Ever since microscopes enabled visualization of the organization of eukaryotic cells, scientists have been fascinated by the function and structure of their largest organelle, the cell nucleus [1,2]. The realization that this is the compartment where the cell stores its genetic material further intensified research on nuclear organization. Studying the in vivo folding of chromosomes long relied predominantly on microscopy. Fluorescence in situ hybridization (FISH) in particular has been, and still is, instrumental for localizing the positions of whole chromosomes, individual loci and actively transcribed genes in nuclei of single cells. Sophisticated FISH strategies revealed that that the nucleus may be structurally and functionally compartmentalized with active and inactive genes adopting different sub-nuclear positions [3]. They also highlighted the probabilistic nature of genome folding, by showing cell-to-cell differences in the positioning of chromosomes and genes [4]. Classical FISH studies however are limited in throughput and resolution. Although resolution continues to improve, FISH cannot provide the fine-structure of chromosomal sub-regions where regulatory sequences communicate with target genes. The technique can be applied to interrogate contacts between more distal chromosomal sites, but only between those selected by the investigator.
The development of chromosome conformation capture (3C) technology [5], ten years ago now, opened up completely new avenues to study genome topology. 3C technology is a biochemical method that uses formaldehyde to fix the three-dimensional organization of chromatin in living cells. DNA is then digested and re-ligated, with ligation taking place under diluted conditions to favor fusions between crosslinked DNA fragments. As a result, a library is generated of ligation products between DNA fragments that were originally physically close together in the nuclear space. In 3C, a number of selected fusion products are analyzed and compared in a quantitative manner by PCR, to provide a measure for their relative interaction frequency in the cell population [5].
3C technology has enabled studying chromosome folding at unprecedented resolution. It was applied to demonstrate that regulatory DNA sequences control transcription by looping to and physically contacting target genes tens or hundreds of kilobases away [6] (see also [7]). 3C studies also revealed that such regulatory contacts can dynamically change during development and differentiation [8] and that they are mediated by transcription factors bound to the genomic sites involved [9,10]. The concept of chromatin loops being involved in the developmental regulation of mammalian gene expression is now well established [11,12].
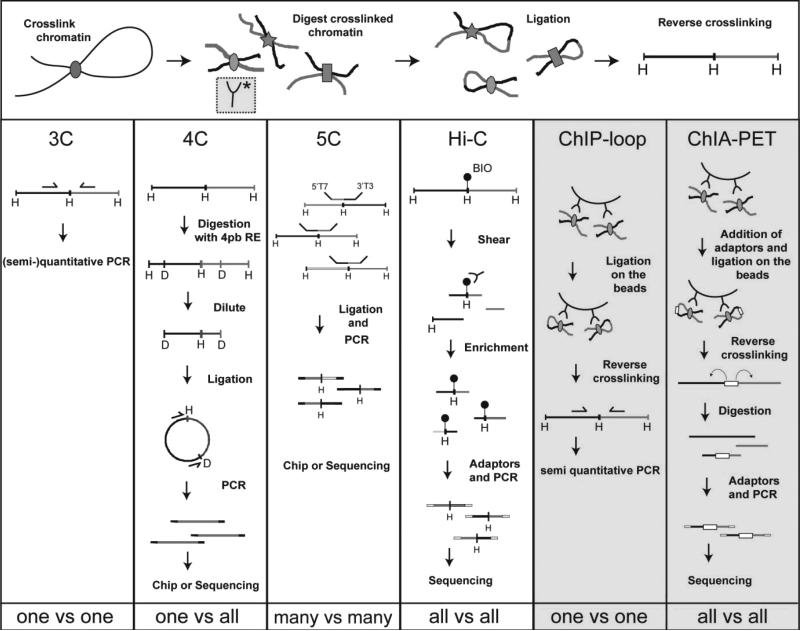
The subsequent development of genomics variants of 3C technology (4C, 5C, Hi-C, ChIA-PET) provides new tools to investigate the shape of the genome in a more systematic and unbiased manner. 4C, 5C and Hi-C technology analyze respectively one-versus-all (4C), many-versus-many (5C), and all-versus-all (Hi-C) contacts in the genome (Figure 1) [13,14]. The method of choice depends on the research question asked, and the resolution required to address it.
Figure 1.
