Abstract
RNAi is conserved and has been studied in a broad cross-section of the fungal kingdom, including Neurospora crassa, Schizosaccharomyces pombe, Cryptococcus neoformans, and Mucor circinelloides. And yet well known species, including the model yeast Saccharomyces cerevisiae and the plant pathogen Ustilago maydis, have lost RNAi, providing insights and opportunities to illuminate benefits conferred both by the presence of RNAi and its loss. Some of the earliest studies of RNAi were conducted in Neurospora, contemporaneously with the elucidation of RNAi in C. elegans. RNAi is a key epigenetic mechanism for maintaining genomic stability and integrity, as well as to defend against viruses, and given its ubiquity was likely present in the last eukaryotic common ancestor. In this review we describe the diversity of RNAi mechanisms found in the fungi, highlighting recent work in Neurospora, S. pombe, and Cryptococcus. Finally we consider frequent, independent losses of RNAi in diverse fungal lineages and both review and speculate on evolutionary forces that may drive the losses or result therefrom.
Keywords: RNAi, Fungi, Quelling, MSUD, Sex-Induced Silencing, miRNA
Introduction
RNAi is a robust gene regulatory system that triggers silencing of transcripts via the detection of double-stranded RNA. In its most basic form, RNAi is driven by two canonical components: a double-strand RNA endonuclease (Dicer) and a small RNA-binding protein (Argonaute). Dicer typically acts on dsRNA to produce small RNAs from 20–30 bases in length that are then loaded onto Argonaute and used to identify complementary messages, triggering further degradation of the target message. Argonaute is the catalytic subunit of what is termed the RNA-induced silencing complex (RISC). In addition, there is a third canonical component, the RNA-dependent RNA polymerase, that can amplify this signal by generating additional dsRNA to trigger Dicer and Argonaute. In some cases RdRP can be essential for RNAi as it generates the initial dsRNA from ssRNA templates or targets. RNAi is involved in a broad spectrum of functions, including genome defense against viral or transposon invasion, and also regulates diverse cellular and developmental processes.
In this review we first describe RNAi-dependent silencing mechanisms in fungi. We will then discuss the biological functions of RNAi in the context of loss and retention of the RNAi pathway across the fungal kingdom. Many of the RNAi phenomena identified in fungi are better understood mechanistically than they are in a functional, biological context. By bringing together these two areas, we hope to emphasize some of the hypotheses for the inherent biological functions that RNAi serves in the highly diverse lineages of the fungal kingdom.
Homology Dependent Gene Silencing
The development of gene transfer methods has allowed transgenic DNA to be integrated into the genome and transmitted as a heritable Mendelian trait in transgenic lines. However, the introduced transgenes are in some case silenced and can also cause silencing of endogenous genes that share sufficient homology. This phenomenon has been termed homology dependent gene silencing (HDGS), co-suppression, or transgene silencing and has been found in myriad organisms including plants, animals, ciliates, and fungi (Matzke et al., 1989, Jorgensen, 1990, Cogoni and Macino, 1999a, Hannon, 2002, Moazed, 2009). HDGS usually occurs when multiple copies of particular sequences are present in the genome. Thus, transposable elements, which are repetitive sequences, are also subject to HDGS. One central mechanism involved in HDGS is RNA interference (RNAi) in which small RNAs (~20 to ~30 nt) homologous to the repetitive sequences are generated and orchestrate gene silencing posttranscriptionally (Catalanotto et al., 2002, Catalanotto et al., 2004) or transcriptionally (Volpe et al., 2002, Zofall and Grewal, 2006, Moazed, 2009), in a sequence-specific manner.
In fungi, quelling is the best-characterized RNAi-dependent HDGS process and the first to be discovered (Romano and Macino, 1992). Quelling occurs in the vegetative tissue of the filamentous fungus Neurospora crassa and is mediated by sRNA (Cogoni et al., 1996). Studies on quelling have uncovered key RNAi components and functions, contributing significantly to advance the RNAi field (Cogoni et al., 1996, Cogoni and Macino, 1997, Cogoni and Macino, 1999a, Cogoni and Macino, 1999c, Dang et al., 2011). Quelling is a post-transcriptional gene silencing (PTGS) process that is induced by aberrant RNA (aRNA) and requires the core RNAi components, including Dicer-like proteins, Argonaute, and RdRP (Cogoni and Macino, 1999a, Lee et al., 2003, Fulci and Macino, 2007). In quelling, the silenced loci can act in trans, leading to silencing of some or all homologous genes.
The first eukaryotic RNAi component identified was the RNA-dependent RNA polymerase QDE-1 and it is required for quelling (Cogoni and Macino, 1999a). It was identified shortly after the silencing effect of injected dsRNA was found in C. elegans and the discovery of QDE-1 contributed to elucidate the role of dsRNAs in RNAi (Fire et al., 1998). The Argonaute protein QDE-2 and the Dicer-like proteins DCL-1 and DCL-2 constitute the main core components of the quelling pathway and are highly conserved among eukaryotes (Catalanotto et al., 2000, Catalanotto et al., 2004). Interestingly, QDE-3 is a RecQ helicase that usually participates in homologous recombination and DNA repair in N. crassa, but also plays an important role in quelling. QDE-3 may recognize transposable elements or transgenes that contain inverted repeats and recruit QDE-1 to generate aberrant RNA (aRNA) that is then transcribed into dsRNAs. The dsRNA is bound by the dicers DCL-1 and DCL-2 and processed into siRNAs of 20–25 nt that are loaded onto RISC, formed by Argonaute QDE-2 and QIP, a QDE-2-interacting exonuclease (Maiti et al., 2007).
DNA-damage also induces the expression of the quelling Argonaute protein QDE-2 and a novel class of QDE-2 associated sRNAs, designated qiRNAs (Lee et al., 2009). These DNA damage-induced sRNAs derive from the highly repetitive sequences of rDNA loci, are shorter in length (~21 nt) than standard siRNA, and require similar components to quelling (QDE-1, QDE-3, and the Dicers), indicating the two pathways are mechanistically related (Lee et al., 2009, Lee et al., 2010a). Moreover, the two pathways share the same initial signal, which is highly repetitive sequences, although for quelling they are exogenous (transposon or viruses) and for qiRNAs they are endogenous (repetitive rDNA). Recent studies showed sRNAs regulate the DNA damage response pathway in rice and the production of qiRNAs from rDNA loci may be a novel mechanism of DNA repair in plants (Wei et al., 2012, Chen et al., 2013). DNA damage-induced sRNAs have been detected in flies and mammals, indicating that this mechanism is conserved across eukaryotes (Francia et al., 2012, Michalik et al., 2012).
DNA damage induces cell-cycle arrest, delays DNA replication, and reduces protein synthesis. The accumulation of qiRNAs indicates that DNA damage may cause stress on DNA replication of repetitive sequences and it may act as a DNA damage checkpoint control by inhibiting protein synthesis. Recent findings demonstrate that homologous recombination and chromatin remodeling factors, including Rad51, Rad52, and Rad54, are essential for qiRNAs and homologous recombination can distinguish between endogenous and exogenous repetitive sequences and activate DNA damage response sRNAs and quelling respectively (Zhang et al., 2013).
