ABSTRACT
The primary methyl group donor S-adenosylmethionine (SAM) is important for a plethora of cellular pathways including methylation of nucleic acids, proteins, and the 5′ cap structure of mRNAs, as well as biosynthesis of phospholipids and polyamines. In addition, because it is the cofactor for chromatin methylation, SAM is an important metabolite for the establishment and maintenance of epigenetic marks. Here, we demonstrate that cells halt proliferation when SAM levels become low. Cell cycle arrest occurs primarily in the G1 phase of the cell cycle and is accompanied by activation of the mitogen-activated protein kinase p38 (MAPK14) and subsequent phosphorylation of MAPK-activated protein kinase-2 (MK2). Surprisingly, Cdk4 activity remains high during cell cycle arrest, whereas Cdk2 activity decreases concomitantly with cyclin E levels. Cell cycle arrest was induced by both pharmacological and genetic manipulation of SAM synthesis through inhibition or downregulation of methionine adenosyltransferase, respectively. Depletion of methionine, the precursor of SAM, from the growth medium induced a similar cell cycle arrest. Unexpectedly, neither methionine depletion nor inhibition of methionine adenosyltransferase significantly affected mTORC1 activity, suggesting that the cellular response to SAM limitation is independent from this major nutrient-sensing pathway. These results demonstrate a G1 cell cycle checkpoint that responds to limiting levels of the principal cellular methyl group donor S-adenosylmethionine. This metabolic checkpoint might play important roles in maintenance of epigenetic stability and general cellular integrity.
KEY WORDS: Cell cycle, MK2, One carbon cycle, S-adenosylmethionine, SAM, p38 MAPK, MAPKAPK2
INTRODUCTION
In order to maintain DNA integrity and faithfully transmit genetic information, mammalian cells have evolved a plethora of cell cycle checkpoints to ensure cell cycle arrest when conditions are not suitable for cell division (Hartwell and Weinert, 1989). Limiting levels of low-molecular-mass nutrients was noted to inhibit cell proliferation more than thirty years ago (Holley and Kiernan, 1974; Pardee, 1974). The simplest explanation of this phenotype is substrate limitation; cells simply cannot grow when nutrient building blocks are short in supply. Alternatively, nutrient levels might be integrated in signaling pathways connected to cell cycle control as a means to protect cellular integrity during nutrient limitation. Metabolic checkpoints that respond to low glucose or low amino acid levels have been reported to be mediated by AMPK (AMP-activated kinase) through the p53 and mTORC1 (mammalian target of rapamycin complex 1) signaling pathways (Gwinn et al., 2008; Jones et al., 2005). Disruption of metabolic checkpoints mediated by the AMPK-mTORC1 pathway during either glucose or amino acid crisis has been shown to decrease cell viability.
Among the countless metabolites in cells, one particularly interesting metabolite with an undefined link to cell cycle is S-adenosylmethionine (SAM). SAM is synthesized from methionine and the adenosine moiety of ATP by methionine adenosyltransferase (MAT). The activated methyl group and the aminopropyl part of SAM are used in numerous cellular reactions including DNA, RNA, protein and lipid methylation, as well as polyamine synthesis, making SAM the most versatile metabolite second to ATP (Loenen, 2006). Most importantly, as the cofactor for all chromatin methylation, SAM is required for faithful maintenance and transmission of epigenetic marks. It is plausible that cells would develop a system that monitors SAM concentrations and stops S phase initiation if SAM levels are too low to support proper chromatin re-methylation. This idea is reminiscent of the SAM checkpoint proposed in the model organism S. cerevisiae (Kaiser et al., 2006). Consistent with this notion, targeted inhibition of MAT with a chemical inhibitor or shRNA knockdown was shown to inhibit leukemia cell proliferation and induce apoptosis (Attia et al., 2008; Jani et al., 2009), suggesting not only the existence of a SAM checkpoint in mammalian cells but also its potential as a therapeutic target. Furthermore, recent reports indicate breast and prostate cancer cells suffer a G1 cell cycle arrest when cultured in medium where methionine is replaced with its metabolic precursor homocysteine (Booher et al., 2012; Lu and Epner, 2000), probably as a consequence of reduced flux through the homocysteine-methionine-SAM metabolic axis, which results in insufficient SAM to support cell cycle progression (Booher et al., 2012).
To understand the signals and mechanism of the SAM checkpoint in mammalian cells, we used methionine-free medium, chemical inhibitor, and genetic tools to decrease intracellular SAM. Here, we demonstrate that SAM limitation induced robust G1 arrest with high Cdk4 and low Cdk2 activity, which was independent from the mTORC1 and polyamine pathways, but depended on p38 MAPK and its downstream checkpoint kinase MAPK-activated protein kinase-2 (MK2, also known as MAPKAPK2).
RESULTS
SAM depletion induces cell cycle arrest in G1
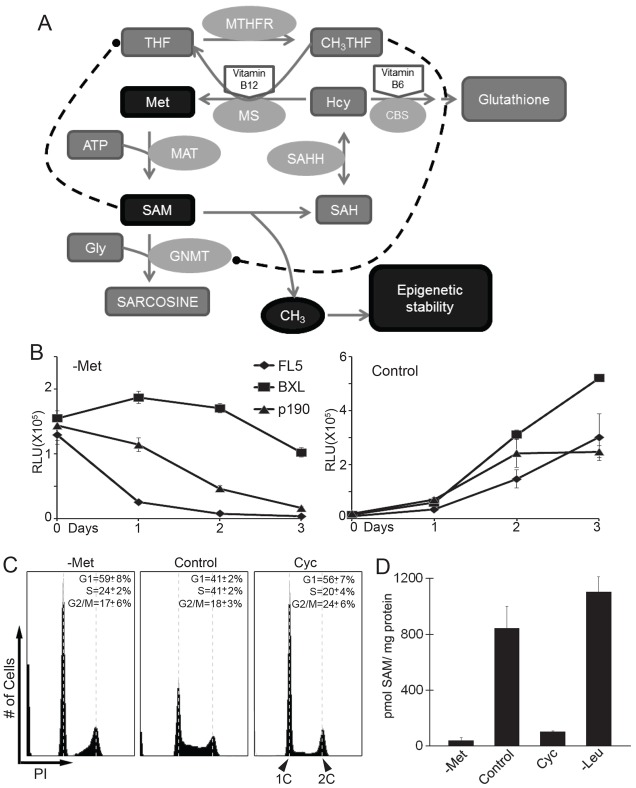
To analyze effects of SAM availability on cell cycle progression we used the IL3-dependent mouse pre-B-cell FL5.12 because they have well described and robust nutrient response pathways (Edinger and Thompson, 2002). In addition, genetically similar FL5.12 derivatives are available that are either tumorigenic owing to stable expression of the oncogenic fusion protein p190 BCR-Abl (p190 cells) (Li et al., 1999), or resistant to induction of apoptosis owing to stable expression of the anti-apoptotic factor Bcl-XL (BXL cells). Whereas the latter remain IL3-dependent, p190 cells can proliferate without IL3 (Neshat et al., 2000). We first tested the effect of methionine depletion on these cell lines. Methionine is the direct metabolic precursor of SAM (Fig. 1A) and its depletion is a convenient and efficient way to reduce intracellular SAM levels. As expected, all cell lines (FL5.12, p190, BXL) stopped proliferation immediately after they were shifted to methionine-free medium, and cell numbers rapidly decreased (Fig. 1B). The decrease in cell number was likely to be caused by apoptosis because BXL cells showed significantly higher viability compared to FL5.12 and p190 cells. Flow cytometric analyses showed that cells were primarily arrested in the G1 phase of the cell cycle with a smaller fraction arrested in G2/M (Fig. 1C). A comparable cell cycle arrest profile was observed when SAM levels were depleted through inhibition of methionine adenosyltransferase (MAT) (Fig. 1C, right panel) with cycloleucine (Lombardini and Talalay, 1970). Measurement of intracellular SAM concentrations revealed that SAM levels dropped rapidly after cells were shifted to methionine-free growth medium and were nearly undetectable after 4 hours (Fig. 1D). A similar rapid drop in cellular SAM was observed after cells were treated with cycloleucine. In contrast, SAM levels were unaffected in cells shifted to leucine-free medium (Fig. 1D), although leucine deprivation induced G1 arrest in cells (data not shown).
Fig. 1.
