Abstract
In spite of decades of research, our knowledge of the mode of interaction of macrolide antibiotics with their ribosomal target and of the mechanism of action of these drugs remain fragmentary. Experimental facts obtained over the last several years question some of the concepts that were viewed as a ‘common knowledge’. This review focuses on certain aspects of binding and action of macrolides that may need re-evaluation in view of the new findings.
Introduction
Six decades after the discovery of macrolides and their introduction into medical practice we still have only a sketchy understanding of how these drugs work. While some aspects of macrolide action have been firmly established, many points which are presumed to be well known are based on tentative models and need to be re-evaluated in view of newer discoveries. Treating speculative evidence as known facts not only slows down the progress of obtaining a true understanding of how macrolides work but also impedes the progress of developing better drugs. In this review, I will touch upon several controversial aspects concerning the interaction of macrolides with the ribosome target and the molecular mechanisms of macrolide action.
Macrolide facts
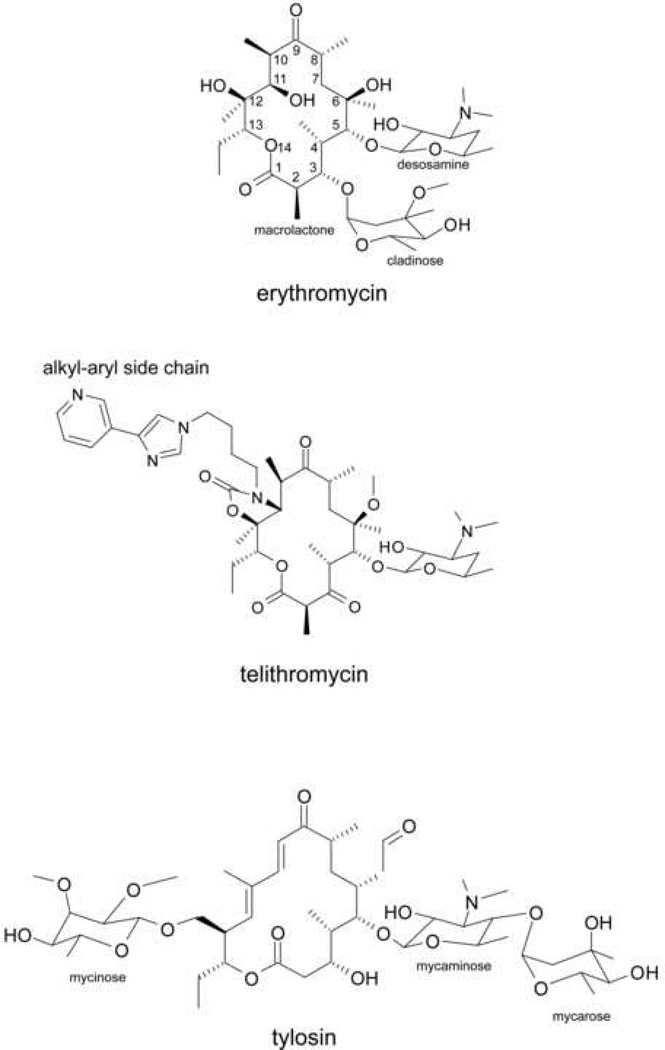
Macrolide antibiotics are composed of a 12- to 16-member macrolactone ring decorated with various amino-sugars (Figure 1). The target of action of macrolide antibiotics is the ribosome. The macrolide binding site is located in the large ribosomal subunit in the upper part of the nascent peptide exit tunnel (Figure 2). The exit tunnel connects the site of polypeptide assembly, the peptidyl transferase center, located at the ‘inner’ (interface) side of the large ribosomal subunit, with the protein release outlet on the ‘back’ (solvent) side of the subunit. The easy passage of the newly-made protein through the tunnel is critical for efficient protein synthesis. Binding of the macrolide antibiotics in the tunnel impedes progression of the nascent peptide and results in a general inhibition of translation.
Figure 1.
Chemical structures of macrolide antibiotics, inhibitors of protein synthesis. The major representatives of clinically-relevant classes of macrolide antibiotics are shown and structural elements relevant to the subject of the review are indicated.