Meiotic Silencing by Unpaired DNA
In addition to quelling, the study of RNAi in N. crassa has revealed surprisingly diverse types of sRNAs with different biogenesis pathways and functions. During sexual development, the presence of unpaired DNA initiates an RNAi-related mechanism termed meiotic silencing by unpaired DNA (MSUD). During the sexual cycle, haploid hyphae of the opposite mating type of N. crassa fuse to produce a transient heterokaryon that, following nuclear fusion to the diploid, undergoes one round of meiosis and one of mitosis to yield 8 ascospores. The presence of unpaired DNA in the prophase of meiosis I initiates silencing of the unpaired DNA, and also of genes homologous to the unpaired DNA in the diploid ascus, although this silencing is not always completely penetrant (Aramayo and Metzenberg, 1996, Shiu et al., 2001). It is interesting that MSUD triggers silencing of all of the genes homologous with the unpaired DNA, even copies that may be paired, indicating an initial trans-sensing step signals the MSUD pathway (Aramayo and Metzenberg, 1996, Shiu et al., 2001). This signal triggers the production of unpaired-DNA-specific aRNA in the nucleus that is converted to dsRNA by SAD-1, an RdRP, SAD-2, a scaffold protein that localizes and physically interacts with SAD-1 in the perinuclear region, and SAD-3, an RNA/DNA helicase (Shiu et al., 2001, Pratt et al., 2004, Chang et al., 2012, Hammond et al., 2013). dsRNA is cleaved by DCL-1, a Dicer-like protein, and the 20–25 nt sRNAs produced then bind SMS-2, an Argonaute homologue, and QIP, an exonuclease, driving posttranscriptional silencing of the genes homologous to the unpaired DNA (Maine et al., 2005, Turner et al., 2005, Lee et al., 2009, Son et al., 2011).
One region of the genome that is naturally unpaired is the mating type locus, at which there are two idiomorphic, unrelated sequences, mata and matA. It is not known if MSUD operates at MAT or whether MAT is in some way protected from MSUD action. While heterozygosity at MAT is necessary for progression through the sexual cycle, it is formally possible that the action of both or even either MAT idiomorph is not required for progression through the stage of meiosis at which MSUD acts. Alternatively, the MAT locus may be uniquely marked in some fashion to render it immune, or resistant to MSUD. Studies to address this could examine the expression levels of MAT encoded genes during the sexual cycle in wild type vs. MSUD mutant strains, or seek to detect whether sRNA accumulates directed against the MAT locus during crosses.
Interestingly, quelling and MSUD function separately and employ different components, with the exception of DCL-1 and QIP, indicating that quelling and MSUD may have a shared ancestry but have diverged over time (Chang et al., 2012). Although the silencing pathway of MSUD has been extensively studied, little is known about the trans-sensing mechanism. There is some evidence that DNA methylation and chromatin structure may direct recognition of unpaired DNA during MSUD (Pratt et al., 2004). In addition, recent studies identified a novel protein, SAD-5, that is essential for MSUD and localizes in the nucleus, indicating a possible role in the initial stages of the pathway (Hammond et al., 2013). Surprisingly, silencing by unpaired DNA during meiosis has been observed in numerous species, including other fungi, C. elegans, D. melanogaster, and mice, indicating a highly conserved method for genome defense against exogenous DNA (Turner et al., 2005, Maine et al., 2005, Son et al., 2011, Duan et al., 2013).
Homology-dependent gene silencing can also operate in an RNAi-independent manner. This has most recently been shown in Aspergillus nidulans, where introduction of a transgenic copy of the matAHMG gene causes silencing of the endogenous matA. Unusually, this silencing is highly penetrant, and does not rely on the RNAi components, including Argonaute. The silencing is also recessive and fails to spread between nuclei within heterokaryons, unlike canonical silencing (Czaja et al., 2013). A number of similar RNAi-independent silencing mechanisms likely exist undiscovered in the fungi.
RNAi in Cryptococcus
The genome of the more evolutionarily distant basidiomycete and human fungal pathogen Cryptococcus neoformans also encodes a fully functional RNAi pathway (Loftus et al., 2005), and introduction of transgene-expressed dsRNA has been used to successfully repress expression of the target genes (Liu et al., 2002, Bose and Doering, 2011). Recently, a quelling like co-suppression phenomenon termed mitotic-induced silencing (MIS) was discovered that occurs during C. neoformans vegetative mitotic asexual growth (Wang et al., 2012). During screens to isolate cpa1 single mutant strains following introduction of a cpa1Δ::ADE2 disruption allele, some transformants that had decreased mRNA levels of both the CPA1 and CPA2 genes were obtained. CPA1 and CPA2 are two homologous genes and both encode cyclophilin A, the target of the immunosuppressive antifungal natural product cyclosporine A (CsA). Because both CPA1 and CPA2 are silenced in these transformants, they exhibit a phenotype similar to cpa1 cpa2 double mutants (resistance to CsA). Like quelling, this silencing process is dependent on the RNAi silencing pathway. First, the RNAi components Rdp1, Ago1, and Dcr2 are required for silencing. Second, siRNAs homologous to both CPA1 and CPA2 (by virtue of the high sequence identity it shares with CPA1) were detected in the silenced strains. The silencing efficiency of the CPA1 and CPA2 genes is correlated with the transgene copy number and reached ~90% in the presence of a >25-copy repeat transgene, while much lower (0.1%) or no silencing occurred in transformants containing the one or three copies of the transgene.
Sex-Induced Silencing
A related silencing process operates during the sexual cycle of C. neoformans. This transgene-induced posttranscriptional silencing mechanism is more active than in vegetative mitotic growth, and hence this phenomenon was named sex-induced silencing (SIS) (Wang et al., 2010). For example, silencing of a tandem insertion of a triplicated SXI2a-URA5 transgene during a-α opposite-sex reproduction occurs at 250-fold higher frequency than in vegetative mitotic asexual growth. Because C. neoformans can produce spores via two distinct mating pathways: a-α opposite sex and α- α unisexual reproduction, a subsequent study tested and demonstrated that SIS is not limited to a-α opposite-sex but also operates during α- α unisexual reproduction (Wang et al., 2013). More importantly, the silencing frequency observed during opposite-sex and unisexual reproduction is comparable. Similar to the mitotic induced co-suppression pathway in C. neoformans, SIS is an RNAi-related process and requires the presence of a multicopy transgene array.
Sequencing of small RNAs in C. neoformans under both mating and vegetative growth conditions revealed that abundant small RNAs map to repetitive transposable elements (Wang et al., 2010, Dumesic et al., 2013). In rdp1 mutant strains, siRNAs generated from these repetitive loci were abolished, supporting a role for RNAi in transposon control in C. neoformans. Furthermore, two lines of evidence may explain why C. neoformans evolved a more robust sex-related silencing mechanism (Wang et al., 2010, Wang et al., 2013). First, transposons and retrotransposons are highly expressed during sexual reproduction of RNAi mutant strains. Second, by employing the FKBP12 encoding gene FRR1 as a transposon trap, a much increased transposition/mutation rate was detected in progeny derived from the rdp1 mutant a-α opposite sex mating and α- α unisexual reproduction than during mitotic growth of the rdp1 mutant (Wang et al., 2010, Wang et al., 2013). This evidence supports models in which an efficient silencing mechanism is necessary to defend genomic integrity and reduce a potential higher mutational burden during sex. Thus, the robust nature of highly efficient SIS could be one strategy that C. neoformans deploys to squelch the activity of selfish DNA elements during mating and effectively guard the genome integrity of the progeny. Most interestingly, the RNAi machinery components are more abundant during mating or under mating growth conditions, supporting a model in which increased expression of RNAi machinery may function to silence potentially overexpressed transposons during mating (Wang et al., 2010). This increase in abundance does not occur at the transcriptional level of the RNAi components and thus may be attributable to either an RNA operon that enhances their coordinate translation or to regulation of protein localization/stability.