Methionine deprivation leads to SAM depletion and a cell proliferation defect. (A) Schematic representation of the transmethylation pathway. (B) FL5.12 cells, FL5.12 cells stably expressing Bcl-xL (BXL), and FL5.12 cells stably expressing p190 BCR-Abl (p190) were shifted to either control or methionine-free media. Cell proliferation was monitored with Cell Titer-Glo (Promega®). (C) p190 cells were shifted to either methionine-free, control or cycloleucine-containing media for 16 hours. Cells were stained with propidium iodide (PI) and analyzed by flow cytometry. (D) p190 cells were shifted to methionine free (-Met), control, cycloleucine-containing (Cyc) or leucine free (-Leu) media for 4 hours and SAM concentrations were measured using reverse-phase HPLC. All data are reported as mean±s.d., n = 3.
SAM depletion blocks entry into S phase despite high Cdk4 activity
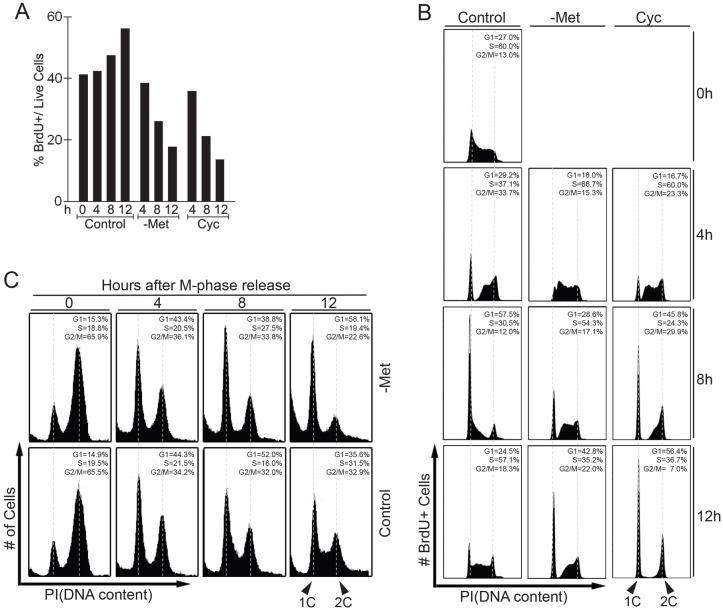
To better characterize the effect of SAM depletion on cell cycle arrest, we monitored the ability of cells to replicate DNA after they were shifted to methionine-free medium or treated with cycloleucine (Fig. 2A). S phase was monitored by pulse-labeling with bromodeoxyuridine (BrdU). Both methionine depletion and inhibition of SAM synthesis with cycloleucine significantly prevented DNA replication (Fig. 2A). Flow cytometric analysis showed that cells accumulated in the G1 phase and there was no noticeable S phase population after SAM depletion (Fig. 1C) suggesting that the drop in BrdU incorporation is caused by arrest at the G1/S transition. To further determine the effect of SAM depletion on cell cycle progression we decided to follow a synchronous cell population. To this end, we pulse-labeled S phase cells with BrdU, then shifted cells to either methionine-free medium or treated them with cycloleucine, and followed the BrdU-labeled cells progressing through different cell cycle phases (Fig. 2B). Consistent with the cell cycle profile of the entire cell population after SAM depletion (Fig. 1C), the majority of cells that were shifted to methionine-depleted or cycloleucine medium progressed through S phase and mitosis, and arrested in G1 (Fig. 2B). Methionine depletion did reduce progression rate through S phase but did not block DNA-replication of cells that had committed to S phase, and a significant fraction of cells reached G1 after 12 hours. Cycloleucine did not affect the rate of S phase progression suggesting that the decreased DNA replication rate is a consequence of methionine depletion. However, SAM depletion did significantly delay cells in G2/M, although most cells eventually finished mitosis and arrested in G1 (Fig. 2B, data not shown). The delay in G2/M is probably due to slow progression from the G2 phase into metaphase, because cells pre-synchronized in metaphase with nocodazole progressed to the next G1 phase without delay when intracellular SAM was depleted (Fig. 2C). Taken together, these data demonstrate that at no point after SAM depletion can G1 cells re-enter S phase, indicating that SAM limitation induces a robust cell cycle arrest at the G1/S transition. We refer to this cell cycle arrest as the SAM checkpoint.
Fig. 2.
SAM depletion decreases the S phase population and induces a robust G1 cell cycle arrest. (A) p190 cells were shifted to control, methionine-free (-Met), or cycloleucine-containing (Cyc) media for the indicated time course then labeled with BrdU for 30 minutes. The proportion of BrdU-positive cells was determined by flow cytometry. (B) To follow the S phase population through the cell cycle, p190 cells were pulse labeled with BrdU for 30 minutes then shifted to control, -Met, or Cyc media without BrdU and incubated for the time intervals indicated before analysis by flow cytometry. Only BrdU-positive cells (i.e. cells that were in S phase at t = 0) are shown throughout the time course. (C) p190 cells were treated with nocodazole for 12 hours to synchronize cells in M phase. Cells were released into the cell cycle in either control or -Met media and cell cycle profiles were monitored by flow cytometry.
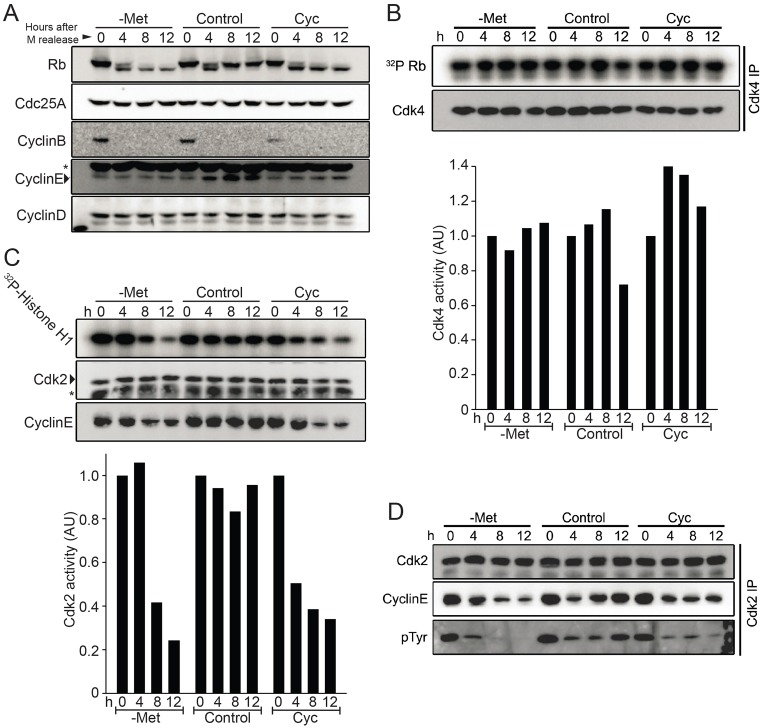
We next monitored some of the regulatory proteins important for G1/S transition. Cells were pre-synchronized in metaphase with a sequential thymidine–nocodazole block and then released into methionine-depleted medium or treated with cycloleucine (Fig. 3A). As expected cyclin B was present in metaphase but rapidly disappeared as cells progressed through the cell cycle. Most strikingly, SAM depletion by both methionine-free culture medium and cycloleucine treatment prevented phosphorylation of the retinoblastoma protein Rb in G1 (Fig. 3A). Two cyclin-dependent kinases (Cdks), Cdk4–cyclin-D and Cdk2–cyclin-E, phosphorylate Rb during G1 to enable entry into S phase (Malumbres and Barbacid, 2009). Levels of cyclin D1 were unaffected by SAM depletion (Fig. 3A). In agreement with constant cyclin D1 levels, Cdk4 activity as measured by Rb phosphorylation in vitro remained unaffected during SAM depletion (Fig. 3B). In contrast, the increase of cyclin E levels observed in control cells during G1, was absent when SAM levels were depleted (Fig. 3A), and accordingly Cdk2 activity in vitro dropped significantly (Fig. 3C). This is in contrast to previous results obtained with MDA-MB468 breast cancer cells where cyclin E levels remained high during methionine stress (Booher et al., 2012). This is probably due to dysregulation of cyclin E in these breast cancer cells owing to mutations in cyclin E regulators.
Fig. 3.
