Abstract
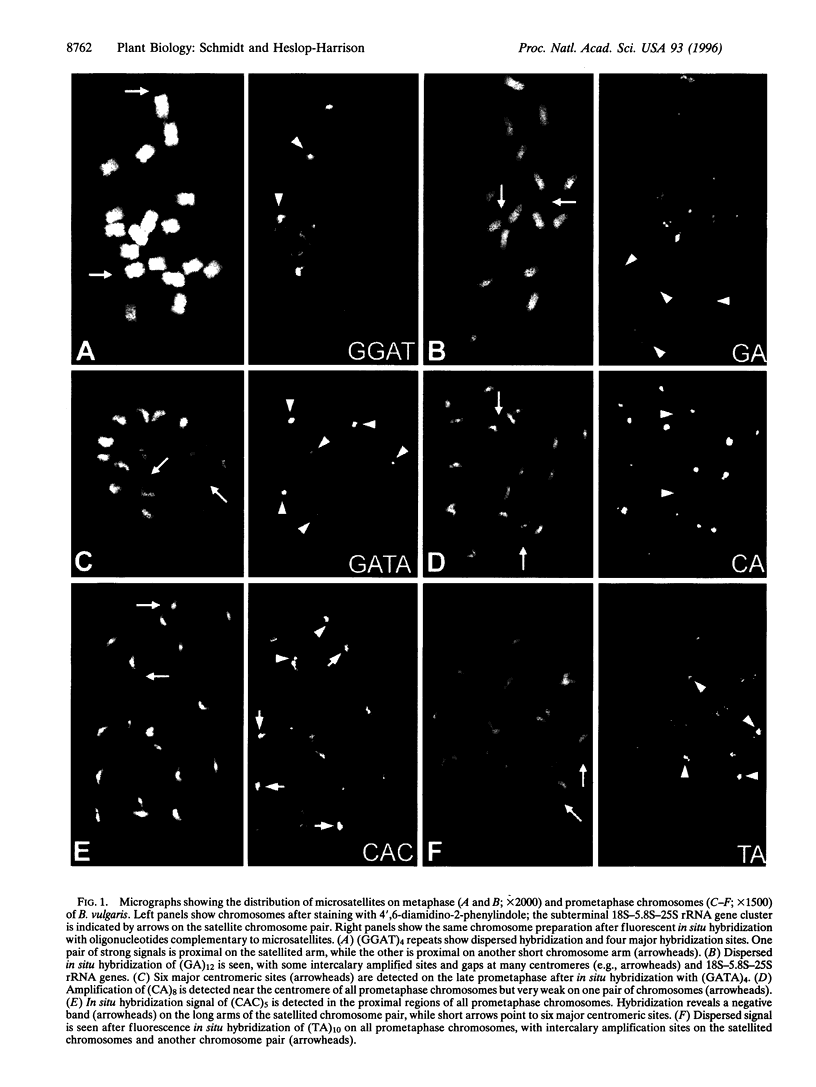
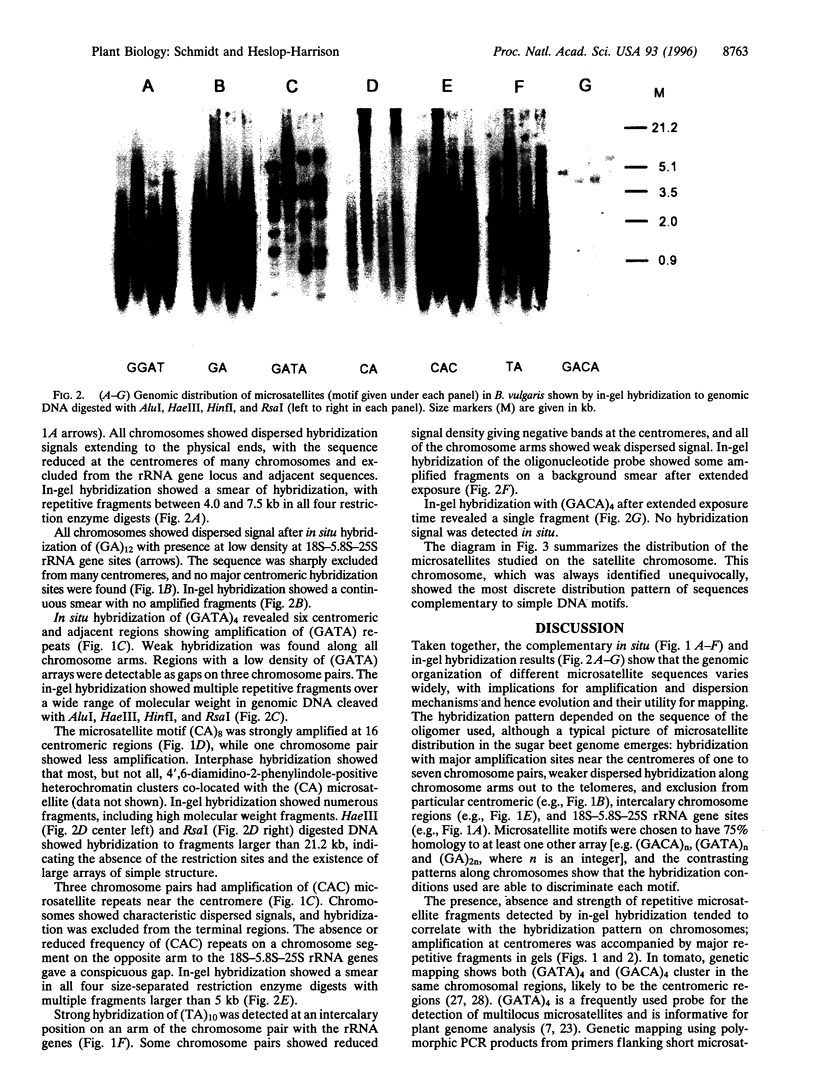
Microsatellites, tandem arrays of short (2-5 bp) nucleotide motifs, are present in high numbers in most eukaryotic genomes. We have characterized the physical distribution of microsatellites on chromosomes of sugar beet (Beta vulgaris L.). Each microsatellite sequence shows a characteristic genomic distribution and motif-dependent dispersion, with site-specific amplification on one to seven pairs of centromeres or intercalary chromosomal regions and weaker, dispersed hybridization along chromosomes. Exclusion of some microsatellites from 18S-5.8S-25S rRNA gene sites, centromeres, and intercalary sites was observed. In-gel and in situ hybridization patterns are correlated, with highly repeated restriction fragments indicating major centromeric sites of microsatellite arrays. The results have implications for genome evolution and the suitability of particular microsatellite markers for genetic mapping and genome analysis.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Akkaya M. S., Bhagwat A. A., Cregan P. B. Length polymorphisms of simple sequence repeat DNA in soybean. Genetics. 1992 Dec;132(4):1131–1139. doi: 10.1093/genetics/132.4.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali S., Müller C. R., Epplen J. T. DNA finger printing by oligonucleotide probes specific for simple repeats. Hum Genet. 1986 Nov;74(3):239–243. doi: 10.1007/BF00282541. [DOI] [PubMed] [Google Scholar]
- Arens P., Odinot P., van Heusden A. W., Lindhout P., Vosman B. GATA- and GACA-repeats are not evenly distributed throughout the tomato genome. Genome. 1995 Feb;38(1):84–90. doi: 10.1139/g95-010. [DOI] [PubMed] [Google Scholar]
- Becker J., Heun M. Barley microsatellites: allele variation and mapping. Plant Mol Biol. 1995 Feb;27(4):835–845. doi: 10.1007/BF00020238. [DOI] [PubMed] [Google Scholar]
- Bell C. J., Ecker J. R. Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics. 1994 Jan 1;19(1):137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- Broun P., Tanksley S. D. Characterization and genetic mapping of simple repeat sequences in the tomato genome. Mol Gen Genet. 1996 Jan 15;250(1):39–49. doi: 10.1007/BF02191823. [DOI] [PubMed] [Google Scholar]