The mechanistic details of the initiation and effectiveness of SIS are not yet understood. A related central question is: how do cells recognize repetitive sequences and produce transgene-specific dsRNA? It was hypothesized in plants that a qualitatively aberrant feature of transgenic DNA or RNA (aDNA or aRNA), can trigger gene silencing (English et al., 1996, Cogoni et al., 1996). If so, stages of the sexual cycle involving the sequence homology search that mediates meiotic recombination may accelerate the formation of aberrant DNA-RNA hybrids in tandem repeat sequences, possibly producing templates for Rdp1, thus activating silencing in C. neoformans. With regard to quelling in N. crassa, it was proposed that large tandem repeats may produce aberrant DNA (aDNA) secondary structures that are recognized by the recQ DNA helicase Qde3 (Cogoni and Macino, 1999c). Then, by an unknown mechanism, Qde1, which can act as both a DNA-dependent RNA polymerase (DdRP) and as an RNA-dependent RNA polymerase, is recruited to the transgene loci and generates dsRNA using the single strand repetitive DNA and the first strand aRNA as the template (Lee et al., 2009). Replication protein A, a ssDNA-binding protein complex, has recently been shown to function during the recruiting steps and promotes dsRNA formation (Lee et al., 2010a). In C. neoformans, two components of the RPA complex, Rpa32 and Rpa70, are also required for transgene silencing (Wang et al., 2012), suggesting similar initiation steps are involved as in quelling (Cogoni and Macino, 1999b). However, a component in C. neoformans that functions like Qde3 to recognize aDNA/aRNA structures has not yet been identified.
Heterochromatin formation
Heterochromatin formation induced by RNAi has been extensively studied in S. pombe over the last decade (Volpe et al., 2002, Chang et al., 2012, Reyes-Turcu and Grewal, 2012). dh and dg repeats are characteristic of the heterochromatin domains at centromeres, subtelomeres, and the MAT locus (Hall et al., 2002, Volpe et al., 2002). Transcripts produced by RNA polymerase II carrying these repetitive sequences are substrates for the RNA-dependent RNA polymerase complex (RDRC), which includes the RNA-dependent RNA polymerase Rdp1 (Motamedi et al., 2004). This enzyme synthesizes the complementary strand to generate dsRNA that is then processed into siRNAs by the Dicer protein Dcr1. The RNA-induced transcriptional silencing (RITS) complex, containing an Argonaute protein (Ago1), binds the siRNA and reinforces the silencing mechanism by facilitating the localization of Rdp1 to the nascent target complementary to the siRNA (Verdel et al., 2004). In addition to RDRC, the histone methyltransferase Clr4 (homologue of mammalian SUV39H) is also recruited to the DNA target where it methylates histone 3 at lysine 9 (H3K9me), a landmark feature of heterochromatin (Motamedi et al., 2004, Zhang et al., 2008, Bayne et al., 2010).
Transient heterochromatin has also been detected over convergent genes during the G1-S phase transition (Gullerova and Proudfoot, 2008). Read-through transcription that occurs frequently during this phase of the cell cycle generates dsRNA from overlapping transcripts at these loci to activate RNAi. The formation of heterochromatin then promotes the recruitment of cohesin, which blocks transcriptional read-through during G2 (Lengronne et al., 2004, Gullerova and Proudfoot, 2008). This transient heterochromatin is a key regulatory mechanism for RNAi genes themselves, as most feature convergent configuration. Perturbation of this orientation dysregulates their expression and causes aberrant cell morphology (Gullerova et al., 2011).
RNAi mechanisms operating in S. pombe go beyond the silencing and maintenance of centromeres, telomeres, or convergent genes. In addition, RNAi suppresses the expression of antisense RNAs at euchromatic loci (Gullerova and Proudfoot, 2008, Zofall et al., 2009), competing with RNA quality control factors such as the exosome, a multi-protein complex involved in RNA degradation (Zhang et al., 2011). RNAi components interact, through Clr4 and Mlo3 (an mRNA export protein (Thakurta et al., 2005)), with the Trf4p/Air2p/Mtr4p polyadenylylation complex (TRAMP) (Zhang et al., 2011), an exosome cofactor that stimulates the degradation of aberrant RNA (Houseley et al., 2006, Zhang et al., 2011). Mlo3, perhaps in cooperation with TRAMP, could be responsible of driving the RNAs into RNAi or exosome pathways (Zhang et al., 2011). This collaborative mechanism is conserved in Drosophila and has thus far been implicated in the silencing of sexual differentiation genes, genes encoding transmembrane proteins, and Tf2 retrotransposons, implying that RNAi may serve as an adaptive response mechanism for development and sensing of environmental signals (Yamanaka et al., 2013).
Interestingly, recent work in Cryptococcus has also begun to forge a link between RNAi and mRNA processing. Delays in splicing trigger RNAi-dependent degradation of the stalled message via a nuclear RNAi complex termed SCANR (Spliceosome-Coupled and Nuclear RNAi) (Dumesic et al., 2013). This complex contains a canonical RdRP, Argonaute, and QIP in association with the Srr1 splicing component. As with the case of exosome studies in S. pombe, this work is beginning to show a previously unsuspected link between mRNA processing and silencing. This may represent the RNA equivalent of mismatch repair, effectively proofreading transcripts as they are produced and processed. Further, it also represents a mechanism for distinguishing “self” from “non-self.” If all native transcripts are selected for efficiently spliced introns, exogenous elements with less optimal, more poorly spliced introns will have a difficult time integrating into the genome and being expressed.
miRNAs
miRNAs are small endogenous non-coding RNAs derived from precursor transcripts with a hairpin structure. Two different Dicer and dsRNA-binding domain (dsRBD) proteins process the precursors, producing 20–24 nt small RNAs that bind to Argonaute and regulate target expression by endonucleolytic cleavage or translational repression of the mRNA (Ghildiyal and Zamore, 2009). miRNAs have been extensively studied in animals, plants, and algae, but fungi were assumed to lack this pathway. However, miRNA-like small RNAs (milRNAs) have been found in N. crassa, and four different classes have been described so far that are distinguished by the mechanism of their biogenesis (Lee et al., 2010b). Different combinations of RNAi proteins are implicated in the production of the four different milRNAs classes, but none is precisely the same mechanism as for conventional miRNAs. milR-2 is Dicer-independent but QDE-2-dependent; milR-3 and milR-4 are completely or partially dependent on Dicer but not QDE-2; milR-1, the most abundant, is dependent on Dicer, the presence of QDE-2, but not its slicer activity, and the exonuclease QIP (Lee et al., 2010b). The maturation of milR-1 is also dependent on the RNA exosome (Xue et al., 2012), which may explain why the QDE-2 nuclease activity is not required. Despite their differences in biogenesis, milRNAs also seem to silence endogenous transcripts with partial complementarity, similar to animal miRNAs (Lee et al., 2010b).
miRNAs have been also recently described in C. neoformans (Jiang et al., 2012). The two miRNA identified, miR1 and miR2, share similarities with conventional miRNAs: 22 and 18 nt long respectively, a promoter of ~70 nt in size, and a preference for a U at the 5′ end of the small RNA. RNAi gene mutations (rdp1, ago1, dcr1, or dcr2) prevent the generation and action of these miRNAs on a reporter gene fused to miR1 or miR2. Also, as their sequences map to transposons and pseudogenes, they may be involved in regulating these elements (Jiang et al., 2012).
nat-siRNAs and disiRNAs
Natural antisense transcripts (NATs) are non-coding RNA molecules that share sequence complementarity with genes encoding RNA transcripts. In plants, stress conditions can induce NAT transcription. These anti-sense transcripts then bind the coding transcript to produce dsRNA, which is processed by RNAi to create NAT-derived siRNAs (nat-siRNAs) (Borsani et al., 2005, Katiyar-Agarwal et al., 2006). Although NATs are widely present in fungi (Ni et al., 2010, Yassour et al., 2010, Donaldson and Saville, 2012, Ehrensberger et al., 2013), NAT-induced RNAi activation has thus far only been observed in convergent gene configurations as described above for S. pombe.