SAM depletion decreased Cdk2 but not Cdk4 activity. (A) p190 cells were synchronized in M phase by thymidine–nocodazole block and released into control, methionine-free (-Met), or cycloleucine-containing (Cyc) media. Cell cycle regulators were detected by immunoblotting. Please note that the cyclin E blot shows a strong nonspecific band (marked by asterisk) above the genuine cyclin E signal, which was confirmed by cyclin E immunopurification followed by immunoblotting. (B,C) Asynchronous p190 cells were shifted to -Met, control, or Cyc media for the indicated time intervals. Cdk2 and Cdk4 complex were immunopurified from cell lysate, washed with kinase buffer and subjected to in vitro kinase activity assays using histone H1 or Rb as substrates as indicated. Kinase activities were quantified by the Bio-Rad Quantity One™ analysis software. The immunopurification was also analyzed by immunoblotting to detect Cdk2 and cyclin E (C) and Cdk4 (B) protein levels. (D) Asynchronous p190 cells were shifted to -Met, control, and Cyc media and collected at indicated time points for Cdk2 immunopurification. Immuno-complexes were analyzed by immunoblotting to detect the level of inhibitory phosphorylation on Tyr15 of Cdk2 and cyclin E. AU, arbitrary units.
During growth factor withdrawal, degradation of the phosphatase Cdc25A causes downregulation of Cdk2 activity owing to the increase in inhibitory phosphorylation on Cdk2 (Khaled et al., 2005). Notably, Cdc25A levels were unchanged during SAM checkpoint activation and inhibitory Cdk2 tyrosine phosphorylation rapidly decreased during SAM depletion (Fig. 3D), indicating that the observed reduction in Cdk2 activity during SAM checkpoint activation (Fig. 3C) is not caused by inhibitory tyrosine phosphorylation. In addition, these results suggest that SAM checkpoint activation is caused by a pathway distinct from growth factor withdrawal, which is characterized by Cdc25A degradation and a concomitant increase in Cdk2 inhibitory tyrosine phosphorylation (Khaled et al., 2005). Consistent with this notion, SAM depletion readily induces G1 cell cycle arrest in the growth-factor-independent cell line p190 (Fig. 2B).
Knock-down of methionine adenosyltransferase MAT2A and MAT2B induces cell cycle arrest
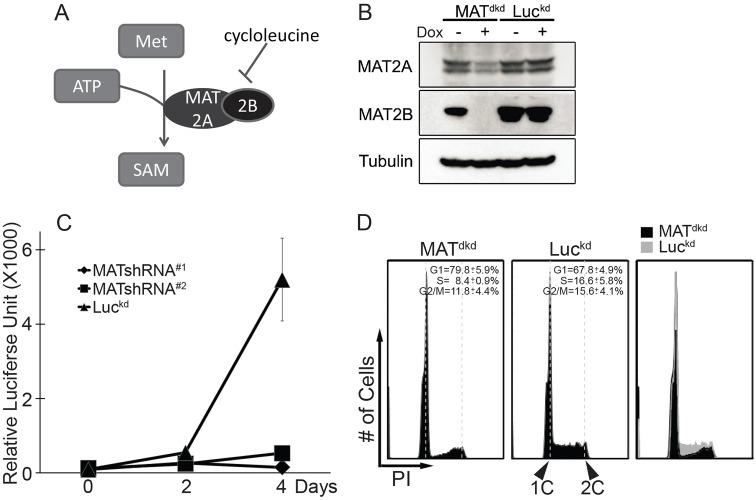
SAM is synthesized from methionine and ATP by methionine adenosyltransferase. Mammalian MAT consists of two subunits, the catalytic subunit MAT2A and the regulatory subunit MAT2B (Fig. 4A). To further support our previous result that SAM depletion induces a SAM checkpoint activation and G1 cell cycle arrest, we generated doxycycline-inducible MAT2A and MAT2B dual knockdown cell lines (MATdkd) to genetically decrease intracellular SAM production. Induction of shRNA expression partially reduced MAT2A levels and dramatically decreased expression of MAT2B (Fig. 4B). To efficiently reduce intracellular SAM, we needed to reduce flux through the SAM synthesis pathway by lowering the methionine concentration to a more physiological level than that in tissue culture medium. Proliferation of control cells was not affected by the lower methionine levels during the time course. However, knockdown of MAT2A and MAT2B together prevented proliferation of cells under these conditions (Fig. 4C), and resulted in the same cell cycle phenotype, namely depletion of the S phase population and arrest in G1, as observed in response to SAM depletion by cycloleucine or methionine-free medium (Fig. 4D). Comparable results were observed with a different pair of shRNAs that also target MAT2A and MAT2B simultaneously (Fig. 4C).
Fig. 4.
S phase inhibition and G1 arrest in response to methionine adenosyltransferase (MAT) knockdown. (A) Mammalian MATII consists of the catalytic subunit MAT2A and regulatory subunit MAT2B. (B) p190 cells were infected with a lentivirus expressing inducible shRNAs directed against both MAT2A and MAT2B to generate double-knockdown cells (MATdkd). Cells expressing doxycycline-inducible luciferase shRNA (Luckd) were used as control. Both cell lines were cultured for 48 hours with or without doxycycline and analyzed by immunoblotting with antibodies against MAT2A, MAT2B and tubulin. (C) Cells harboring two different inducible pairs of MAT2A and MAT2B shRNAs (MATshRNA#1, MATshRNA#2), or luciferase shRNA (Luckd) were shifted to media with 2.5 µM methionine for three days. The number of viable cells was analyzed with Cell Titer-Glo reagent. Data are represented as mean±s.d., n = 3. (D) MATdkd and Luckd cells were incubated with doxycycline for 48 hours then shifted to medium with 2.5 µM methionine plus doxycycline for 24 hours before analysis of cell cycle profiles by flow cytometry.
SAM-depletion-induced cell cycle arrest is independent of mTORC1 signaling
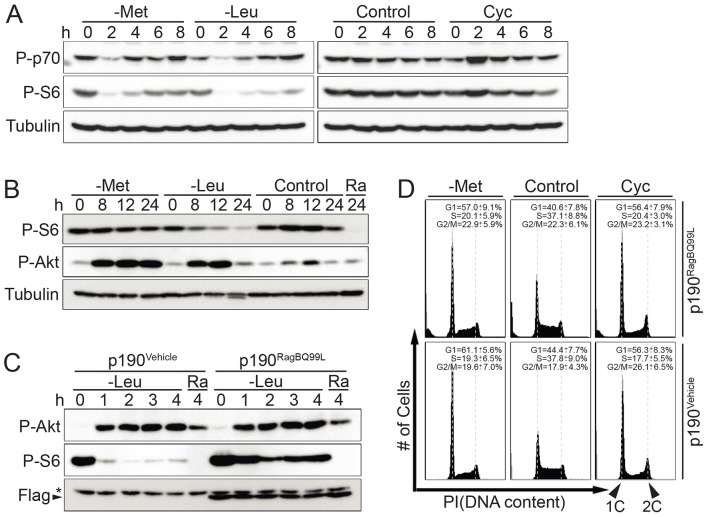
The mTORC1 complex controls one of the major nutrient signaling pathways in eukaryotes. This pathway is particularly important in sensing amino acid availability. As expected, when cells were transferred to leucine-depleted growth medium, mTORC1 was inhibited, as demonstrated by reduced phosphorylation of the ribosomal subunit S6 (Fig. 5A). Surprisingly, depletion of intracellular SAM by culturing cells in methionine-free medium only transiently inhibited mTORC1 signaling. Moreover, inhibition of SAM synthesis with cycloleucine had no effect on S6 or S6 kinase phosphorylation, indicating that metabolic pathways around SAM are not monitored by mTORC1.
Fig. 5.
The mTORC1 signaling pathway is not involved in SAM checkpoint activation. (A) p190 cells were shifted to -Met, -Leu, control or Cyc media. Cells were harvested at indicated time and analyzed by immunoblotting using antibodies directed against phospho-p70S6K (P-p70), phospho-S6 (P-S6) and tubulin. (B), p190 cells were shifted to -Met, -Leu or control media, or medium with 20 nM rapamycin (Ra) and analyzed as for A, and also for phospho-Akt Ser473 (P-Akt). (C) p190 cells were infected with a retrovirus carrying vehicle (p190Vehicle) or a Flag–RagBQ99L (p190RagBQ99L) expression cassette and selected with puromycin for stable expression. Selected cells were shifted to -Leu or rapamycin-containing media for the time intervals indicated and analyzed by immunoblotting to detect phospho-S6 and Flag–RagBQ99L protein levels. (D) p190Vehicle and p190RagBQ99L cells were shifted to -Met, control, and Cyc media for 12 hours and cell cycle profiles were analyzed by flow cytometry.