- Charlesworth B., Sniegowski P., Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994 Sep 15;371(6494):215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- Dietrich W. F., Miller J., Steen R., Merchant M. A., Damron-Boles D., Husain Z., Dredge R., Daly M. J., Ingalls K. A., O'Connor T. J. A comprehensive genetic map of the mouse genome. Nature. 1996 Mar 14;380(6570):149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- Lagercrantz U., Ellegren H., Andersson L. The abundance of various polymorphic microsatellite motifs differs between plants and vertebrates. Nucleic Acids Res. 1993 Mar 11;21(5):1111–1115. doi: 10.1093/nar/21.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenhaupt K., Rich A., Pardue M. L. Nonrandom distribution of long mono- and dinucleotide repeats in Drosophila chromosomes: correlations with dosage compensation, heterochromatin, and recombination. Mol Cell Biol. 1989 Mar;9(3):1173–1182. doi: 10.1128/mcb.9.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore G., Devos K. M., Wang Z., Gale M. D. Cereal genome evolution. Grasses, line up and form a circle. Curr Biol. 1995 Jul 1;5(7):737–739. doi: 10.1016/s0960-9822(95)00148-5. [DOI] [PubMed] [Google Scholar]
- Morell V. The puzzle of the triple repeats. Science. 1993 Jun 4;260(5113):1422–1423. doi: 10.1126/science.8502986. [DOI] [PubMed] [Google Scholar]
- Nanda I., Zischler H., Epplen C., Guttenbach M., Schmid M. Chromosomal organization of simple repeated DNA sequences used for DNA fingerprinting. Electrophoresis. 1991 Feb-Mar;12(2-3):193–203. doi: 10.1002/elps.1150120216. [DOI] [PubMed] [Google Scholar]
- Nowak R. Mining treasures from 'junk DNA'. Science. 1994 Feb 4;263(5147):608–610. doi: 10.1126/science.7508142. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Involvement of transposition in dispersion of tandem repeat sequences (TrsA) in rice genomes. Mol Gen Genet. 1994 Nov 15;245(4):449–455. doi: 10.1007/BF00302257. [DOI] [PubMed] [Google Scholar]
- Paterson A. H., Lin Y. R., Li Z., Schertz K. F., Doebley J. F., Pinson S. R., Liu S. C., Stansel J. W., Irvine J. E. Convergent domestication of cereal crops by independent mutations at corresponding genetic Loci. Science. 1995 Sep 22;269(5231):1714–1718. doi: 10.1126/science.269.5231.1714. [DOI] [PubMed] [Google Scholar]
- Pedersen C., Linde-Laursen I. Chromosomal locations of four minor rDNA loci and a marker microsatellite sequence in barley. Chromosome Res. 1994 Jan;2(1):65–71. doi: 10.1007/BF01539456. [DOI] [PubMed] [Google Scholar]
- Rafalski J. A., Tingey S. V. Genetic diagnostics in plant breeding: RAPDs, microsatellites and machines. Trends Genet. 1993 Aug;9(8):275–280. doi: 10.1016/0168-9525(93)90013-8. [DOI] [PubMed] [Google Scholar]