Natural overlapping transcripts in N. crassa can also produce dicer-independent small interfering RNAs (disiRNA). Despite having canonical siRNA characteristics (22 nt long and a 5′ uridine bias), biogenesis of disiRNAs is independent of all canonical RNAi components (Dicer, Argonaute, and RNA-dependent RNA polymerase) (Lee et al., 2010b), suggesting an unknown RNAi-independent sRNA production mechanism remains to be discovered.
Other endogenous sRNAs
The first endogenous sRNAs described as regulators of gene expression through RNAi in fungi were the exonic-siRNAs (ex-siRNAs) found in M. circinelloides (Nicolas et al., 2010). A canonical RNAi pathway had been previously studied in this organism as a response to non-integrated transgenes (Nicolas et al., 2003). Dcl2, Ago1, and RdRP2 are the main proteins implicated in this RNA silencing, and RdRP1 is responsible for dsRNA production from sense transgenes (de Haro et al., 2009, Calo et al., 2012, Cervantes et al., 2013). However, ex-siRNAs produced endogenously from exons are generated by 4 different mechanisms, involving different combinations of RNAi proteins, similar to N. crassa miRNAs (Nicolas et al., 2010). Classes 1 and 2 have a bias for uracil as the 5′ nucleotide and are DCL2-dependent. These two classes differ in their requirement for RdRP1 (class 1) or RdRP2 (class 2). On the other hand, class 3 is dependent on both RdRPs and the Dicer proteins are redundant for the production of these ex-siRNAs, while class 4 depends on only Dcl1 (Nicolas et al., 2010). All 4 classes are dependent on the Ago1 protein but only classes 1 and 2 actually bind to it (Cervantes et al., 2013), indicating that the biogenesis of some of these ex-siRNAs may not follow the canonical RNAi pathway.
Similar results have been obtained in the fungi Trichoderma atroviride and Magnaporthe oryzae (Nunes et al., 2011, Carreras-Villasenor et al., 2013). In T. atroviride, RNAi has been implicated in the control of development and metabolism based on defects in conidiation or morphological alterations exhibited by RNAi mutants. Transcriptional profiles from dcr mutants corroborated those observations (Carreras-Villasenor et al., 2013). In the case of M. oryzae, deep sequencing revealed endogenous sRNAs (esRNAs) derived from protein coding genes, intergenic regions, and repetitive elements (Nunes et al., 2011). However, the tissue specialized in plant invasion, the appressorium, showed enrichment in sRNA derived from tRNA. The involvement of RNAi in the production of these sRNAs has not been experimentally proven yet, but it has been proposed to restrict protein translation as a mechanism to direct cellular metabolism and thereby promote infection (Nunes et al., 2011).
Conservation of RNAi
RNAi is found in each of the main groups of fungi, including the Ascomycota (Cogoni and Macino, 1999a), Basidiomycota (Wang et al., 2010), and Zygomycota (Nicolas et al., 2003) phyla. In fact, it appears likely that RNAi, at least in a basic form, was found in the last common ancestor to all eukaryotes (Shabalina and Koonin, 2008). However, in addition to the considerable diversity of mechanisms and functions described earlier in this review, there are a number of fungal species that have lost RNAi completely. The model yeast S. cerevisiae has lost RNAi although the pathway can be resurrected by introducing the Dicer and Argonaute genes from the closely related ascomycete S. castellii (Drinnenberg et al., 2009). In addition, the molecular subtype (VGII) of the pathogenic fungus Cryptococcus gattii responsible for an ongoing outbreak in the Pacific Northwest has also lost its RNAi pathway, while the rest of the Cryptococcus species complex has retained RNAi (Wang et al., 2010, D’Souza et al., 2011). Notably, the fungi that have lost RNAi have all lost it relatively recently (Drinnenberg et al., 2011). For example, the RNAi-deficient VGII subtype of C. gattii is estimated to be 12.4 million years diverged from the RNAi-proficient VGI subtype (D’Souza et al., 2011). This raises an important question: under what conditions is RNAi disadvantageous and dispensable instead of highly important? In the remainder of this review we will discuss two possible answers, including viral defense and hyper-mutability. It is also possible that RNAi loss in neutral, and simply tolerated by the strains that lose it. This seems unlikely, given the number of species that have lost multiple components of the pathway and the degree to which this loss has become fixed in these species. Some selective mechanism seems necessary. Additional hypotheses for RNAi losses are examined in more detail elsewhere (Nicolás et al., 2013).
Viral Defense
While RNAi is typically considered an asset for defense against viruses, there is at least one exception to this rule in the case of advantageous viruses. Viruses are typically thought of as deleterious for the infected host, but at least in the case of S. cerevisiae, this is not always true. S. cerevisiae can harbor a double-stranded RNA virus called killer virus (Welsh and Leibowitz, 1982). This element is transmitted either vertically from mother to daughter cell or horizontally by infecting the zygote during mating, but typically does not exist outside the host cell as an infectious particle. Killer viruses were described classically in yeast genetics because of a distinct phenotype: killer-positive strains inhibit the growth of neighboring killer-negative strains and species. This provides a clear growth advantage in a competitive environment. This enhanced competitiveness conferred by killer is inherently incompatible with the presence of an RNAi pathway and in fact reintroduction of RNAi results in the silencing of the killer viruses and loss of clearing of unrelated neighbors (Drinnenberg et al., 2011). The presence of killer-like viruses across the fungi kingdom correlates inversely with the presence of RNAi in four of the nine documented cases, suggesting that this type of advantageous virus may provide at least a partial explanation for loss of RNAi (Drinnenberg et al., 2011).
However viruses are also often bad for fungi. Unlike the beneficial viruses of S. cerevisiae, Cryphonectria parasitica, the causative agent of Chestnut Blight, is plagued by a hypovirus (Choi and Nuss, 1992). When infected with this dsRNA element, C. parasitica is substantially less infectious to chestnut trees, resulting in infections that the plant can resolve on its own, instead of causing substantial damage. Mutation of the RNAi pathway renders C. parasitica substantially less resistant to these hypoviruses (Segers et al., 2007). This suggests that viral defense via RNAi may be a balancing act: fungal lineages that have many successful viral pathogens may benefit by retaining RNAi. In contrast, the presence of an advantageous virus may push the balance towards loss of RNAi. In the short term this strategy may prove highly advantageous but could also render the organism susceptible to new viruses in the long term, as well as transposons and repetitive sequences. In the case of S. cerevisiae, it has been posited that the balance has swung towards loss of RNAi because of killer viruses, suggesting that the fungi that have lost RNAi may represent an evolutionary bubble waiting to burst (Drinnenberg et al., 2011).