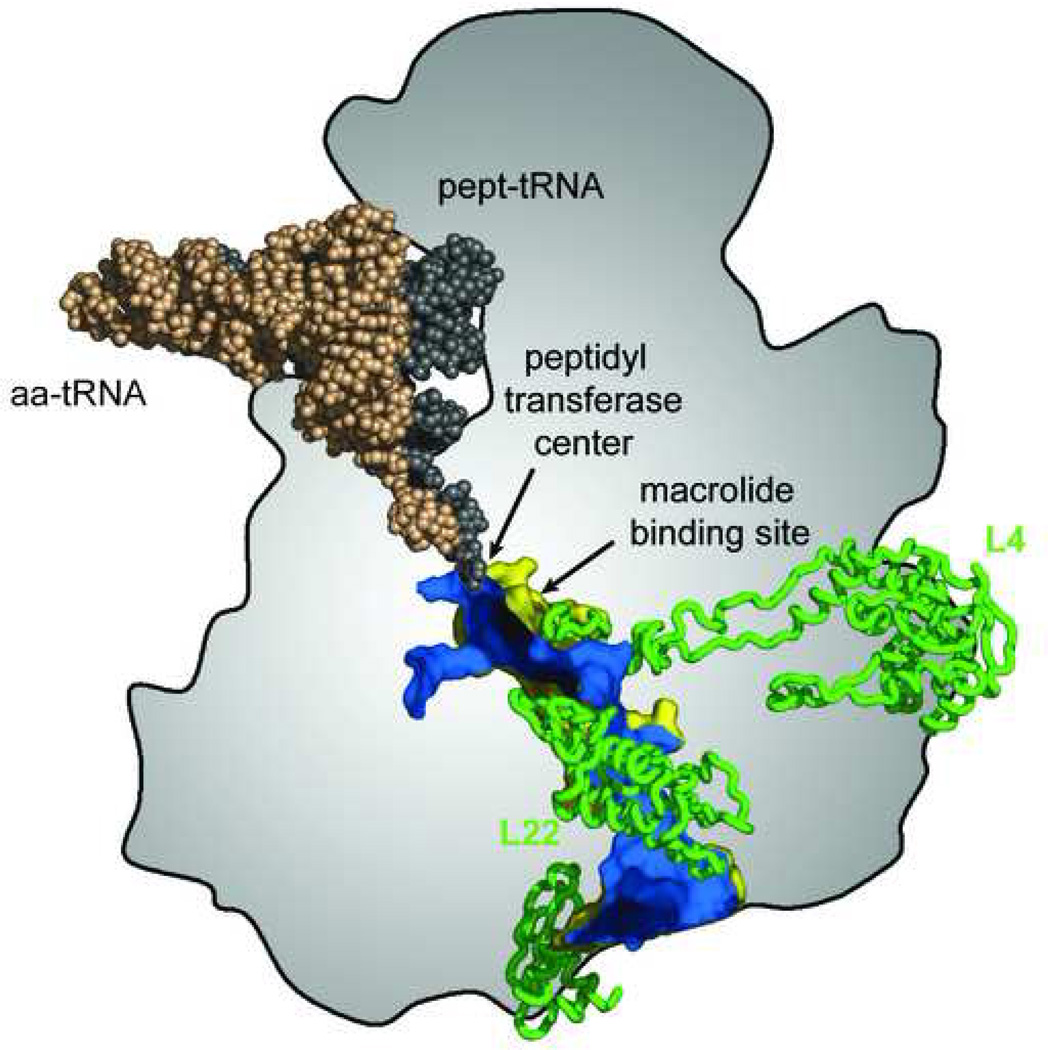
Figure 2.
The nascent peptide exit tunnel. 3’ ends of aminoacyl- and peptidyl-tRNAs converge in the peptidyl transferase center where assembly of amino acids into a polypeptide takes place. The nascent peptide traverses the body of the large ribosomal subunit through the exit tunnel whose outside surface is shown in yellow and inside surface is shown in blue. The tunnel walls are formed primarily by segments of rRNA but extended loops of ribosomal proteins L4 and L22 reach the tunnel near its constriction close to where the macrolide binding site is located. The location of the peptidyl transferase center and of the macrolide binding site near the tunnel constriction are indicated. Reproduced with minor changes from [36].
Genetic and biochemical analyses showed that 23S rRNA – the main structural and functional component of the large ribosomal subunit – is intimately involved in macrolide binding [1–6]. Crystallographic studies of ribosome-macrolide complexes confirmed that binding of macrolides to their ribosomal target site depends primarily on their interaction with rRNA [7–10]. Though some discrepancy remains between the published crystallographic structures, an overall consensus appears to emerge. The lactone ring binds to the wall of the tunnel due primarily to the hydrophobic interactions which involve, among others, residues A2058 and A2059 (E. coli numbering, here and throughout) (Figure 3). The C5-linked sugars (desosamine, in the case of erythromycin, clarithromycin, azithromycin and telithromycin or mycaminose-mycarose in the case of 16-member ring macrolides) project towards the peptidyl transferase center. Its hydrophobic and hydrogen bonding interactions with the 23S rRNA residues A2058 and A2059 contribute significantly to the binding energy of the drug. Contacts of the macrolactone ring and the C5-linked sugar residues involve exclusively rRNA and account for a significant portion of the drugs’ binding energy. Steric complementarity of the antibiotic molecule with the surface of the tunnel wall formed by residues A2058 and A2059 explains why the chemical structure of these nucleobases is critical for efficient binding of macrolides to their target: mutations at these residues or N6 dimethylation of A2058 by Erm-type methyltransferases negatively affects interactions with the drug [4,8,10,11].
