.S. Gov’t Research Support, U.S. Gov’t, Non-P.H.S. Research Support, U.S. Gov’t, P.H.S.] 2003 Oct 24;302(5645):639–42. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 33.Delacote F, Han M, Stamato TD, Jasin M, Lopez BS. An xrcc4 defect or Wortmannin stimulates homologous recombination specifically induced by double-strand breaks in mammalian cells. Nucleic Acids Res. 2002 Aug 1;30(15):3454–63. doi: 10.1093/nar/gkf452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol. 2007 Aug;9(8):978–81. doi: 10.1038/ncb1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kruse V, Rottey S, De Backer O, Van Belle S, Cocquyt V, Denys H. PARP inhibitors in oncology: a new synthetic lethal approach to cancer therapy. Acta Clin Belg. 2011 Jan-Feb;66(1):2–9. doi: 10.2143/ACB.66.1.2062507. [DOI] [PubMed] [Google Scholar]
- 36.de Olano N, Koo CY, Monteiro LJ, Pinto PH, Gomes AR, Aligue R, et al. The p38 MAPK-MK2 Axis Regulates E2F1 and FOXM1 Expression after Epirubicin Treatment. Mol Cancer Res. 2012 Aug 20; doi: 10.1158/1541-7786.MCR-11-0559. doi:10.1158/541-7786.MCR-11-0559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palmieri C, Krell J, James CR, Harper-Wynne C, Misra V, Cleator S, et al. Rechallenging with anthracyclines and taxanes in metastatic breast cancer. Nat Rev Clin Oncol. [Review] 2010 Oct;7(10):561–74. doi: 10.1038/nrclinonc.2010.122. [DOI] [PubMed] [Google Scholar]
- 38.Park YY, Jung SY, Jennings N, Rodriguez-Aguayo C, Peng G, Lee SR, et al. FOXM1 mediates Dox resistance in breast cancer by enhancing DNA repair. Carcinogenesis. 2012 May 10; doi: 10.1093/carcin/bgs167. doi:10.1093/carcin/bgs167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gong Z, Kim JE, Leung CC, Glover JN, Chen J. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol Cell. 2010 Feb 12;37(3):438–46. doi: 10.1016/j.molcel.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998 Mar 6;273(10):5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 41.Garinis GA, Mitchell JR, Moorhouse MJ, Hanada K, de Waard H, Vandeputte D, et al. Transcriptome analysis reveals cyclobutane pyrimidine dimers as a major source of UV-induced DNA breaks. EMBO J. 2005 Nov 16;24(22):3952–62. doi: 10.1038/sj.emboj.7600849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Myatt SS, Lam EW. Targeting FOXM1. Nat Rev Cancer. 2008 Mar;8(3):242. doi: 10.1038/nrc2223-c2. [DOI] [PubMed] [Google Scholar]
- 43.Hegde NS, Sanders DA, Rodriguez R, Balasubramanian S. The transcription factor FOXM1 is a cellular target of the natural product thiostrepton. Nat Chem. 2011 Sep;3(9):725–31. doi: 10.1038/nchem.1114. [DOI] [PubMed] [Google Scholar]
- 44.Laoukili J, Kooistra MR, Bras A, Kauw J, Kerkhoven RM, Morrison A, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. [Research Support, Non-U.S. Gov’t] 2005 Feb;7(2):126–36. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 45.Bennardo N, Cheng A, Huang N, Stark JM. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. [Comparative Study Research Support, N.I.H., Extramural] 2008 Jun;4(6):e1000110. doi: 10.1371/journal.pgen.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weinstock DM, Nakanishi K, Helgadottir HR, Jasin M. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006;409:524–40. doi: 10.1016/S0076-6879(05)09031-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988 Mar;175(1):184–91. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 48.Kwok JM, Myatt SS, Marson CM, Coombes RC, Constantinidou D, Lam EW. Thiostrepton selectively targets breast cancer cells through inhibition of forkhead box M1 expression. Mol Cancer Ther. 2008 Jul;7(7):2022–32. doi: 10.1158/1535-7163.MCT-08-0188. [DOI] [PubMed] [Google Scholar]
- 49.Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques. 2005 Nov;39(5):715–25. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- 50.Peck B, Chen CY, Ho KK, Di Fruscia P, Myatt SS, Coombes RC, et al. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol Cancer Ther. [Research Support, Non-U.S. Gov’t] 2010 Apr;9(4):844–55. doi: 10.1158/1535-7163.MCT-09-0971. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.