Mutator States
RNAi, as described above, plays a role in controlling transposable elements in fungi (Moazed, 2009, Yamanaka et al., 2013). Loss of RNAi therefore allows more rampant transposon movement, as has been observed in Cryptococcus (Janbon et al., 2010, Wang et al., 2010). Likewise, quelling in N. crassa has been suggested to play a role in transposon control, at least ancestrally (Nolan et al., 2005), while over time the introduction of repeat-induced point-mutation has rendered the remaining transposons inoperative. Therefore, loss of RNAi could conceivably contribute to a hypermutator phenotype. These phenotypes are often advantageous in organisms attempting to adapt rapidly to a new environment, as typified by the high prevalence of hypermutator Pseudomonas aeruginosa isolates in the lungs of cystic fibrosis patients (Oliver et al., 2000). This is particularly poignant in the case of VGII C. gattii, which is the subtype responsible for the outbreak in the Pacific Northwest. C. gattii is normally considered a tropical or sub-tropical pathogen and thus its spread to the temperate climate of the Pacific Northwest may reflect a certain amount of adaptation. Experiments are currently underway exploring whether the loss of RNAi in the outbreak C. gattii isolates may have contributed to this spread into a new formerly inhospitable environment. As in the case of the viral defense example, mutators tend to be beneficial only in the short term. Bacteria can switch from a stable genome to a mutable one by inducing the SOS response, and then return to stability by turning off the SOS pathway (Fijalkowska et al., 1997). Loss of the genes encoding an entire pathway is essentially irreversible. This could also explain why there are no large groups of fungal species that have lost RNAi. Instead what is observed is frequent and independent loss of RNAi in diverse fungal species throughout the fungal tree of life.
One other possible source of diversity is the link between RNAi and aneuploidy observed in S. pombe. RNAi dysfunction leads to a higher incidence of chromosome missegregation in S. pombe and this stems from a role of RNAi in proper centromere function (Volpe et al., 2003). In the context of multicellular eukaryotes, aneuploidy is typically considered maladaptive, but in fungi there are important advantages to aneuploidy in the right environment. In C. albicans, generation of an aneuploid with an isochromosome containing additional copies of the ERG11 gene can provide protection against azole antifungals (Selmecki et al., 2006). Similarly, C. neoformans can also acquire drug resistance by becoming disomic for chromosome 1, which contains the C. neoformans ERG11 (Sionov et al., 2010). In addition, C. neoformans can generate advantageous de novo phenotypic diversity via generation of aneuploids during opposite-sex or unisexual reproduction (Ni et al., 2013). It remains to be seen whether RNAi contributes to centromere function in fungi outside of S. pombe, but if so, an increased frequency of aneuploidy is an intriguing possible hypothesis for the loss of RNAi.
The fungal kingdom represents an amazing diversity of RNAi pathways and functions. This makes fungi ideal model systems for elucidating the mechanisms of RNAi present in other eukaryotes. Recent studies in Cryptococcus have revealed a link between intron splicing and RNAi (Dumesic et al., 2013), while S. pombe has been shown to have a mechanism for selecting between the exosome and RNAi for message degradation (Yamanaka et al., 2013), either of which may prove to represent general mechanisms for RNAi. Continued work on RNAi in the fungi is essential, not just for delineating mechanisms but also for understanding the basic biological roles of RNAi. This is most clearly illustrated by the frequent losses of RNAi that confer both known and as yet unknown selective benefits involving acquisition of beneficial dsRNA viruses, and mechanisms of genome stability and instability.
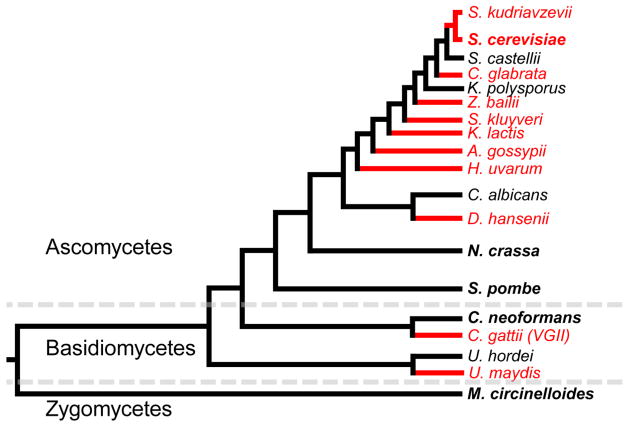
Figure 1.
Above is a phylogeny of selected fungal species, intended to display the broad loss and retention of RNAi components in the fungal kingdom. Species names in red are those that have lost RNAi, while those in black have retained it. In addition, the names of species where RNAi has been more extensively studied are in bold, including the model RNAi loss of S. cerevisiae.
Abbreviations
- aRNA
aberrant RNA
- CsA
cyclosporine A
- disiRNA
dicer-independent small interfering RNAs
- dsRNA
double-strand RNA
- ex-siRNAs
exonic small interfering RNAs
- HDGS
homology dependent gene silencing
- MAT
mating type locus
- miRNA
microRNA
- milRNAs
miRNA-like small RNAs
- MSUD
meiotic silencing by unpaired DNA
- NAT
natural antisense transcripts
- PTGS
post-transcriptional gene silencing
- qiRNA
20–21 nt quelling-associated small RNAs
- rDNA
ribosomal DNA sequence
- RDRC
RNA-dependent RNA polymerase complex
- RdRP
RNA-dependent RNA polymerase
- RISC
RNA-induced silencing complex
- RITS
RNA-induced transcriptional silencing
- RNAi
RNA interference
- SCANR
spliceosome-coupled and nuclear RNAi
- SIS
sex-induced silencing
- siRNA
small interfering RNA
- sRNA
small RNA
- ssRNA
single-strand RNA
- TRAMP
Trf4p/Air2p/Mtr4p polyadenylylation complex
References
- ARAMAYO R, METZENBERG RL. Meiotic transvection in fungi. Cell. 1996;86:103–13. doi: 10.1016/s0092-8674(00)80081-1. [DOI] [PubMed] [Google Scholar]
- BAYNE EH, WHITE SA, KAGANSKY A, BIJOS DA, SANCHEZ-PULIDO L, HOE KL, KIM DU, PARK HO, PONTING CP, RAPPSILBER J, ALLSHIRE RC. Stc1: a critical link between RNAi and chromatin modification required for heterochromatin integrity. Cell. 2010;140:666–77. doi: 10.1016/j.cell.2010.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BORSANI O, ZHU J, VERSLUES PE, SUNKAR R, ZHU JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–91. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOSE I, DOERING TL. Efficient implementation of RNA interference in the pathogenic yeast Cryptococcus neoformans. J Microbiol Methods. 2011;86:156–9. doi: 10.1016/j.mimet.2011.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CALO S, NICOLAS FE, VILA A, TORRES-MARTINEZ S, RUIZ-VAZQUEZ RM. Two distinct RNA-dependent RNA polymerases are required for initiation and amplification of RNA silencing in the basal fungus Mucor circinelloides. Mol Microbiol. 2012;83:379–94. doi: 10.1111/j.1365-2958.2011.07939.x. [DOI] [PubMed] [Google Scholar]
- CARRERAS-VILLASENOR N, ESQUIVEL-NARANJO EU, VILLALOBOS-ESCOBEDO JM, ABREU-GOODGER C, HERRERA-ESTRELLA A. The RNAi machinery regulates growth and development in the filamentous fungus Trichoderma atroviride. Mol Microbiol. 2013;89:96–112. doi: 10.1111/mmi.12261. [DOI] [PubMed] [Google Scholar]