An extended timecourse confirmed that methionine-depleted medium had no persisting effects on S6 phosphorylation, suggesting that mTORC1 signaling is largely unaffected (Fig. 5B). In contrast, as expected, leucine deprivation dramatically reduced the phosphorylation of the mTORC1 downstream target ribosomal subunit S6 (Fig. 5B). mTORC1 is known to be activated by growth factor and nutrients through an IRS1–PI3K–PDK1–Akt pathway and feedback inhibits Akt activity through S6K-mediated IRS1 phosphorylation and proteasomal degradation (Easton et al., 2006). Accordingly, decreasing mTORC1 activity by leucine deprivation attenuated this feedback inhibition, which in turn resulted in Akt activation. Surprisingly, the phosphorylation of Akt Ser473 was also significantly increased during SAM depletion, despite constant mTORC1 activity. These results suggest that under these conditions other signaling pathways contribute to Akt activation.
The nutrient-insensitive RagBQ99L mutant cannot suppress SAM-depletion-induced cell cycle arrest
The Rag family small GTPases interact with mTORC1 in an amino-acid-dependent manner and are required for mTORC1 activation in response to intracellular amino acid levels (Sancak et al., 2008). Methionine, besides being the immediate metabolic precursor for SAM synthesis, is also an essential amino acid. Our previous result showed that depletion of SAM in methionine-free medium does not persistently affect mTORC1 activity (Fig. 5A,B). However, to further exclude the involvement of mTORC1 in the SAM checkpoint, we generated a cell line stably expressing RagBQ99L, a GTPase mutant that renders mTORC1 insensitive to amino acid levels (Sancak et al., 2008). As previously demonstrated (Sancak et al., 2008), phosphorylation levels of the mTORC1 downstream target ribosomal protein S6 was only slightly sensitive to leucine deprivation in RagBQ99L-expressing cells, while parental cells exhibit dramatic reduction of phosphorylated S6 (Fig. 5C). Next, we tested the cell cycle response of RagBQ99L cells to SAM depletion. RagBQ99L expressing cells showed robust cell cycle arrest in the G1 phase after SAM depletion with either methionine-free medium or cycloleucine (Fig. 5D). Together these results strongly suggest that the SAM checkpoint is independent of the major nutrient signaling pathway, which is controlled by mTORC1.
The SAM-checkpoint-induced G1 arrest in FL5.12 cells is distinct from the polyamine-depletion phenotype
In addition to serving as a cofactor for methylation reactions, SAM is also required for polyamine synthesis. Numerous studies have connected polyamine synthesis and increased expression of relevant enzymes in this pathway, such as ornithine decarboxylase (ODC) and S-adenosylmethionine decarboxylase (SAMDC or AdoMetDC), with tumor growth (Casero and Marton, 2007; Meyskens and Gerner, 1999; Seidenfeld et al., 1986). AdoMetDC consumes SAM to produce aminopropyl groups for polyamine synthesis. It was thus possible that SAM depletion decreases polyamine pools and induces polyamine-depletion phenotypes, which are characterized by Akt activation, p27-dependent G1 arrest and p53-dependent apoptosis (Koomoa et al., 2009; Koomoa et al., 2008; Wallick et al., 2005). However, we did not detect any significant upregulation of p21, p27 or p53 levels during SAM checkpoint activation (supplementary material Fig. S1A), nor could spermidine supplementation suppress the cell cycle arrest induced by SAM depletion (supplementary material Fig. S2A). In addition, FL5.12 cells treated with the ODC inhibitor difluoromethylornithine (DFMO), to block polyamine production, did not mimic the cell cycle arrest phenotype induced by SAM depletion (supplementary material Fig. S2B). Taken together, these results suggest that SAM depletion in FL5.12 cells induces a cell cycle arrest that is distinct from the effects of polyamine depletion.
The p38–MK2 pathway is required for SAM-depletion-induced cell cycle arrest
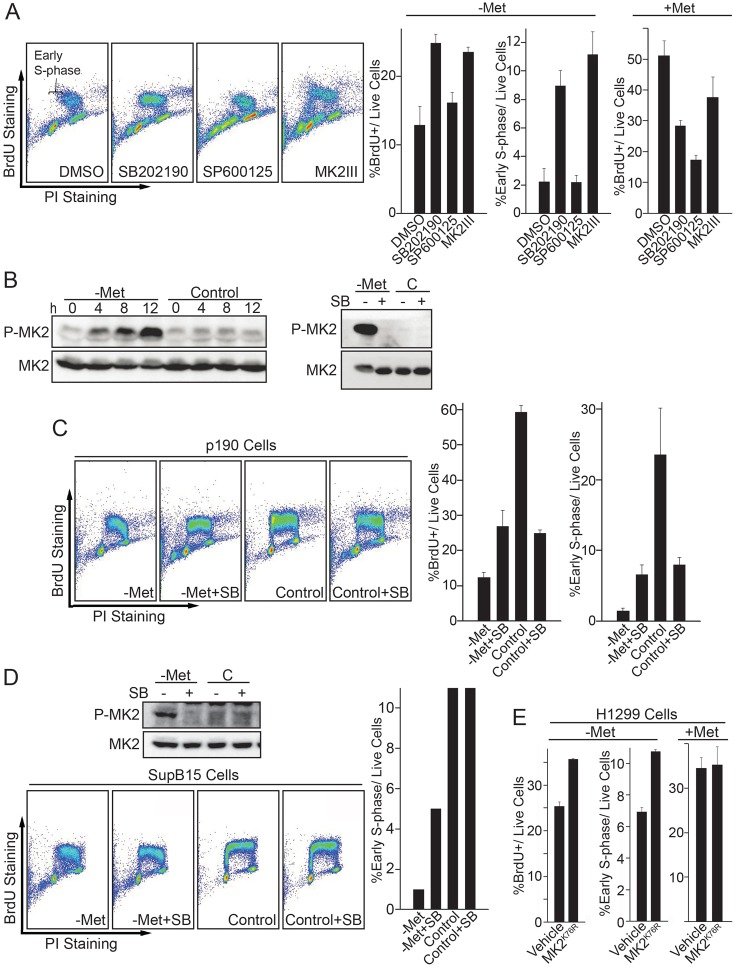
In addition to mTORC1, AMP-activated protein kinase (AMPK) regulates cellular energy homeostasis (Hardie et al., 2012). Recently, AMPK has been linked to cell cycle regulation, through mTORC1, p53, p27 and p38, in response to cellular metabolic cues (Gwinn et al., 2008; Jones et al., 2005; Liang et al., 2007; Sengupta et al., 2007; Zhuang and Miskimins, 2008). Our results have largely excluded mTORC1 as a crucial component of the SAM checkpoint pathway. Furthermore, p53 and p27 levels only increased after very extended periods of SAM depletion, long after cells had arrested at G1 (data not shown). Thus, p53 and p27 might contribute to the maintenance of cell cycle arrest after SAM depletion, but are probably not involved in the cell cycle arrest initiation in response to SAM depletion. These results prompted us to investigate whether the AMPK–p38 signaling pathway is activated during SAM depletion. To this end, we treated cells with MAPK inhibitors during SAM depletion induced by either culture in methionine-free medium or cycloleucine and monitored S phase entry by BrdU pulse labeling (Fig. 6A). Both p38 inhibitor SB202190 and MK2 inhibitor MK2III significantly restored S phase entry after SAM depletion, which is particularly clear when the cells were gated for early S phase (Fig. 6A). In contrast, even though the JNK inhibitor SP600125 also increased the total S phase population (Fig. 6A), this was an indirect effect due to slower DNA replication. Consequently, JNK inhibition did not suppress the block in G1/S transition, as evident by loss of early S phase cells (Fig. 6A). MK2 is a direct substrate for p38 phosphorylation and functions as a checkpoint kinase in parallel to Chk1 and Chk2 during UV-induced DNA damage (Manke et al., 2005). Our data suggests p38–MK2 signaling as an important component of SAM checkpoint activation. To test whether p38 is activated during SAM depletion, we monitored the p38-specific phosphorylation site Thr334 on MK2 after shifting cells to methionine-free medium. MK2 was rapidly phosphorylated on Thr334 during SAM depletion (Fig. 6B). MK2 phosphorylation was blocked by the p38 inhibitor SB202190, confirming p38 dependency (Fig. 6B). We have previously demonstrated that there are decreased levels of the DNA replication factor Cdc6 as a result of SAM checkpoint activation in the human breast cancer cell line MDA-MB 468 (Booher et al., 2012). Consistent with that study, Cdc6 levels were also decreased in FL5.12 cells during SAM depletion, and this phenotype could be partially reversed by treatment with the p38 inhibitor SB202190 (supplementary material Fig. S1A,B).