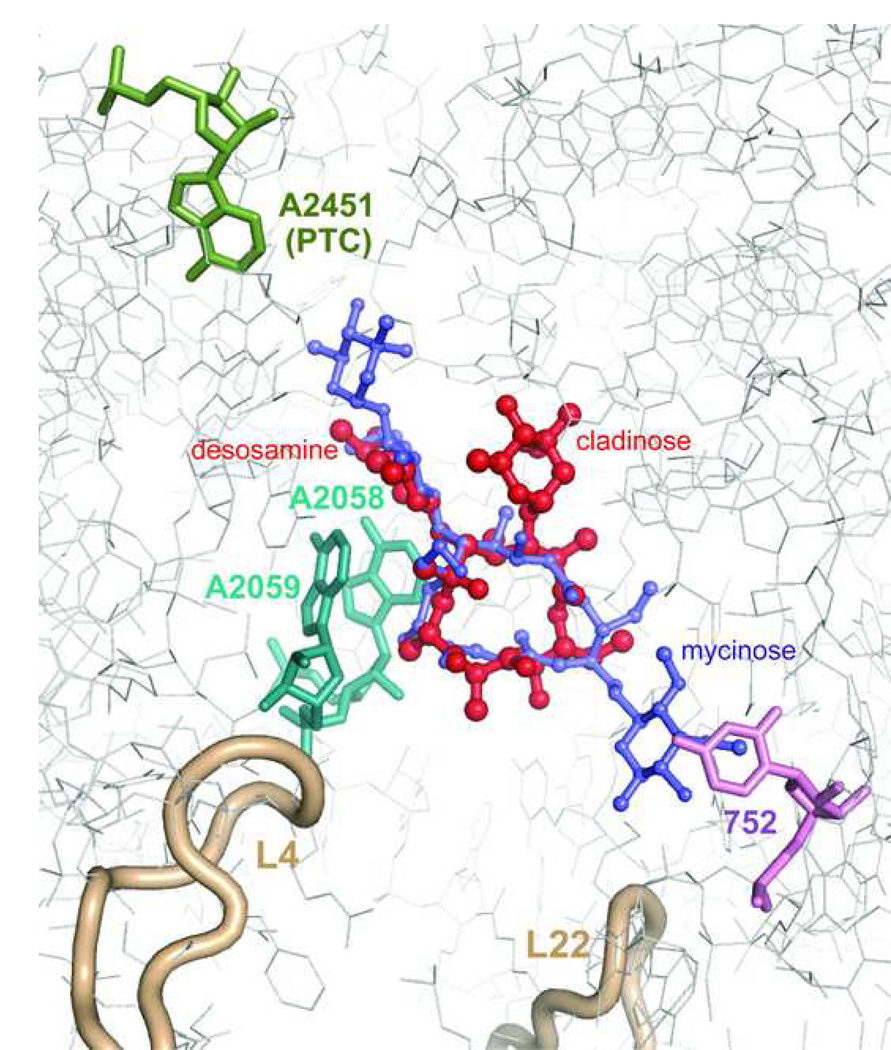
Figure 3.
Interactions of macrolides with the ribosome. Erythromycin (red) and tylosin (purple) are shown within their binding sites in the H. marismortui large ribosomal subunit [9,10]. A2451 which marks the location of the peptidyl transferase active site (PTC) is shown in green. Nucleotide residues A2058 and A2059 involved in interactions with desosamine sugar of erythromycin or mycaminose-mycarose disaccharide of tylosin (A2058 and A2059) are shown in cyan. U752 in H. marismortui (A752 in most pathogenic bacteria) interacting with the mycinose sugar of tylosin and likely involved in interactions with alkyl-aryl side chains of ketolides in some bacteria is shown in violet. Loops of ribosomal proteins L4 and L22 approaching the macrolide binding site are shown in beige.
Other sugar residues found in some of the macrolide antibiotics lend additional contacts with the ribosomal target. In particular, the C14-linked mycinose residue of tylosin projects down the tunnel, away from the peptidyl transferase center and interacts with the loop of helix 35 in domain II of 23S rRNA and with ribosomal protein L22. The extended alkyl-aryl side chain of clinically-relevant ketolides bound to the bacterial ribosome appears to project in the same direction and likely makes similar contacts although the controversy about placement of this important pharmacophore remains unresolved (see below) [5,6,12].
In spite of the fact that different macrolide antibiotics bind to the same site in the ribosome, their mode of action critically depends on the structure of the drug. The C5-linked disaccharides of 16-member ring macrolides are oriented similarly to the desosamine of 14- and 15-member ring macrolides. The C5-disaccharide reach into the peptidyl transferase center and can directly inhibit peptide bond formation [9,13]. Macrolides with a shorter (desosamine) C5-side chain do not interfere with peptide bond formation but, because of steric hindrance, block elongation of longer nascent peptides [14]. Overall, the inhibition of protein synthesis by macrolides likely results from the rapid drop-off of the peptidyl-tRNA from the ribosome during early rounds of translation [15–17]. In addition, similar to many other protein synthesis inhibitors, macrolides interfere with ribosome assembly [18].
Macrolide myths
Myth 1. Macrolides bind in the same way to ribosomes from different species
Ribosomal RNA, the main structural and functional component of the ribosome, shows an extremely high degree of evolutionary conservation among species. Many rRNA residues, especially those located in important functional centers of the ribosome, and thus, in the sites of action of many ribosome-targeting antibiotics, are invariable among bacteria. Because of the conservation of ribosomal structure and function, it is often assumed that drugs bind in the same, or in very similar ways to ribosomes isolated from different bacterial species. As a consequence, crystallographic, genetic and biochemical data obtained with laboratory models of drug-ribosome complexes are (sometimes indiscriminately) used to guide development of the drugs that would act upon ribosomes of ‘real life’ pathogenic microorganisms.