- CATALANOTTO C, AZZALIN G, MACINO G, COGONI C. Gene silencing in worms and fungi. Nature. 2000;404:245. doi: 10.1038/35005169. [DOI] [PubMed] [Google Scholar]
- CATALANOTTO C, AZZALIN G, MACINO G, COGONI C. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev. 2002;16:790–5. doi: 10.1101/gad.222402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CATALANOTTO C, PALLOTTA M, REFALO P, SACHS MS, VAYSSIE L, MACINO G, COGONI C. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol Cell Biol. 2004;24:2536–45. doi: 10.1128/MCB.24.6.2536-2545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CERVANTES M, VILA A, NICOLAS FE, MOXON S, DE HARO JP, DALMAY T, TORRES-MARTINEZ S, RUIZ-VAZQUEZ RM. A single argonaute gene participates in exogenous and endogenous RNAi and controls cellular functions in the basal fungus. Mucor circinelloides. PLOS One. 2013;8:e69283. doi: 10.1371/journal.pone.0069283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHANG SS, ZHANG Z, LIU Y. RNA interference pathways in fungi: mechanisms and functions. Annu Rev Microbiol. 2012;66:305–23. doi: 10.1146/annurev-micro-092611-150138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN H, KOBAYASHI K, MIYAO A, HIROCHIKA H, YAMAOKA N, NISHIGUCHI M. Both OsRecQ1 and OsRDR1 are required for the production of small RNA in response to DNA-damage in rice. PLOS One. 2013;8:e55252. doi: 10.1371/journal.pone.0055252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHOI GH, NUSS DL. Hypovirulence of chestnut blight fungus conferred by an infectious viral cDNA. Science. 1992;257:800–3. doi: 10.1126/science.1496400. [DOI] [PubMed] [Google Scholar]
- COGONI C, IRELAN JT, SCHUMACHER M, SCHMIDHAUSER TJ, SELKER EU, MACINO G. Transgene silencing of the al-1 gene in vegetative cells of Neurospora is mediated by a cytoplasmic effector and does not depend on DNA-DNA interactions or DNA methylation. EMBO J. 1996;15:3153–63. [PMC free article] [PubMed] [Google Scholar]
- COGONI C, MACINO G. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. PNAS. 1997;94:10233–8. doi: 10.1073/pnas.94.19.10233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- COGONI C, MACINO G. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999a;399:166–9. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- COGONI C, MACINO G. Homology-dependent gene silencing in plants and fungi: a number of variations on the same theme. Curr Opin Microbiol. 1999b;2:657–62. doi: 10.1016/s1369-5274(99)00041-7. [DOI] [PubMed] [Google Scholar]
- COGONI C, MACINO G. Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science. 1999c;286:2342–4. doi: 10.1126/science.286.5448.2342. [DOI] [PubMed] [Google Scholar]
- CZAJA W, MILLER KY, MILLER BL. Novel sexual-cycle-specific gene silencing in Aspergillus nidulans. Genetics. 2013;193:1149–62. doi: 10.1534/genetics.112.147546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’SOUZA CA, KRONSTAD JW, TAYLOR G, WARREN R, YUEN M, HU G, JUNG WH, SHAM A, KIDD SE, TANGEN K, LEE N, ZEILMAKER T, SAWKINS J, MCVICKER G, SHAH S, GNERRE S, GRIGGS A, ZENG Q, BARTLETT K, LI W, WANG X, HEITMAN J, STAJICH JE, FRASER JA, MEYER W, CARTER D, SCHEIN J, KRZYWINSKI M, KWON-CHUNG KJ, VARMA A, WANG J, BRUNHAM R, FYFE M, OUELLETTE BF, SIDDIQUI A, MARRA M, JONES S, HOLT R, BIRREN BW, GALAGAN JE, CUOMO CA. Genome variation in Cryptococcus gattii, an emerging pathogen of immunocompetent hosts. mBio. 2011;2:e00342–10. doi: 10.1128/mBio.00342-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DANG Y, YANG Q, XUE Z, LIU Y. RNA interference in fungi: pathways, functions, and applications. Eukaryotic Cell. 2011;10:1148–55. doi: 10.1128/EC.05109-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DE HARO JP, CALO S, CERVANTES M, NICOLAS FE, TORRES-MARTINEZ S, RUIZ-VAZQUEZ RM. A single dicer gene is required for efficient gene silencing associated with two classes of small antisense RNAs in Mucor circinelloides. Eukaryotic Cell. 2009;8:1486–97. doi: 10.1128/EC.00191-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DONALDSON ME, SAVILLE BJ. Natural antisense transcripts in fungi. Mol Microbiol. 2012;85:405–17. doi: 10.1111/j.1365-2958.2012.08125.x. [DOI] [PubMed] [Google Scholar]
- DRINNENBERG IA, FINK GR, BARTEL DP. Compatibility with killer explains the rise of RNAi-deficient fungi. Science. 2011;333:1592. doi: 10.1126/science.1209575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DRINNENBERG IA, WEINBERG DE, XIE KT, MOWER JP, WOLFE KH, FINK GR, BARTEL DP. RNAi in budding yeast. Science. 2009;326:544–50. doi: 10.1126/science.1176945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DUAN G, SAINT RB, HELLIWELL CA, BEHM CA, WANG MB, WATERHOUSE PM, GORDON KH. C. elegans RNA-dependent RNA polymerases rrf-1 and ego-1 silence Drosophila transgenes by differing mechanisms. Cell Mol Life Sci. 2013;70:1469–81. doi: 10.1007/s00018-012-1218-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DUMESIC PA, NATARAJAN P, CHEN C, DRINNENBERG IA, SCHILLER BJ, THOMPSON J, MORESCO JJ, YATES JR, III, BARTEL DP, MADHANI HD. Stalled spliceosomes are a signal for RNAi-mediated genome defense. Cell. 2013;152:957–68. doi: 10.1016/j.cell.2013.01.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EHRENSBERGER KM, MASON C, CORKINS ME, ANDERSON C, DUTROW N, CAIRNS BR, DALLEY B, MILASH B, BIRD AJ. Zinc-dependent regulation of the adh1 antisense transcript in fission yeast. J Biol Chem. 2013;288:759–69. doi: 10.1074/jbc.M112.406165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENGLISH JJ, MUELLER E, BAULCOMBE DC. Suppression of virus accumulation in transgenic plants exhibiting silencing of nuclear genes. The Plant Cell. 1996;8:179–88. doi: 10.1105/tpc.8.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FIJALKOWSKA IJ, DUNN RL, SCHAAPER RM. Genetic requirements and mutational specificity of the Escherichia coli SOS mutator activity. J Bacteriol. 1997;179:7435–45. doi: 10.1128/jb.179.23.7435-7445.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FIRE A, XU S, MONTGOMERY MK, KOSTAS SA, DRIVER SE, MELLO CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- FRANCIA S, MICHELINI F, SAXENA A, TANG D, DE HOON M, ANELLI V, MIONE M, CARNINCI P, D’ADDA DI FAGAGNA F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–5. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FULCI V, MACINO G. Quelling: post-transcriptional gene silencing guided by small RNAs in Neurospora crassa. Curr Opin Microbiol. 2007;10:199–203. doi: 10.1016/j.mib.2007.03.016. [DOI] [PubMed] [Google Scholar]