Fig. 6.
The p38–MK2 pathway is required for SAM checkpoint activation. (A) FL5.12 cells were shifted to -Met media with either DMSO, 20 µM p38 inhibitor SB202190, 20 µM JNK inhibitor SP600125 or 20 µM MK2 inhibitor MK2III. After 12 hours, cells were labeled with BrdU for 30 minutes and analyzed by BrdU immunostaining and flow cytometry. BrdU-positive cells with one-third of the 2C DNA content were gated and presented as early S phase cells to indicate the fraction of cells entering S phase. (B) FL5.12 cells were shifted to either -Met or control media for the time intervals indicated and analyzed by immunoblotting with antibodies against phospho-MK2 and total MK2 (left panel). To test p38-dependence of MK2 phosphorylation a similar experiment was performed with and without 20 µM p38 inhibitor SB202190 for 12 hours (right panel). (C) p190 cells were shifted to -Met or control media with or without p38 inhibitor SB202190 for 12 hours, labeled with BrdU for 30 minutes and analyzed by flow cytometry. (D) Human SupB15 cells were shifted to -Met or control media with or without p38 inhibitor SB202190 for 24 hours. Cells were either harvested for MK2 immunoblot analysis or labeled with BrdU for flow cytometric analysis. (E) Human H1299 cells (p53-null) were infected by a retrovirus carrying vehicle (pQCXIP) or the dominant-negative mutant MK2K76R expression cassette. Cells were shifted to -Met media for 24 hours, labeled with BrdU for 30 minutes and analyzed by flow cytometry. All data are reported as mean±s.d., n = 3.
The p38 inhibitor experiments described above were performed in IL3-dependent murine pre-B cells, FL5.12. Similar results were observed with leukemic, IL3-independent FL5.12 cells that stably express BCR-Abl p190 (p190 cells). SAM depletion in p190 cells efficiently blocked S phase entry, and inhibition of p38 allowed a significant portion of the cells to overcome this block (Fig. 6C). Interestingly, inhibition of p38 to override the SAM checkpoint increased the proportion of sub-G1 cells, suggesting that cell cycle progression in low SAM levels is deleterious to cells and that the SAM checkpoint response is important for the maintenance of cellular integrity in both non-tumorigenic and tumorigenic cells (Fig. 6A,C). Accordingly, p38 inhibition during induction of the SAM checkpoint with methionine-free medium or the MAT inhibitor cycloleucine significantly increased cell death in a dose-dependent manner (supplementary material Fig. S2C).
We next asked whether the p38–MK2 pathway is also important for SAM checkpoint activation in human cells. To this end, we used human SupB15 cells, an acute lymphoblastic leukemia (ALL) cell line with BCR-Abl (p190) expression. Similar to murine cells, SAM depletion in SupB15 cells induced p38 activity as indicated by MK2 phosphorylation on Thr334 (Fig. 6D), and MK2 phosphorylation was blocked upon p38 inhibition with SB202190 (Fig. 6D). Importantly, inhibition of p38 allowed a significant number of SupB15 cells to enter S phase after SAM depletion (Fig. 6D). Finally, we further confirmed the importance of this pathway by extending our analyses to adherent cells. SAM depletion in human small cell lung carcinoma H1299 cells (p53-null) induced a G1 arrest (data not shown), and inhibition of MK2 by expression of the dominant kinase-dead MK2K76R mutant (Winzen et al., 1999) significantly increased the number of cells entering S phase during SAM checkpoint conditions (Fig. 6E). H1299 cells lack p53 tumor suppressor function, suggesting that the SAM checkpoint is independent of p53.
DISCUSSION
Cell cycle checkpoint activation in response to various environmental challenges is essential for the maintenance of cellular, genetic and epigenetic integrity of eukaryotic cells (Bartek and Lukas, 2003). Disruption of checkpoints due to inherited or acquired mutations has been recognized as an important contributor to human cancer. Since introduction of the concept of cell cycle checkpoints by Hartwell and Weinert (Hartwell and Weinert, 1989), research has largely been focused on molecular dissection of DNA damage, S phase and spindle checkpoints. In comparison, our understanding of checkpoints that monitor important cellular metabolites is in its infancy. In this study, we explore a pathway that monitors levels of the primary methyl-group donor SAM and its connections to cell cycle control. Conceptually, this pathway might define a checkpoint that is important for epigenetic stability because failing to stop cell cycle progression when SAM levels are insufficient to support chromatin methylation could lead to loss of chromatin methylation marks. Similar checkpoints monitoring other key metabolites are likely to exist.
Recently, metabolic checkpoints have emerged as potential therapeutic cancer targets (Gwinn et al., 2008; Laplante and Sabatini, 2012; Shackelford and Shaw, 2009). Tumor cells often adapt and reprogram their metabolism to fuel cell growth and proliferation and concomitantly acquire metabolic addictions that distinguish cancer cells from normal cells (Hanahan and Weinberg, 2011). Interestingly, addiction of cancer cells to a high flux through the methionine metabolism pathway has been known for over 35 years (Halpern et al., 1974). Recent studies in breast cancer cells suggest that SAM rather than methionine might be the limiting metabolite defining addiction to this metabolic pathway (Booher et al., 2012). Consistent with this idea, targeting methionine adenosyltransferase (MAT) with either a small molecule inhibitor or shRNA induces apoptosis in leukemia cells (Attia et al., 2008; Jani et al., 2009). The mechanisms of how cells induce cell cycle arrest and apoptosis in response to SAM limitation is currently unknown, but experiments in the yeast and breast cancer cell models indicate loss of pre-replication complexes from chromatin as a contributing factor (Booher et al., 2012; Su et al., 2005).
In this study, we define a cell cycle checkpoint induced by SAM limitation and identify p38 MAPK and its downstream checkpoint kinase MK2 as part of the signaling pathway for the mammalian SAM checkpoint. It has been proposed that the p38–MK2 pathway is a third cell cycle checkpoint module, parallel to ATR–Chk1 and ATM–Chk2, and it has been shown that it is essential for the survival of p53-null cells after DNA damage (Manke et al., 2005; Reinhardt et al., 2007; Reinhardt and Yaffe, 2009). DNA damage induces degradation of the G1/S-transition driver phosphatase Cdc25A in order to halt cell proliferation in G1 (Hassepass et al., 2003; Mailand et al., 2000). Inhibition of MK2 prevents degradation of Cdc25A after DNA damage, by an unknown mechanism, resulting in sustained cell proliferation despite DNA damage (Manke et al., 2005). Interestingly, p38-mediated degradation of Cdc25A has been suggested to control cell cycle arrest in response to growth factor withdrawal (Khaled et al., 2005). Whether MK2 is involved is unknown. Our study implicates the p38–MK2 module in G1 checkpoint arrest in response to SAM depletion. However, the p38–MK2 pathway appears to function through a distinct mechanism because Cdc25A levels remained constant throughout SAM checkpoint activation, Cdk2 phosphorylation of Tyr15 did not increase, and expression of the degradation-resistant Cdc25AS76A (Hassepass et al., 2003) did not override SAM-checkpoint-induced cell cycle arrest (data not shown). Furthermore, SAM checkpoint activation remains robust in growth-factor-independent cells expressing BCR-Abl p190, indicating that signaling pathways responding to growth factor withdrawal are different from those in SAM checkpoint activation.
Interestingly, p38 has previously been indirectly connected to cell cycle arrest in response to SAM depletion. Arsenic is an environmental toxin, a carcinogen and a cancer drug (de Thé and Chen, 2010; Kitchin and Conolly, 2010; Tseng, 2008). One of the main detoxification pathways for arsenic is methylation by human SAM-dependent arsenite methyltransferase, which consumes SAM and glutathione to neutralize intracellular arsenicals (Hayakawa et al., 2005). Exposure to arsenite decreases intracellular SAM levels (Reichard et al., 2007) and induces p38-dependent cell cycle arrest (Kim et al., 2002). It is thus conceivable that arsenite inhibits cell proliferation through the p38–MK2-activated SAM checkpoint we describe here.