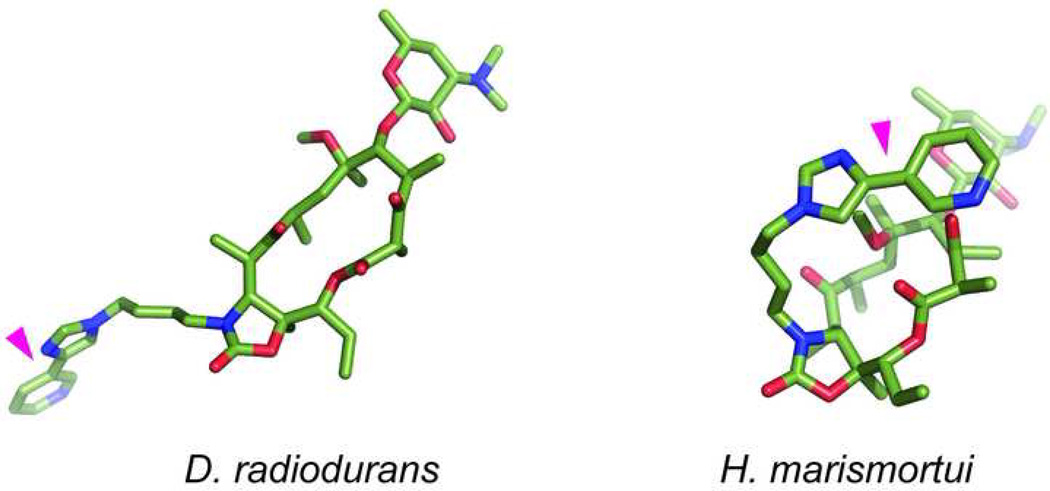
Although we can be fairly certain that the general location of the macrolide binding site is the same in ribosomes of different bacteria, this does not necessarily mean that all the molecular interactions of the drugs with the ribosome, or even the conformation of the ribosome-bound antibiotic is preserved. A rather striking example of species-specific interactions of macrolides with the ribosome is revealed by analyzing the binding of ketolides, which represent the newest generation of macrolide antibiotics. Ketolides derive their name from the C3-linked keto group which replaces the cladinose residue found in the 14- and 15-member ring macrolides of the previous generations (Figure 1). In addition, therapeutically-active ketolides possess 11,12-linked carbamate and, importantly, an extended alkyl-aryl side chain which is important for the binding and action of ketolides. All macrolides, when bound to the ribosome, universally protect two residues, A2058 and A2059 in domain V of 23S rRNA from chemical modification. Footprinting studies carried out with the E. coli ribosome showed that ketolides in addition protect A752 in the loop of helix 35 in domain II of 23S rRNA. Such protection, which strictly depends on the presence of an alkyl-aryl side chain, was interpreted as an indication of a direct interaction of the side chain with the A752 residue and implies that the alkyl-aryl side chain stretches down the exit tunnel away from the peptidyl transferase center [5,6,19,20]. Subsequent crystallographic studies of the Deinococcus radiodurans ribosome complexed with ketolides confirmed such general orientation of the side chain. However, no direct contact with A752 was observed [7,12]. Instead, the side chains of the studied ketolides closely approached the nucleotide residue 790 located at a distance of 11 Å from A752. When the structure of telithromycin complexed with the ribosome of halophilic archaeon Haloarcula marismortui was solved, the placement of the side chain was totally different from that observed in the D. radiodurans complex [10]. In the telithromycin molecule bound to the archaeal ribosome, the side chain was folded over the plain of the macrolactone ring oriented in the direction opposite to that observed in the bacterial ribosome (Figure 4). Simultaneous footprinting studies of telithromycin complexed with the ribosomes isolated form E. coli, Staphylococcus aureus, D. radiodurans and Halobacterium halobium (a close relative of H. marismortui) corroborated crystallographic structures and confirmed species-specific interactions of the drug: telithromycin protected A752 in the E. coli and S. aureus ribosomes, whereas no protection of the position 752 was observed when the drugs were complexed to the D. radiodurans or H. halobium ribosomes (Xiong and Mankin, unpublished). All of these observations consistently point that the placement of the pharmacophoric side chain of ketolides varies when the drug binds to ribosomes from different species.
Figure 4.
Different orientation of the alkyl-aryl side chain (indicated by magenta triangle) of telithromycin bound to the D. radiodurans (left) or H. marismortui (right) large ribosomal subunit.
The same general conclusion pointing to species-specific interactions of macrolides with the ribosome follows also from analysis of resistance mutations: similar mutations in different bacteria may confer somewhat different resistance profiles [4,21].
Thus, it is important to keep in mind that macrolide binding data are not always portable: one should exercise considerable caution when extrapolating facts obtained with laboratory model organisms for developing drugs that target ribosomes of ‘real world’ pathogens.