Overview of the 3C technologies
4C (one-versus-all) studies [15,16] (see also [17]) revealed that individual genes in cell populations form many contacts with loci elsewhere in the genome, mostly in cis (on their own chromosome), but also in trans (with other chromosomes). Selected active genes were shown to preferentially cluster with other active genes; they can switch to inactive chromatin environments in tissues where their transcription is silenced [15]. These 4C results show that co-expressed genes cluster together. Furthermore, there are some indications that co-regulated genes, i.e. genes regulated by the same factors, from across the genome, or even from different chromosomes, preferentially come together in the nuclear space. This was found in Drosophila for loci silenced by the polycomb group proteins [18,19] and for tissue-specific genes in mouse erythroid cells [20]; the generality of nuclear clustering of co-regulated genes still needs to be demonstrated though. 4C strategies recently also provided insight into the enigmatic phenomenon of partner selection during chromosomal rearrangements in diseases such as cancer. It was shown that the three-dimensional contact profile of a given locus determines its unscheduled selection of chromosomal rearrangement partners after double-strand break formation [21,22]. Whereas original 4C protocols only enabled moderate resolution analysis of contacts between gene loci, higher resolution versions of 4C technology are now available and used to uncover the regulatory sequences that contact genes of interest [23,24].
5C (many-versus-many) technology [25] was the first 3C-based method to employ deep-sequencing and the first method to simultaneously analyze DNA interactions between multiple combinations of selected chromosomal sequences. This offers the advantage that a given interaction profile can be interpreted in the context of interaction profiles of other, surrounding, sequences. The resulting contact matrices have served as input for 3D modeling of chromosomal regions [26,27]. A recent comprehensive 5C study led to the discovery of topologically associating domains (TADs) [28]. TADs are analogous to the domains that were simultaneously discovered by Hi-C (see below) [29]: they represent chromosomal segments in the megabase size range within which sequences preferentially interact with each other. Presumably, TADs encompass genes and their cognate regulatory sequences and serve to ensure that they efficiently find each other. Finally, 5C, like 4C, can be used to identify contacts between regulatory sequences and gene promoters. In a recent systematic 5C analysis, the regulatory interactions of transcriptional start sites in 1% of the human genome were uncovered [30].
Hi-C is a completely unbiased method that analyzes contacts between all fragments in the genome [31]. Hi-C showed that spatial separation of active and inactive chromatin occurs throughout the genome. Analysis of Hi-C contact profiles has suggested that at the scale of several megabases chromatin is spatially organized into fractal globules, a densely packed but unknotted folding state [31]. Hi-C studies have provided detailed genome-wide contact maps of the yeast [32,33] and Drosophila genomes [34], showing that similarly typed chromatin regions preferentially cluster in the nuclear space. Hi-C results led to the forementioned discovery of TADs; both the mammalian [29] and Drosophila [34] genome seem to adopt such topological domains.
A fundamentally different branch of 3C-based methods combines chromatin immunoprecipitation (ChIP) with chromosome conformation capture technology. The aim of this combination of techniques is to direct analysis of DNA contacts exclusively to chromosomal sites that are bound by a protein of interest. The original chromatin immunoprecipitation-combined loop assays combined ChIP with the traditional 3C method to analyze DNA interactions between selected chromosomal sites bound by the protein of interest [35]. ChIA-PET (chromatin interaction analysis by paired-end tag sequencing) is effectively the unbiased high-throughput Hi-C variant of the ChIP-loop assay and searches for contacts between any pair of sites in the genome bound by a protein of interest [36]. ChIA-PET uncovered DNA interaction networks between sites bound by respectively the estrogen receptor, CTCF and RNA polymerase II [36-38]. In principle, ChIA-PET should be able to discover the DNA interaction networks of all DNA-associated proteins.
Collectively this shows how important the original development of 3C technology and of its subsequent genomics variants has been for our understanding of the structure and function of the genome. The methods are rapidly becoming standard research tools in many laboratories that study chromatin structure, gene expression and genome function. In a future not far from now, we expect that the systematic application of 4C, 5C, Hi-C and ChIA-PET technologies, combined with ultra-deep sequencing, will lead to a thorough understanding of how every gene is physically and functionally wired to its regulatory sequences, how this wiring changes between cell types and what the protein factors are that set up this three-dimensional regulatory circuit. This understanding is not only relevant to crack the code of how our genome functions, but also to interpret the relevance of disease-associated genetic variants, of which many localize to sequences outside the genes [39].