In this study, we identify and characterize the cell cycle checkpoint response to limiting levels of SAM, and discovered the p38–MK2 pathway as an important link between SAM homeostasis and cell cycle arrest. Molecular mechanisms for how SAM levels are monitored are currently unknown. Our results suggest that the cellular response to SAM limitation is independent from the mTORC1 pathway and might define a distinct metabolite monitoring system. This is surprising because the mTORC1 pathway is the major molecular link between the metabolic state of cells and cellular functions. mTORC1 integrates many extracellular and intracellular inputs, including growth factors, stress, energy status, oxygen level and amino acids, with protein translation, lipid synthesis, cell growth, cell cycle and autophagy (Laplante and Sabatini, 2012). Interestingly, methionine depletion had only marginal effects on mTORC1 activity (Fig. 5A), and rendering mTORC1 insensitive to amino acid levels by expression of RagBQ99L did not bypass the induction of G1 cell cycle arrest in response to methionine depletion (Fig. 5C,D). These results suggest that intracellular methionine levels might be monitored through the downstream metabolite SAM, and integrated with cell cycle progression through the SAM checkpoint.
Methionine stress hypersensitivity is an almost universal metabolic characteristic of cancer cells and tumors (Guo et al., 1993; Kreis, 1979; Mecham et al., 1983; Poirson-Bichat et al., 1997; Stern et al., 1984; Tisdale et al., 1983) and was recently suggested to reflect a hypersensitive SAM checkpoint in cancer cells (Booher et al., 2012). Therefore, further understanding of the pathways connecting levels of SAM with cell proliferation will provide molecular insight into integration of metabolite homeostasis with cell cycle regulation and might lead to discovery of new cancer drug targets.
MATERIALS AND METHODS
Cell lines and reagents
Human SupB15, murine pre-B cells FL5.12, and FL5.12 derivatives are kind gifts from David Fruman (Molecular Biology & Biochemistry, University of California, Irvine) and Aimee Edinger (Develomental & Cell Biology, University of California, Irvine). Human H1299 cells were kind gifts from Wen-Hwa Lee (Biological Chemistry, University of California, Irvine). Cells were maintained in RPMI 1640 medium (R8999, US Biological, Swampscott, MA, USA) with 10% dialyzed FBS (Gemini, West Sacramento, CA, USA), 10 µM folic acid, 2.5 µM vitamin B12 (Fisher Scientific, Pittsburgh, PA, USA), 10 mM HEPES (Mediatech, Manassas, VA, USA), 500 pg/ml murine recombinant IL-3 (BD Pharmingen, San Jose, CA, USA) and 1% Penicillin-Streptomycin-Amphoterincin B solution (Mediatech). In the case of methionine-free medium, RPMI 1640 medium was replaced with RPMI 1640 medium deficient in methionine, cysteine, and glutamine (R9016, US biological) and was supplemented with L-cysteine and L-glutamine at the same concentration as in standard RPMI 1640 medium. Medium supplemented with spermidine contained 1 mM aminoguanidine to inhibit serum polyamine oxidase (Koomoa et al., 2008). MK2 inhibitor MK2III was from Calbiochem (EMD Millipore, KGaA, Darmstadt, Germany). Cell viability was measured with CellTiter-Glo® Luminescent Cell Viability Assay (Promega, Madison, WI); 2000 cells/well were used per assay. Antibodies against phospho-S6 (no. 5364), phosphor-Akt (no. 9271), phospho-p70 S6K (no. 9234), phospho-MK2 (no. 3041), total MK2 (no. 3042), Cyclin D1 (no. 2926) and Cdc25A (no. 3652) were from Cell Signaling Technology (Boston, MA, USA). Antibodies against α-tubulin and FLAG® were obtained from Sigma-Aldrich (Ontario, CA, USA). The retinoblastoma antibody was from BD Pharmingen (San Jose, CA, USA). Antibodies against cyclin B, cyclin E, Cdk2 (sc-163) and Cdk4 (sc-260) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Immunoblots were imaged on a Fuji LAS-4000 imaging system (Fujifilm, Tokyo, Japan).
BrdU labeling
Cells were shifted to conditional medium for indicated time period before labeling with 25 µM BrdU for 25 minutes. Cells were harvested, fixed with ice-cold 70% ethanol, and stored overnight at 4°C. Cells were denatured with 2 M HCl, neutralized with 0.1 M NaBr (pH 8.0), and blocked with PBS containing 5% FBS and 1% BSA before incubation with anti-BrdU antibodies (GTX26326, Genetex, Irvine, CA, USA) overnight. For BrdU-chase experiments, cells were labeled with BrdU prior to being shifted to conditional medium. BrdU immunostained cells were analyzed on a BD FACS Calibur Flow Cytometer (BD Biosciences, San Jose, CA, USA) and analyzed with Flowjo software (Tree Star Inc., Ashland, OR, USA).
Cdk2 and Cdk4 kinase activity assay
Cells were lysed in kinase extraction buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 2.5 mM EDTA, 0.2% Triton X-100, 10% glycerol, 10 mM β-glycerophosphate, 2 mM NaF, 2 mM Na3VO4, 10 µg/ml leupeptin and 0.1 M phenylmethylsulfonyl fluoride) and incubated on a rotator at 4°C for 30 minutes. A total of 300 µg of total protein extracts were used per reaction. Kinases were immunopurified with either anti-Cdk2 or -Cdk4 antibodies and protein G-sepharose beads at 4°C overnight. Immuncomplexes were washed two times with kinase extraction buffer and twice with kinase buffer (50 mM HEPES pH 7.5, 10 mM MgCl2, 20 µM ATP). Immunocomplexes were resuspended in 25 µl kinase buffer, and 4 µg of the substrates histone H1 (Cdk2 kinase assay) or Rb (CDK4 kinase assay; a kind gift from Yu-Mei Chen, Biological Chemistry, University of California, Irvine), and 5 µCi of [γ-32P] ATP. The reactions were incubated at 30°C for 20 minutes. The reaction was stopped by adding 25 µl 2× concentrated Laemmli sample buffer, and products were separated by SDS-PAGE (10% gel). Gels were dried and kinase activity was detected by phosphoimaging and quantified by Bio-Rad Quantity One.
shRNA
The pSLIK lentiviral vectors for knockdown of MAT were constructed as described previously (Shin et al., 2006). Briefly, oligonucleotides designed with BfuAI compatible protrusion (lower cased) were annealed and cloned into the BfuAI site in pEN_TGmirRc3 (ATCC, Manassas, VA, USA). Subsequent recombinations with pSLIKpuro were catalyzed with LR clonase (Invitrogen, Carlsbad, CA, USA). pSLIKpuro was generated from pSLIKhygro (Shin et al., 2006) by replacing the hygromycin resistance marker with a puromycin resistance cassette. The miR-shRNA sequence were designed by the RNAi codex algorithm (http://cancan.cshl.edu/RNAi_central/RNAi.cgi?type = shRNA).
The sequences used for shRNA are as follows (the mRNA-targeting sequence is underlined): luciferase (shRNA), 5′-agcgCCCGCCTGAAGTCTCTGATTAATAGTGAAGCCACAGA TGTATTAATCAGAGACTTCAGGCGGTtgcc-3′; mouse MAT2A (shRNA), 5′-agcgCGCAGTCACTCTAATCAATAACTAGTGAAGCCACAGATGTAGTTATTGATTAGAGTGACTGCAtgcc-3′; mouse MAT2B (shRNA), 5′-agcgCTCTTACTAAGTGATGTTTCATTAGTGAAGCCACAGATGTAATGAAACATCACTTAGTAAGATtgcc-3′.
Lentiviruses were generated by transfecting HEK293T cells with the pSLIK vectors together with packaging vectors pMDG, pCMVdeltaR8.91. Lentiviruses were collected 24 and 48 hours post-transfection for target cell infection. Cells were selected with 1 ug/ml puromycin 48 hours post viral infection.