Myth 2. Selectivity of macrolides is determined by the nature of the 2058 residue in the large ribosomal subunit rRNA
Macrolides exert their inhibitory action on protein synthesis and cell growth in bacteria, but not in archaea and eukaryotes. Such selectivity correlates with the tight binding of erythromycin to bacterial ribosomes and its negligible binding to the ribosomes isolated from archaea or the cytoplasm of eukaryotic cells [10,19,20,22]. In their binding site, erythromycin and other macrolides establish intimate interactions with the A2058 of 23S rRNA. Adenosine at the position 2058 is almost universally conserved in bacteria whereas in archaeal and eukaryotic mitochondrial ribosomes this position is occupied by G. The identity of the residue at position 2058 in 23S rRNA has been viewed as the key factor that determines selectivity of macrolide binding and action. Indeed, mutation of A2058 to G render bacteria resistant to macrolides [4], whereas replacement of G2058 in archaeal ribosomes with A notably increases their sensitivity to erythromycin [10] (Xiong and Mankin, unpublished). This conclusion does not, however, hold true for at least some eukaryotic cells. Recent experiments of Zengel and coworkers [22] showed that mutant yeast cells in which protein synthesis was carried out by ribosomes containing adenosine at the rRNA position equivalent to the bacterial 2058 remain resistant to erythromycin. Furthermore, in the binding assay, the mutant (A2058) yeast ribosomes did not show appreciably higher affinity for the drug compared to the wild type (G2058) ribosomes. Thus, the identity of the 2058 residue is not the factor, or at least not the only factor, that determines the selectivity of action of macrolide antibiotics. In the absence of a high-resolution structure of eukaryotic ribosomes, we have no means of knowing how different the placement of nucleotide residues in the macrolide binding site in ribosomes of bacteria are from those of human cytoplasm. Even in spite of the conservation of most nucleotides constituting the binding site of macrolide antibiotics, their exact placement may critically depend on interactions with the other, less conserved rRNA residues or ribosomal proteins.
Myth 3. Macrolides plug the tunnel
Macrolides bind in the upper chamber of the exit tunnel near the constriction formed by the extended loops of proteins L4 and L22 (Figure 2). An important question is: does the bound molecule of a macrolide antibiotic completely plug the tunnel and prevent the passage of a nascent peptide or is it ‘just’ an obstacle which narrows the tunnel opening while leaving enough room for the nascent peptide to squeeze through? The initial prevailing view was that macrolides form an impassable barrier. This followed from the fact that homopolymeric nascent peptides synthesized by the ribosome in the presence of the drug were very short: 2–5 amino acids long [14,16]. Furthermore, when the first crystallographic structures of ribosome-macrolide complexes were unveiled, the impression was that the tunnel opening left by the antibiotic bound in the tunnel was too narrow for the passage of the nascent peptide [8,9].
The distance between the peptidyl transferase active site, where formation of peptide bonds takes place, and the macrolide binding site is only 10 Å. A 3–4 amino acid long nascent peptide should already reach the bound antibiotic (Figure 5). Yet, accurate measurements showed that peptidyl-tRNA dropped-off of the erythromycin-bound ribosome carried nascent peptides which were 6–8 amino acids long. Ribosomes with bound telithromycin could polymerize even longer peptides – 9–10 amino acid residues long - prior to dissociation of peptidyl-tRNA [17]. There does not appear to be enough room in the tunnel segment between the peptidyl transferase center and antibiotic to house such a long nascent peptide!
Figure 5.
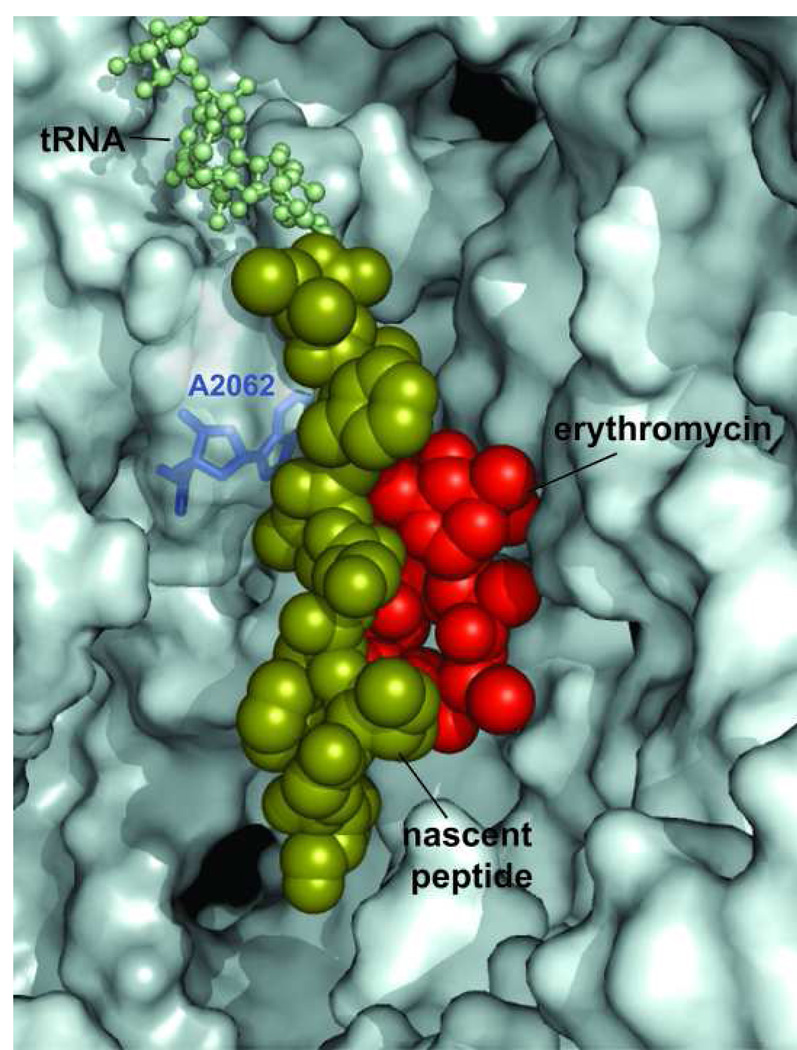
The bound molecule of erythromycin leaves sufficient space in the exit tunnel for the nascent peptide to squeeze by. The 9 amino acid-long nascent peptide (fMG-I-F-S-I-F-V-I) encoded in the ermCL regulatory ORF was modeled in the exit tunnel of the H. marismortui large ribosomal subunit complexed with erythromycin [10,24]. The erythromycin molecule is shown in red, the nascent peptide is shown in a space-fill representation in olive and CCA end of peptidyl-tRNA in the ribosomal P site is shown as balls-and-sticks (light-green). The A2062 residue of 23S rRNA, which needs to lie flat against the tunnel wall in order to let the nascent peptide slide by the bound macrolide molecule, is shown in blue.