- GHILDIYAL M, ZAMORE PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GULLEROVA M, MOAZED D, PROUDFOOT NJ. Autoregulation of convergent RNAi genes in fission yeast. Genes Dev. 2011;25:556–68. doi: 10.1101/gad.618611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GULLEROVA M, PROUDFOOT NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell. 2008;132:983–95. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- HALL IM, SHANKARANARAYANA GD, NOMA K, AYOUB N, COHEN A, GREWAL SI. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–7. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- HAMMOND TM, XIAO H, BOONE EC, DECKER LM, LEE SA, PERDUE TD, PUKKILA PJ, SHIU PK. Novel proteins required for meiotic silencing by unpaired DNA and siRNA generation in Neurospora crassa. Genetics. 2013;194:91–100. doi: 10.1534/genetics.112.148999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HANNON GJ. RNA interference. Nature. 2002;418:244–51. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- HOUSELEY J, LACAVA J, TOLLERVEY D. RNA-quality control by the exosome. Nat Rev Mol Cell Biol. 2006;7:529–39. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- JANBON G, MAENG S, YANG DH, KO YJ, JUNG KW, MOYRAND F, FLOYD A, HEITMAN J, BAHN YS. Characterizing the role of RNA silencing components in Cryptococcus neoformans. Fungal Genet Biol. 2010;47:1070–80. doi: 10.1016/j.fgb.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JIANG N, YANG Y, JANBON G, PAN J, ZHU X. Identification and functional demonstration of miRNAs in the fungus Cryptococcus neoformans. PLOS One. 2012;7:e52734. doi: 10.1371/journal.pone.0052734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JORGENSEN R. Altered gene expression in plants due to trans interactions between homologous genes. Trends Biotechnol. 1990;8:340–4. doi: 10.1016/0167-7799(90)90220-r. [DOI] [PubMed] [Google Scholar]
- KATIYAR-AGARWAL S, MORGAN R, DAHLBECK D, BORSANI O, VILLEGAS A, JR, ZHU JK, STASKAWICZ BJ, JIN H. A pathogen-inducible endogenous siRNA in plant immunity. PNAS. 2006;103:18002–7. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE DW, PRATT RJ, MCLAUGHLIN M, ARAMAYO R. An argonaute-like protein is required for meiotic silencing. Genetics. 2003;164:821–28. doi: 10.1093/genetics/164.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE HC, AALTO AP, YANG Q, CHANG SS, HUANG G, FISHER D, CHA J, PORANEN MM, BAMFORD DH, LIU Y. The DNA/RNA-dependent RNA polymerase QDE-1 generates aberrant RNA and dsRNA for RNAi in a process requiring replication protein A and a DNA helicase. PLOS Biology. 2010a;8:e1000496. doi: 10.1371/journal.pbio.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE HC, CHANG SS, CHOUDHARY S, AALTO AP, MAITI M, BAMFORD DH, LIU Y. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009;459:274–7. doi: 10.1038/nature08041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEE HC, LI L, GU W, XUE Z, CROSTHWAITE SK, PERTSEMLIDIS A, LEWIS ZA, FREITAG M, SELKER EU, MELLO CC, LIU Y. Diverse pathways generate microRNA-like RNAs and Dicer-independent small interfering RNAs in fungi. Mol Cell. 2010b;38:803–14. doi: 10.1016/j.molcel.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENGRONNE A, KATOU Y, MORI S, YOKOBAYASHI S, KELLY GP, ITOH T, WATANABE Y, SHIRAHIGE K, UHLMANN F. Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature. 2004;430:573–8. doi: 10.1038/nature02742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU H, COTTRELL TR, PIERINI LM, GOLDMAN WE, DOERING TL. RNA interference in the pathogenic fungus Cryptococcus neoformans. Genetics. 2002;160:463–70. doi: 10.1093/genetics/160.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOFTUS BJ, FUNG E, RONCAGLIA P, ROWLEY D, AMEDEO P, BRUNO D, VAMATHEVAN J, MIRANDA M, ANDERSON IJ, FRASER JA, ALLEN JE, BOSDET IE, BRENT MR, CHIU R, DOERING TL, DONLIN MJ, D’SOUZA CA, FOX DS, GRINBERG V, FU J, FUKUSHIMA M, HAAS BJ, HUANG JC, JANBON G, JONES SJM, KOO HL, KRZYWINSKI MI, KWON-CHUNG JK, LENGELER KB, MAITI R, MARRA MA, MARRA RE, MATHEWSON CA, MITCHELL TG, PERTEA M, RIGGS FR, SALZBERG SL, SCHEIN JE, SHVARTSBEYN A, SHIN H, SHUMWAY M, SPECHT CA, SUH BB, TENNEY A, UTTERBACK TR, WICKES BL, WORTMAN JR, WYE NH, KRONSTAD JW, LODGE JK, HEITMAN J, DAVIS RW, FRASER CM, HYMAN RW. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–4. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAINE EM, HAUTH J, RATLIFF T, VOUGHT VE, SHE X, KELLY WG. EGO-1, a putative RNA-dependent RNA polymerase, is required for heterochromatin assembly on unpaired DNA during C. elegans meiosis. Curr Biol. 2005;15:1972–8. doi: 10.1016/j.cub.2005.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAITI M, LEE HC, LIU Y. QIP, a putative exonuclease, interacts with the Neurospora argonaute protein and facilitates conversion of duplex siRNA into single strands. Genes Dev. 2007;21:590–600. doi: 10.1101/gad.1497607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATZKE MA, PRIMIG M, TRNOVSKY J, MATZKE AJ. Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. EMBO J. 1989;8:643–9. doi: 10.1002/j.1460-2075.1989.tb03421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MICHALIK KM, BOTTCHER R, FORSTEMANN K. A small RNA response at DNA ends in Drosophila. Nucleic Acids Res. 2012;40:9596–603. doi: 10.1093/nar/gks711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOAZED D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–20. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOTAMEDI MR, VERDEL A, COLMENARES SU, GERBER SA, GYGI SP, MOAZED D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- NI M, FERETZAKI M, LI W, FLOYD-AVERETTE A, MIECZKOWSKI P, DIETRICH FS, HEITMAN J. Unisexual and heterosexual meiotic reproduction generate aneuploidy and phenotypic diversity de novo in the yeast Cryptococcus neoformans. PLOS Biology. 2013;11:e1001653. doi: 10.1371/journal.pbio.1001653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NI T, TU K, WANG Z, SONG S, WU H, XIE B, SCOTT KC, GREWAL SI, GAO Y, ZHU J. The prevalence and regulation of antisense transcripts in Schizosaccharomyces pombe. PLOS One. 2010;5:e15271. doi: 10.1371/journal.pone.0015271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NICOLÁS FE, MOXON S, DE HARO JP, CALO S, GRIGORIEV IV, TORRES-MARTINEZ S, MOULTON V, RUIZ-VAZQUEZ RM, DALMAY T. Endogenous short RNAs generated by Dicer 2 and RNA-dependent RNA polymerase 1 regulate mRNAs in the basal fungus Mucor circinelloides. Nucleic Acids Res. 2010;38:5535–41. doi: 10.1093/nar/gkq301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NICOLÁS FE, TORRES-MARTINEZ S, RUIZ-VAZQUEZ RM. Two classes of small antisense RNAs in fungal RNA silencing triggered by non-integrative transgenes. EMBO J. 2003;22:3983–91. doi: 10.1093/emboj/cdg384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NICOLÁS FE, TORRES-MARTÍNEZ S, RUIZ-VÁZQUEZ RM. Loss and retention of RNA interference in fungi and parasites. PLOS Pathog. 2013;9:e1003089. doi: 10.1371/journal.ppat.1003089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NOLAN T, BRACCINI L, AZZALIN G, DE TONI A, MACINO G, COGONI C. The post-transcriptional gene silencing machinery functions independently of DNA methylation to repress a LINE1-like retrotransposon in Neurospora crassa. Nucleic Acids Res. 2005;33:1564–73. doi: 10.1093/nar/gki300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NUNES CC, GOWDA M, SAILSBERY J, XUE M, CHEN F, BROWN DE, OH Y, MITCHELL TK, DEAN RA. Diverse and tissue-enriched small RNAs in the plant pathogenic fungus, Magnaporthe oryzae. BMC Genomics. 2011;12:288. doi: 10.1186/1471-2164-12-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OLIVER A, CANTÓN R, CAMPO P, BAQUERO F, BLÁZQUEZ J. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science. 2000;288:1251–3. doi: 10.1126/science.288.5469.1251. [DOI] [PubMed] [Google Scholar]