Measurement of cellular SAM levels
SAM concentrations were measured following a protocol adapted from Wang et al. (Wang et al., 2001). Briefly, a Waters HPLC system with UV spectrometer was employed to carry mobile phases and detect SAM. Separation was performed on a PartiSphere C18 reverses phase analytical column (Waters, Milford, MA, USA). The mobile phase consisted of two solvents: solvent A, 8 mM octanesulfonic acid sodium salt, with pH adjusted with phosphoric acid to 3.0; and solvent B, 100% methanol. The column was equilibrated with 80% A and 20% B. Separation was obtained using a step gradient: 8 minutes at equilibration condition, 30 seconds to increase to 40% B, 12.5 minutes at 40% B, 30 seconds to decrease B to equilibration condition, 10 minutes at the equilibration condition. The flow rate was 1 ml/minutes and the detection was set at 254 nm. HPLC was performed at room temperature. SAM was identified by its retention time (17 minutes) and the co-chromatography with the standard. Quantification was based on integration of the area under the curve and was compared to a SAM standard curve from 0 to 10,000 pmol. Cell samples from 15-cm dishes were extracted with 500 µl 0.4 M HClO4 at room temperature, centrifuged at 16,000 g for 15 minutes, and 200 µl supernatant was used for each injection. The resulting pellets after centrifugation were lysed in Urea buffer (8 M Urea, 50 mM Tris-HCl pH 7.5, 150 mM NaCl) and protein content was measured with Bradford protein assay (#23236, Thermo Fisher Scientific Pierce, Rockford, IL, USA). Cellular SAM content was expressed as pmol SAM per mg protein.
Supplementary Material
Acknowledgments
We thank A. Edinger, D. Fruman, E. Lee and W. H. Lee for reagents and helpful suggestions. We are grateful to K. Yokomori and S. White for access to microscopes and HPLC, respectively. We thank H. Piwnica-Worms for Cdc25A and GSK3β constructs, M. Gaestel for MK2 constructs.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
D.W.L. is responsible for execution and design of most experiments. B.P.C. contributed to execution of some experiments. P.K. is responsible for research direction, contributed to experimental design and wrote and revised the manuscript and figures, together with D.W. L.
Funding
This work was partially supported by the National Institute of Health [grant number GM66164]; and the University of California Cancer Research Coordination Committee (to P.K.). D.L. is supported by the California Institute for Regenerative Medicine ‘UCI CIRM Training Grant II’ [grant number TG2-01152]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.127811/-/DC1
References
- Attia R. R., Gardner L. A., Mahrous E., Taxman D. J., Legros L., Rowe S., Ting J. P., Geller A., Kotb M. (2008). Selective targeting of leukemic cell growth in vivo and in vitro using a gene silencing approach to diminish S-adenosylmethionine synthesis. J. Biol. Chem. 283, 30788–30795 10.1074/jbc.M804159200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J., Lukas J. (2003). Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 3, 421–429 10.1016/S1535-6108(03)00110-7 [DOI] [PubMed] [Google Scholar]
- Booher K., Lin D. W., Borrego S. L., Kaiser P. (2012). Downregulation of Cdc6 and pre-replication complexes in response to methionine stress in breast cancer cells. Cell Cycle 11, 4414–4423 10.4161/cc.22767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casero R. A., Jr, Marton L. J. (2007). Targeting polyamine metabolism and function in cancer and other hyperproliferative diseases. Nat. Rev. Drug Discov. 6, 373–390 10.1038/nrd2243 [DOI] [PubMed] [Google Scholar]
- de Thé H., Chen Z. (2010). Acute promyelocytic leukaemia: novel insights into the mechanisms of cure. Nat. Rev. Cancer 10, 775–783 10.1038/nrc2943 [DOI] [PubMed] [Google Scholar]
- Easton J. B., Kurmasheva R. T., Houghton P. J. (2006). IRS-1: auditing the effectiveness of mTOR inhibitors. Cancer Cell 9, 153–155 10.1016/j.ccr.2006.02.027 [DOI] [PubMed] [Google Scholar]
- Edinger A. L., Thompson C. B. (2002). Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol. Biol. Cell 13, 2276–2288 10.1091/mbc.01-12-0584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Lishko V. K., Herrera H., Groce A., Kubota T., Hoffman R. M. (1993). Therapeutic tumor-specific cell cycle block induced by methionine starvation in vivo. Cancer Res. 53, 5676–5679 [PubMed] [Google Scholar]
- Gwinn D. M., Shackelford D. B., Egan D. F., Mihaylova M. M., Mery A., Vasquez D. S., Turk B. E., Shaw R. J. (2008). AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226 10.1016/j.molcel.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpern B. C., Clark B. R., Hardy D. N., Halpern R. M., Smith R. A. (1974). The effect of replacement of methionine by homocystine on survival of malignant and normal adult mammalian cells in culture. Proc. Natl. Acad. Sci. USA 71, 1133–1136 10.1073/pnas.71.4.1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hardie D. G., Ross F. A., Hawley S. A. (2012). AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat. Rev. Mol. Cell Biol. 13, 251–262 10.1038/nrm3311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Weinert T. A. (1989). Checkpoints: controls that ensure the order of cell cycle events. Science 246, 629–634 10.1126/science.2683079 [DOI] [PubMed] [Google Scholar]
- Hassepass I., Voit R., Hoffmann I. (2003). Phosphorylation at serine 75 is required for UV-mediated degradation of human Cdc25A phosphatase at the S-phase checkpoint. J. Biol. Chem. 278, 29824–29829 10.1074/jbc.M302704200 [DOI] [PubMed] [Google Scholar]
- Hayakawa T., Kobayashi Y., Cui X., Hirano S. (2005). A new metabolic pathway of arsenite: arsenic-glutathione complexes are substrates for human arsenic methyltransferase Cyt19. Arch. Toxicol. 79, 183–191 10.1007/s00204-004-0620-x [DOI] [PubMed] [Google Scholar]
- Holley R. W., Kiernan J. A. (1974). Control of the initiation of DNA synthesis in 3T3 cells: low-molecular weight nutrients. Proc. Natl. Acad. Sci. USA 71, 2942–2945 10.1073/pnas.71.8.2942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani T. S., Gobejishvili L., Hote P. T., Barve A. S., Joshi-Barve S., Kharebava G., Suttles J., Chen T., McClain C. J., Barve S. (2009). Inhibition of methionine adenosyltransferase II induces FasL expression, Fas-DISC formation and caspase-8-dependent apoptotic death in T leukemic cells. Cell Res. 19, 358–369 10.1038/cr.2008.314 [DOI] [PubMed] [Google Scholar]
- Jones R. G., Plas D. R., Kubek S., Buzzai M., Mu J., Xu Y., Birnbaum M. J., Thompson C. B. (2005). AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell 18, 283–293 10.1016/j.molcel.2005.03.027 [DOI] [PubMed] [Google Scholar]
- Kaiser P., Su N-Y., Yen J. L., Ouni I., Flick K. (2006). The yeast ubiquitin ligase SCFMet30: connecting environmental and intracellular conditions to cell division. Cell Div. 1, 16 10.1186/1747-1028-1-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaled A. R., Bulavin D. V., Kittipatarin C., Li W. Q., Alvarez M., Kim K., Young H. A., Fornace A. J., Durum S. K. (2005). Cytokine-driven cell cycling is mediated through Cdc25A. J. Cell Biol. 169, 755–763 10.1083/jcb.200409099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. Y., Choi J. A., Kim T. H., Yoo Y. D., Kim J. I., Lee Y. J., Yoo S. Y., Cho C. K., Lee Y. S., Lee S. J. (2002). Involvement of p38 mitogen-activated protein kinase in the cell growth inhibition by sodium arsenite. J. Cell. Physiol. 190, 29–37 10.1002/jcp.10049 [DOI] [PubMed] [Google Scholar]
- Kitchin K. T., Conolly R. (2010). Arsenic-induced carcinogenesis—oxidative stress as a possible mode of action and future research needs for more biologically based risk assessment. Chem. Res. Toxicol. 23, 327–335 10.1021/tx900343d [DOI] [PubMed] [Google Scholar]