The inducible expression of some macrolide-resistance genes require macrolide-dependent ribosome stalling at a regulatory open reading frame (ORF) preceding the resistance cistron. In the best studied case, ermC, stalling takes place when the ribosome, with erythromycin bound in the tunnel, reaches the ninth codon of the regulatory ORF ermCL [23] indicating that a 9-amino acid long nascent peptide can be accommodated in the drug-bound ribosome. Furthermore, in the mutant versions of the ermCL ORF, erythromycin-dependent stalling was observed at codons 10, 11 and even 12 (!) [24] reflecting the general capacity of the drug-bound ribosome to synthesize peptides of sizes unlikely to fit in the space left between the peptidyl transferase center and the bound macrolide molecule. Thus it is likely that at least some nascent peptides are able to squeeze through the opening left by the macrolide molecule in the exit tunnel. Indeed, modeling studies show that the aperture of the tunnel, when erythromycin or a similar drug is bound, is sufficient for accommodating an unfolded peptide chain (Figure 5) [10,24].
It is important to keep in mind that the exit tunnel is a dynamic structure. Reorientation of some nucleotide residues in response to antibiotic binding or specific nascent peptide sequences may significantly change both the geometry and volume of the tunnel. One of such flexible rRNA residues is A2062. In the presence of 16-member ring macrolides, the A2062 base, which in the absence of the drug lies flat against the tunnel wall, rotates into the tunnel lumen thus occluding the aperture of the tunnel to the extent that the progression of the nascent peptide past the bound antibiotic becomes impossible [9]. The drug-induced narrowing of the tunnel may in part account for the fact that the ribosome with the bound 16-member ring macrolide antibiotics drops off peptidyl-tRNA when the nascent peptide is only 2 to 4 amino acids long [17]. In the ribosome complexed with 14- and 15-member ring macrolides, A2062 lies parallel to the tunnel wall leaving enough space for the nascent peptide to slither past the bound drug (Figure 5).
The geometry of the tunnel and the ability of the nascent peptide to sneak by the bound antibiotics may be further influenced by the proteins L22 and L4 whose extended loops form parts of the wall at the tunnel constriction. Though the segments of the protein loops emerge in the tunnel down from the bound antibiotics (Figure 2), they may apparently influence the general geometry of the tunnel [10,25,26].
Even if macrolides do not completely plug the tunnel, they certainly hinder progression of the nascent peptide. So even if the nascent peptide can reach beyond the bound antibiotic, its synthesis will be imminently aborted through the peptidyl-tRNA drop-off mechanism before the nascent peptide reaches any substantially large size. Altogether, however, we can conclude that the ability of macrolides to block the passage of the nascent peptide through the tunnel is not absolute.
Myth 4. Macrolides affect synthesis of all proteins in the same way
The general view of the action of macrolides, as well as of most other ribosome-bound antibiotics, is that by blocking one of the ribosome functions they block production of all and any of the polypeptides equally well. Such a view, however, may be too simplistic. Bound to the ribosome, most of the antibiotics interact not only with the ribosome components but also with the ligands of protein synthesis: aminoacyl- and peptidyl-tRNAs. Such interactions can be influenced by the chemical nature of the RNA- and peptide moieties of the ligand and thus may vary for different polypeptides synthesized by the ribosome [27]. This is especially true when it comes to macrolides which, when bound in the ribosome tunnel, can interact with several amino acids of the growing polypeptide chain (Figure 5) and whose effect, therefore, can critically depend on the sequence of protein being synthesized.
The most profound example of the sequence-specific effect of macrolides on protein synthesis is the ribosome stalling at the regulatory ORFs of inducible erythromycin-resistance genes (already mentioned previously in this review). Only when the nascent peptide with a specific amino acid sequence is synthesized by the drug-bound ribosome does the stalling takes place [28]. Importantly, regulatory ORFs of various macrolide resistance genes encode substantially different stalling peptides [29] (Ramu, Mankin and Vazquez-Laslop, in preparation) indicating that a variety of specific nascent peptides can facilitate drug-dependent ribosome stalling. Therefore, the ribosome stalling whilst translating cellular proteins which fortuitously carry stalling sequences at their N-termini should result in unequal inhibitory actions of macrolides on production of different cellular polypeptides.
Sequence-specific effects of macrolides on protein synthesis is further revealed by the observation that nascent peptides with certain amino acid sequences can evict the drug from the ribosome [30–32]. Due to this effect, overproduction of specific short peptides in the cell can increase the overall fraction of drug-free ribosomes and confer macrolide resistance [30,33]. By extrapolating this finding one would expect that synthesis of some of the natural polypeptides, which carry the ‘macrolide-evicting’ sequences at their N-termini will be refractory to macrolide action [34,35]. Indeed, experimental investigation of the proteins synthesized in E. coli cells treated with erythromycin and telithromycin revealed that certain polypeptides continue to be produced when synthesis of most other polypeptides has been completely shut down (Vazquez-Laslop and Mankin, unpublished).