- Röder M. S., Plaschke J., König S. U., Börner A., Sorrells M. E., Tanksley S. D., Ganal M. W. Abundance, variability and chromosomal location of microsatellites in wheat. Mol Gen Genet. 1995 Feb 6;246(3):327–333. doi: 10.1007/BF00288605. [DOI] [PubMed] [Google Scholar]
- Schlötterer C., Tautz D. Slippage synthesis of simple sequence DNA. Nucleic Acids Res. 1992 Jan 25;20(2):211–215. doi: 10.1093/nar/20.2.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt T., Heslop-Harrison J. S. Variability and evolution of highly repeated DNA sequences in the genus Beta. Genome. 1993 Dec;36(6):1074–1079. doi: 10.1139/g93-142. [DOI] [PubMed] [Google Scholar]
- Schmidt T., Kubis S., Heslop-Harrison J. S. Analysis and chromosomal localization of retrotransposons in sugar beet (Beta vulgaris L.): LINEs and Ty1-copia-like elements as major components of the genome. Chromosome Res. 1995 Sep;3(6):335–345. doi: 10.1007/BF00710014. [DOI] [PubMed] [Google Scholar]
- Schmidt T., Metzlaff M. Cloning and characterization of a Beta vulgaris satellite DNA family. Gene. 1991 May 30;101(2):247–250. doi: 10.1016/0378-1119(91)90418-b. [DOI] [PubMed] [Google Scholar]
- Tautz D., Renz M. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 1984 May 25;12(10):4127–4138. doi: 10.1093/nar/12.10.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., Trick M., Dover G. A. Cryptic simplicity in DNA is a major source of genetic variation. Nature. 1986 Aug 14;322(6080):652–656. doi: 10.1038/322652a0. [DOI] [PubMed] [Google Scholar]
- Valdes A. M., Slatkin M., Freimer N. B. Allele frequencies at microsatellite loci: the stepwise mutation model revisited. Genetics. 1993 Mar;133(3):737–749. doi: 10.1093/genetics/133.3.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenbach J., Gyapay G., Dib C., Vignal A., Morissette J., Millasseau P., Vaysseix G., Lathrop M. A second-generation linkage map of the human genome. Nature. 1992 Oct 29;359(6398):794–801. doi: 10.1038/359794a0. [DOI] [PubMed] [Google Scholar]
- Wu K. S., Tanksley S. D. Abundance, polymorphism and genetic mapping of microsatellites in rice. Mol Gen Genet. 1993 Oct;241(1-2):225–235. doi: 10.1007/BF00280220. [DOI] [PubMed] [Google Scholar]
- Zischler H., Nanda I., Schäfer R., Schmid M., Epplen J. T. Digoxigenated oligonucleotide probes specific for simple repeats in DNA fingerprinting and hybridization in situ. Hum Genet. 1989 Jun;82(3):227–233. doi: 10.1007/BF00291160. [DOI] [PubMed] [Google Scholar]