- Koomoa D. L., Yco L. P., Borsics T., Wallick C. J., Bachmann A. S. (2008). Ornithine decarboxylase inhibition by alpha-difluoromethylornithine activates opposing signaling pathways via phosphorylation of both Akt/protein kinase B and p27Kip1 in neuroblastoma. Cancer Res. 68, 9825–9831 10.1158/0008-5472.CAN-08-1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koomoa D. L., Borsics T., Feith D. J., Coleman C. C., Wallick C. J., Gamper I., Pegg A. E., Bachmann A. S. (2009). Inhibition of S-adenosylmethionine decarboxylase by inhibitor SAM486A connects polyamine metabolism with p53-Mdm2-Akt/protein kinase B regulation and apoptosis in neuroblastoma. Mol. Cancer Ther. 8, 2067–2075 10.1158/1535-7163.MCT-08-1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis W. (1979). Tumor therapy by deprivation of L-methionine: rationale and results. Cancer Treat. Rep. 63, 1069–1072 [PubMed] [Google Scholar]
- Laplante M., Sabatini D. M. (2012). mTOR signaling in growth control and disease. Cell 149, 274–293 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Ilaria R. L., Jr, Million R. P., Daley G. Q., Van Etten R. A. (1999). The P190, P210, and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J. Exp. Med. 189, 1399–1412 10.1084/jem.189.9.1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Shao S. H., Xu Z. X., Hennessy B., Ding Z., Larrea M., Kondo S., Dumont D. J., Gutterman J. U., Walker C. L. et al. (2007). The energy sensing LKB1-AMPK pathway regulates p27(kip1) phosphorylation mediating the decision to enter autophagy or apoptosis. Nat. Cell Biol. 9, 218–224 10.1038/ncb1537 [DOI] [PubMed] [Google Scholar]
- Loenen W. A. (2006). S-adenosylmethionine: jack of all trades and master of everything? Biochem. Soc. Trans. 34, 330–333 10.1042/BST20060330 [DOI] [PubMed] [Google Scholar]
- Lombardini J. B., Talalay P. (1970). Formation, functions and regulatory importance of S-adenosyl-L-methionine. Adv. Enzyme Regul. 9, 349–384 10.1016/S0065-2571(71)80054-7 [DOI] [PubMed] [Google Scholar]
- Lu S., Epner D. E. (2000). Molecular mechanisms of cell cycle block by methionine restriction in human prostate cancer cells. Nutr. Cancer 38, 123–130 10.1207/S15327914NC381_17 [DOI] [PubMed] [Google Scholar]
- Mailand N., Falck J., Lukas C., Syljuâsen R. G., Welcker M., Bartek J., Lukas J. (2000). Rapid destruction of human Cdc25A in response to DNA damage. Science 288, 1425–1429 10.1126/science.288.5470.1425 [DOI] [PubMed] [Google Scholar]
- Malumbres M., Barbacid M. (2009). Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer 9, 153–166 10.1038/nrc2602 [DOI] [PubMed] [Google Scholar]
- Manke I. A., Nguyen A., Lim D., Stewart M. Q., Elia A. E., Yaffe M. B. (2005). MAPKAP kinase-2 is a cell cycle checkpoint kinase that regulates the G2/M transition and S phase progression in response to UV irradiation. Mol. Cell 17, 37–48 10.1016/j.molcel.2004.11.021 [DOI] [PubMed] [Google Scholar]
- Mecham J. O., Rowitch D., Wallace C. D., Stern P. H., Hoffman R. M. (1983). The metabolic defect of methionine dependence occurs frequently in human tumor cell lines. Biochem. Biophys. Res. Commun. 117, 429–434 10.1016/0006-291X(83)91218-4 [DOI] [PubMed] [Google Scholar]
- Meyskens F. L., Jr, Gerner E. W. (1999). Development of difluoromethylornithine (DFMO) as a chemoprevention agent. Clin. Can. Res. 5, 945–951 [PubMed] [Google Scholar]
- Neshat M. S., Raitano A. B., Wang H. G., Reed J. C., Sawyers C. L. (2000). The survival function of the Bcr-Abl oncogene is mediated by Bad-dependent and -independent pathways: roles for phosphatidylinositol 3-kinase and Raf. Mol. Cell. Biol. 20, 1179–1186 10.1128/MCB.20.4.1179-1186.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardee A. B. (1974). A restriction point for control of normal animal cell proliferation. Proc. Natl. Acad. Sci. USA 71, 1286–1290 10.1073/pnas.71.4.1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirson-Bichat F., Gonfalone G., Bras-Gonçalves R. A., Dutrillaux B., Poupon M. F. (1997). Growth of methionine-dependent human prostate cancer (PC-3) is inhibited by ethionine combined with methionine starvation. Br. J. Cancer 75, 1605–1612 10.1038/bjc.1997.274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichard J. F., Schnekenburger M., Puga A. (2007). Long term low-dose arsenic exposure induces loss of DNA methylation. Biochem. Biophys. Res. Commun. 352, 188–192 10.1016/j.bbrc.2006.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt H. C., Yaffe M. B. (2009). Kinases that control the cell cycle in response to DNA damage: Chk1, Chk2, and MK2. Curr. Opin. Cell Biol. 21, 245–255 10.1016/j.ceb.2009.01.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt H. C., Aslanian A. S., Lees J. A., Yaffe M. B. (2007). p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell 11, 175–189 10.1016/j.ccr.2006.11.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., Bar-Peled L., Sabatini D. M. (2008). The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501 10.1126/science.1157535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidenfeld J., Block A. L., Komar K. A., Naujokas M. F. (1986). Altered cell cycle phase distributions in cultured human carcinoma cells partially depleted of polyamines by treatment with difluoromethylornithine. Cancer Res. 46, 47–53 [PubMed] [Google Scholar]
- Sengupta T. K., Leclerc G. M., Hsieh-Kinser T. T., Leclerc G. J., Singh I., Barredo J. C. (2007). Cytotoxic effect of 5-aminoimidazole-4-carboxamide-1-beta-4-ribofuranoside (AICAR) on childhood acute lymphoblastic leukemia (ALL) cells: implication for targeted therapy. Mol. Cancer 6, 46 10.1186/1476-4598-6-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackelford D. B., Shaw R. J. (2009). The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat. Rev. Cancer 9, 563–575 10.1038/nrc2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin K. J., Wall E. A., Zavzavadjian J. R., Santat L. A., Liu J., Hwang J. I., Rebres R., Roach T., Seaman W., Simon M. I. et al. (2006). A single lentiviral vector platform for microRNA-based conditional RNA interference and coordinated transgene expression. Proc. Natl. Acad. Sci. USA 103, 13759–13764 10.1073/pnas.0606179103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern P. H., Wallace C. D., Hoffman R. M. (1984). Altered methionine metabolism occurs in all members of a set of diverse human tumor cell lines. J. Cell. Physiol. 119, 29–34 10.1002/jcp.1041190106 [DOI] [PubMed] [Google Scholar]
- Su N. Y., Flick K., Kaiser P. (2005). The F-box protein Met30 is required for multiple steps in the budding yeast cell cycle. Mol. Cell. Biol. 25, 3875–3885 10.1128/MCB.25.10.3875-3885.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale M. J., Jack G. W., Eridani S. (1983). Differential sensitivity of normal and leukaemic haemopoietic cells to methionine deprivation by L-methioninase. Leuk. Res. 7, 269–277 10.1016/0145-2126(83)90017-6 [DOI] [PubMed] [Google Scholar]
- Tseng C. H. (2008). Cardiovascular disease in arsenic-exposed subjects living in the arseniasis-hyperendemic areas in Taiwan. Atherosclerosis 199, 12–18 10.1016/j.atherosclerosis.2008.02.013 [DOI] [PubMed] [Google Scholar]
- Wallick C. J., Gamper I., Thorne M., Feith D. J., Takasaki K. Y., Wilson S. M., Seki J. A., Pegg A. E., Byus C. V., Bachmann A. S. (2005). Key role for p27Kip1, retinoblastoma protein Rb, and MYCN in polyamine inhibitor-induced G1 cell cycle arrest in MYCN-amplified human neuroblastoma cells. Oncogene 24, 5606–5618 10.1038/sj.onc.1208808 [DOI] [PubMed] [Google Scholar]
- Wang W., Kramer P. M., Yang S., Pereira M. A., Tao L. (2001). Reversed-phase high-performance liquid chromatography procedure for the simultaneous determination of S-adenosyl-L-methionine and S-adenosyl-L-homocysteine in mouse liver and the effect of methionine on their concentrations. J. Chromatogr. B Biomed. Sci. Appl. 762, 59–65 10.1016/S0378-4347(01)00341-3 [DOI] [PubMed] [Google Scholar]
- Winzen R., Kracht M., Ritter B., Wilhelm A., Chen C. Y., Shyu A. B., Müller M., Gaestel M., Resch K., Holtmann H. (1999). The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 18, 4969–4980 10.1093/emboj/18.18.4969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Y., Miskimins W. K. (2008). Cell cycle arrest in Metformin treated breast cancer cells involves activation of AMPK, downregulation of cyclin D1, and requires p27Kip1 or p21Cip1. J. Mol. Signal. 3, 18 10.1186/1750-2187-3-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.