The spectrum of proteins that can escape the inhibitory action of an antibiotic as well as those whose synthesis is hypersusceptible to macrolide inhibition may be drug-specific and may have a profound effect on the kinetics of inhibition of cell growth as well as the bactericidal versus bacteriostatic effect of the drug.
Concluding remarks
Our attempts to understand how drugs, including the important class of macrolides, inhibit protein synthesis is a dynamic process. The models, which accounted for the experimental facts known yesterday need to be re-evaluated today to account for new facts and the resulting new models may fail to explain the observations that will be gained tomorrow. It is important, therefore, to be able to critically evaluate concepts, which are sometimes considered ‘common knowledge’, in order to successfully move on with gaining deeper understanding of the mechanisms of drug action and developing new successful drugs. This review was an attempt to do exactly that!
Acknowledgements
The work in the author’s laboratory is supported by grants from the National Science Foundation (MCB-0515934) and National Institutes of Health (AI072445). I am grateful to Nora Vazquez-Laslop for comments and to Tanel Tenson for communicating the unpublished results.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Moazed D, Noller HF. Chloramphenicol, erythromycin, carbomycin and vernamycin B protect overlapping sites in the peptidyl transferase region of 23S ribosomal RNA. Biochimie. 1987;69:879–884. doi: 10.1016/0300-9084(87)90215-x. [DOI] [PubMed] [Google Scholar]
- 2.Sigmund CD, Morgan EA. Erythromycin resistance due to mutation in a ribosomal RNA operon of Escherichia coli. Proc Nal Acad Sci USA. 1982;79:5602–5606. doi: 10.1073/pnas.79.18.5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vester B, Garrett RA. A plasmid-coded and site-directed mutation in Escherichia coli 23S RNA that confers resistance to erythromycin: implications for the mechanism of action of erythromycin. Biochimie. 1987;69:891–900. doi: 10.1016/0300-9084(87)90217-3. [DOI] [PubMed] [Google Scholar]
- 4.Vester B, Douthwaite S. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob Agents Chemother. 2001;45:1–12. doi: 10.1128/AAC.45.1.1-12.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen LH, Mauvais P, Douthwaite S. The macrolide-ketolide antibiotic binding site is formed by structures in domains II and V of 23S ribosomal RNA. Mol Microbiol. 1999;31:623–632. doi: 10.1046/j.1365-2958.1999.01202.x. [DOI] [PubMed] [Google Scholar]
- 6.Xiong L, Shah S, Mauvais P, Mankin AS. A ketolide resistance mutation in domain II of 23S rRNA reveals proximity of hairpin 35 to the peptidyl transferase centre. Mol Microbiol. 1999;31:633–639. doi: 10.1046/j.1365-2958.1999.01203.x. [DOI] [PubMed] [Google Scholar]
- 7.Schlunzen F, Harms JM, Franceschi F, Hansen HA, Bartels H, Zarivach R, Yonath A. Structural basis for the antibiotic activity of ketolides and azalides. Structure. 2003;11:329–338. doi: 10.1016/s0969-2126(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 8. Schlunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. The first crystallographic structure of macrolide-ribosome complexes.
- 9.Hansen JL, Ippolito JA, Ban N, Nissen P, Moore PB, Steitz TA. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol Cell. 2002;10:117–128. doi: 10.1016/s1097-2765(02)00570-1. [DOI] [PubMed] [Google Scholar]
- 10. Tu D, Blaha G, Moore PB, Steitz TA. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. Currently, the highest resolution structures of macrolide antibiotics complexed to the ribosomal subunit.
- 11.Weisblum B. Erythromycin resistance by ribosome modification. Antimicrob Agents Chemother. 1995;39:577–585. doi: 10.1128/AAC.39.3.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berisio R, Harms J, Schluenzen F, Zarivach R, Hansen HA, Fucini P, Yonath A. Structural insight into the antibiotic action of telithromycin against resistant mutants. J Bacteriol. 2003;185:4276–4279. doi: 10.1128/JB.185.14.4276-4279.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poulsen SM, Kofoed C, Vester B. Inhibition of the ribosomal peptidyl transferase reaction by the mycarose moiety of the antibiotics carbomycin, spiramycin and tylosin. J Mol Biol. 2000;304:471–481. doi: 10.1006/jmbi.2000.4229. [DOI] [PubMed] [Google Scholar]
- 14.Mao JC-H, Robishaw EE. Effects of macrolides on peptide-bond formation and translocation. Biochemistry. 1971;10:2054–2061. doi: 10.1021/bi00787a014. [DOI] [PubMed] [Google Scholar]
- 15.Menninger JR, Otto DP. Erythromycin, carbomycin, and spiramycin inhibit protein synthesis by stimulating the dissociation of peptidyl-tRNA from ribosomes. Antimicrob Agents Chemother. 1982;21:810–818. doi: 10.1128/aac.21.5.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Otaka T, Kaji A. Release of (oligo) peptidyl-tRNA from ribosomes by erythromycin A. Proc Natl Acad Sci USA. 1975;72:2649–2652. doi: 10.1073/pnas.72.7.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tenson T, Lovmar M, Ehrenberg M. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J Mol Biol. 2003;330:1005–1014. doi: 10.1016/s0022-2836(03)00662-4. The accurate characterization of peptidyl-tRNAs dissociated from the ribosome due to macrolide-induced drop-off.