- PRATT RJ, LEE DW, ARAMAYO R. DNA methylation affects meiotic trans-sensing, not meiotic silencing, in Neurospora. Genetics. 2004;168:1925–35. doi: 10.1534/genetics.104.031526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROMANO N, MACINO G. Quelling: transient inactivation of gene expression in Neurospora crassa by transformation with homologous sequences. Mol Microbiol. 1992;6:3343–53. doi: 10.1111/j.1365-2958.1992.tb02202.x. [DOI] [PubMed] [Google Scholar]
- REYES-TURCU FE, GREWAL SI. Different means, same end-heterochromatin formation by RNAi and RNAi-independent RNA processing factors in fission yeast. Curr Opin Genet Dev. 2012;22:156–63. doi: 10.1016/j.gde.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SEGERS GC, ZHANG X, DENG F, SUN Q, NUSS DL. Evidence that RNA silencing functions as an antiviral defense mechanism in fungi. PNAS. 2007;104:12902–6. doi: 10.1073/pnas.0702500104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SELMECKI A, FORCHE A, BERMAN J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–70. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHABALINA SA, KOONIN EV. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evolut. 2008;23:578–87. doi: 10.1016/j.tree.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHIU PK, RAJU NB, ZICKLER D, METZENBERG RL. Meiotic silencing by unpaired DNA. Cell. 2001;107:905–16. doi: 10.1016/s0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- SIONOV E, LEE H, CHANG YC, KWON-CHUNG KJ. Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLOS Pathog. 2010;6:e1000848. doi: 10.1371/journal.ppat.1000848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SON H, MIN K, LEE J, RAJU NB, LEE YW. Meiotic silencing in the homothallic fungus Gibberella zeae. Fungal Biol. 2011;115:1290–302. doi: 10.1016/j.funbio.2011.09.006. [DOI] [PubMed] [Google Scholar]
- THAKURTA AG, GOPAL G, YOON JH, KOZAK L, DHAR R. Homolog of BRCA2-interacting Dss1p and Uap56p link Mlo3p and Rae1p for mRNA export in fission yeast. EMBO J. 2005;24:2512–23. doi: 10.1038/sj.emboj.7600713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TURNER JM, MAHADEVAIAH SK, FERNANDEZ-CAPETILLO O, NUSSENZWEIG A, XU X, DENG CX, BURGOYNE PS. Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet. 2005;37:41–7. doi: 10.1038/ng1484. [DOI] [PubMed] [Google Scholar]
- VERDEL A, JIA S, GERBER S, SUGIYAMA T, GYGI S, GREWAL SI, MOAZED D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–6. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOLPE T, SCHRAMKE V, HAMILTON GL, WHITE SA, TENG G, MARTIENSSEN RA, ALLSHIRE RC. RNA interference is required for normal centromere function in fission yeast. Chromosome Res. 2003;11:137–46. doi: 10.1023/a:1022815931524. [DOI] [PubMed] [Google Scholar]
- VOLPE TA, KIDNER C, HALL IM, TENG G, GREWAL SI, MARTIENSSEN RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–7. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- WANG X, DARWICHE S, HEITMAN J. Sex-induced silencing operates during opposite-sex and unisexual reproduction in Cryptococcus neoformans. Genetics. 2013;193:1163–74. doi: 10.1534/genetics.113.149443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG X, HSUEH YP, LI W, FLOYD A, SKALSKY R, HEITMAN J. Sex-induced silencing defends the genome of Cryptococcus neoformans via RNAi. Genes Dev. 2010;24:2566–82. doi: 10.1101/gad.1970910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG X, WANG P, SUN S, DARWICHE S, IDNURM A, HEITMAN J. Transgene induced co-suppression during vegetative growth in Cryptococcus neoformans. PLOS Genet. 2012;8:e1002885. doi: 10.1371/journal.pgen.1002885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEI W, BA Z, GAO M, WU Y, MA Y, AMIARD S, WHITE CI, RENDTLEW DANIELSEN JM, YANG YG, QI Y. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–12. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- WELSH JD, LEIBOWITZ MJ. Localization of genes for the double-stranded RNA killer virus of yeast. PNAS. 1982;79:786–9. doi: 10.1073/pnas.79.3.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XUE Z, YUAN H, GUO J, LIU Y. Reconstitution of an Argonaute-dependent small RNA biogenesis pathway reveals a handover mechanism involving the RNA exosome and the exonuclease QIP. Mol Cell. 2012;46:299–310. doi: 10.1016/j.molcel.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAMANAKA S, MEHTA S, REYES-TURCU FE, ZHUANG F, FUCHS RT, RONG Y, ROBB GB, GREWAL SI. RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature. 2013;493:557–60. doi: 10.1038/nature11716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YASSOUR M, PFIFFNER J, LEVIN JZ, ADICONIS X, GNIRKE A, NUSBAUM C, THOMPSON DA, FRIEDMAN N, REGEV A. Strand-specific RNA sequencing reveals extensive regulated long antisense transcripts that are conserved across yeast species. Genome Biol. 2010;11:R87. doi: 10.1186/gb-2010-11-8-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG K, FISCHER T, PORTER RL, DHAKSHNAMOORTHY J, ZOFALL M, ZHOU M, VEENSTRA T, GREWAL SI. Clr4/Suv39 and RNA quality control factors cooperate to trigger RNAi and suppress antisense RNA. Science. 2011;331:1624–7. doi: 10.1126/science.1198712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG K, MOSCH K, FISCHLE W, GREWAL SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–8. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- ZHANG Z, CHANG SS, ZHANG Z, XUE Z, ZHANG H, LI S, LIU Y. Homologous recombination as a mechanism to recognize repetitive DNA sequences in an RNAi pathway. Genes Dev. 2013;27:145–50. doi: 10.1101/gad.209494.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZOFALL M, FISCHER T, ZHANG K, ZHOU M, CUI B, VEENSTRA TD, GREWAL SI. Histone H2A.Z cooperates with RNAi and heterochromatin factors to suppress antisense RNAs. Nature. 2009;461:419–22. doi: 10.1038/nature08321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZOFALL M, GREWAL SI. RNAi-mediated heterochromatin assembly in fission yeast. Cold Spring Harb Symp Quant Biol. 2006;71:487–96. doi: 10.1101/sqb.2006.71.059. [DOI] [PubMed] [Google Scholar]