- 18.Champney WS, Burdine R. Macrolide antibiotics inhibit 50S ribosomal subunit assembly in Bacillus subtilis and Staphylococcus aureus. Antimicrob Agents Chemother. 1995;39:2141–2144. doi: 10.1128/aac.39.9.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Douthwaite S, Hansen LH, Mauvais P. Macrolide-ketolide inhibition of MLS-resistant ribosomes is improved by alternative drug interaction with domain II of 23S rRNA. Mol Microbiol. 2000;36:183–193. doi: 10.1046/j.1365-2958.2000.01841.x. [DOI] [PubMed] [Google Scholar]
- 20.Xiong L, Korkhin Y, Mankin AS. Binding site of the bridged macrolides in the Escherichia coli ribosome. Antimicrob Agents Chemother. 2005;49:281–288. doi: 10.1128/AAC.49.1.281-288.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pfister P, Corti N, Hobbie S, Bruell C, Zarivach R, Yonath A, Bottger EC. 23S rRNA base pair 2057–2611 determines ketolide susceptibility and fitness cost of the macrolide resistance mutation 2058A-->G. Proc Natl Acad Sci USA. 2005;102:5180–5185. doi: 10.1073/pnas.0501598102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bommakanti AS, Lindahl L, Zengel JM. Mutation from guanine to adenine in 25S rRNA at the position equivalent to E. coli A2058 does not confer erythromycin sensitivity in Sacchromyces cerevisae. RNA. 2008;14:460–464. doi: 10.1261/rna.786408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayford M, Weisblum B. ermC leader peptide. Amino acid sequence critical for induction by translational attenuation. J Mol Biol. 1989;206:69–79. doi: 10.1016/0022-2836(89)90524-x. [DOI] [PubMed] [Google Scholar]
- 24. Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Mol Cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. Identification of ribosome elements required for the formation of macrolide-dependent stalled translation complex.
- 25.Gregory ST, Dahlberg AE. Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23 S ribosomal RNA. J Mol Biol. 1999;289:827–834. doi: 10.1006/jmbi.1999.2839. [DOI] [PubMed] [Google Scholar]
- 26.Gabashvili IS, Gregory ST, Valle M, Grassucci R, Worbs M, Wahl MC, Dahlberg AE, Frank J. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol Cell. 2001;8:181–188. doi: 10.1016/s1097-2765(01)00293-3. [DOI] [PubMed] [Google Scholar]
- 27.Tenson T, Mankin AS. Antibiotics and the ribosome. Mol Microbiol. 2006;59:1664–1677. doi: 10.1111/j.1365-2958.2006.05063.x. [DOI] [PubMed] [Google Scholar]
- 28.Weisblum B. Insights into erythromycin action from studies of its activity as inducer of resistance. Antimicrob Agents Chemother. 1995;39:797–805. doi: 10.1128/aac.39.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwon AR, Min YH, Yoon EJ, Kim JA, Shim MJ, Choi EC. ErmK leader peptide : amino acid sequence critical for induction by erythromycin. Arch Pharm Res. 2006;29:1154–1157. doi: 10.1007/BF02969307. [DOI] [PubMed] [Google Scholar]
- 30.Tenson T, DeBlasio A, Mankin A. A functional peptide encoded in the Escherichia coli 23S rRNA. Proc Natl Acad Sci USA. 1996;93:5641–5646. doi: 10.1073/pnas.93.11.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tripathi S, Kloss PS, Mankin AS. Ketolide resistance conferred by short peptides. J Biol Chem. 1998;273:20073–20077. doi: 10.1074/jbc.273.32.20073. [DOI] [PubMed] [Google Scholar]
- 32.Lovmar M, Nilsson K, Vimberg V, Tenson T, Nervall M, Ehrenberg M. The molecular mechanism of peptide-mediated erythromycin resistance. J Biol Chem. 2006;281:6742–6750. doi: 10.1074/jbc.M511918200. [DOI] [PubMed] [Google Scholar]
- 33.Tenson T, Xiong L, Kloss P, Mankin AS. Erythromycin resistance peptides selected from random peptide libraries. J Biol Chem. 1997;272:17425–17430. doi: 10.1074/jbc.272.28.17425. [DOI] [PubMed] [Google Scholar]
- 34.Lovmar M, Tenson T, Ehrenberg M. Kinetics of macrolide action: The josamycin and erythromycin cases. J Biol Chem. 2004;279:53506–53515. doi: 10.1074/jbc.M401625200. [DOI] [PubMed] [Google Scholar]
- 35.Macvanin M, Gonzalez de Valdivia EI, Ardell DH, Isaksson LA. Transient erythromycin resistance phenotype associated with peptidyl-tRNA drop-off on early UGG and GGG codons. J Bacteriol. 2007;189:8993–9000. doi: 10.1128/JB.01004-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jenni S, Ban N. The chemistry of protein synthesis and voyage through the ribosomal tunnel. Curr Opin Struct Biol. 2003;13:212–219. doi: 10.1016/s0959-440x(03)00034-4. [DOI] [PubMed] [Google Scholar]