Several general overviews critically have been published that evaluate the technical pitfalls, necessary controls and data interpretation issues of the 3C technologies [40,41]. This volume of Methods provides detailed and up-to-date lab protocols of the many 3C methodologies, provided by expert groups that often first pioneered these technologies. It includes general protocols for 3C technology [42], for 3C technology in plants [43], for 3C and related technologies in bacteria [44], for 4C technology in mammalian cells [45] and in Drosophila [46], protocols for the ChIP-loop assay [47], for 5C [48] and Hi-C in mammalian cells [49] and in yeast [50] and for ChIA-PET [51]. In addition, strategies are described for the modeling of 3C-based data [52] and a conceptual framework is presented to understand chromosome folding [53]. The detailed protocols provide troubleshooting paragraphs and discuss the technical and theoretical issues that need to be considered. Collectively, we expect this Methods volume will be very useful to both expert labs, who will find new shortcuts, tricks and considerations to further optimize their running protocols, and non-expert labs, who will be enabled to set up and add these technologies to their routine tool box for studying chromatin, gene expression and genome function in the living cell.
REFERENCES
- 1.Cajal S. Trab Lab Investig Biol Univ Madr. 1903;2:129–221. [Google Scholar]
- 2.Heitz E. 1 Jahrb wiss Bot. 1928;69:762–818. [Google Scholar]
- 3.Fraser P, Bickmore W. Nature. 2007;447:413–7. doi: 10.1038/nature05916. [DOI] [PubMed] [Google Scholar]
- 4.Bolzer A, Kreth G, Solovei I, Koehler D, Saracoglu K, Fauthm C, Muller S, Eils R, Cremer C, Speicher MR, Cremer T. PLoS Biol. 2005;3:e157. doi: 10.1371/journal.pbio.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dekker J, Rippe K, Dekker M, Kleckner N. Science. 2002;295:1306–11. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 6.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Mol Cell. 2002;10:1453–65. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 7.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Nat Genet. 2002;32:623–6. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 8.Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. Nat Genet. 2003;35:190–4. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 9.Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. Genes Dev. 2004;18:2485–90. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Mol Cell. 2005;17:453–62. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 11.Splinter E, de Laat W. EMBO J. 2011;30:4345–55. doi: 10.1038/emboj.2011.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hou C, Corces VG. Chromosoma. 2012;121:107–16. doi: 10.1007/s00412-011-0352-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Steensel B, Dekker J. Nat Biotechnol. 2010;28:1089–95. doi: 10.1038/nbt.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Wit E, de Laat W. Genes Dev. 2012;26:11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nat Genet. 2006;38:1348–54. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Z, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Nat Genet. 2006;38:1341–7. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 17.Würtele H, Chartrand P. Chromosome Res. 2006;14:477–95. doi: 10.1007/s10577-006-1075-0. [DOI] [PubMed] [Google Scholar]
- 18.Tolhuis B, Blom M, Kerkhoven RM, Pagie L, Teunissen H, Nieuwland M, Simonis M, de Laat W, van Lohuizen M, van Steensel B. PLoS Genet. 2011;7:e1001343. doi: 10.1371/journal.pgen.1001343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bantignies F, Roure V, Comet I, Leblanc B, Schuettengruber B, Bonnet J, Tixier V, Mas A, Cavalli G. Cell. 2011;144:214–26. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 20.Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P. Nat Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y, McCord RP, Ho YJ, Lajoie BR, Hildebrand DG, Simon AC, Becker MS, Alt FW, Dekker J. Cell. 2012;148:908–21. doi: 10.1016/j.cell.2012.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hakim O, Resch W, Yamane A, Klein I, Kieffer-Kwon KR, Jankovic M, Oliveira T, Bothmer A, Voss TC, Ansarah-Sobrinho C, Mathe E, Liang G, Cobell J, Nakahashi H, Robbiani DF, Nussenzweig A, Hager GL, Nussenzweig MC, Casellas R. Nature. 2012;484:69–74. doi: 10.1038/nature10909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lower KM, Hughes JR, De Gobbi M, Henderson S, Viprakasit V, Fisher C, Goriely A, Ayyub H, Sloane-Stanley J, Vernimmen D, Langford C, Garrick D, Gibbons RJ, Higgs DR. Proc Natl Acad Sci USA. 2009;106:21771–6. doi: 10.1073/pnas.0909331106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van de Werken HJ, Landan G, Holwerda SJ, Hoichman M, Klous P, Chachik R, Splinter E, Valdes-Quezada C, Oz Y, Bouwman BA, Verstegen MJ, de Wit E, Tanay A, de Laat W. Nat Methods. 2012;9:969–72. doi: 10.1038/nmeth.2173. [DOI] [PubMed] [Google Scholar]
- 25.Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. Genome Res. 2006;16:1299–309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baù D, Sanyal A, Lajoie BR, Capriotti E, Byron M, Lawrence JB, Dekker J, Marti-Renom MA. Nat Struct Mol Biol. 2011;18:107–14. doi: 10.1038/nsmb.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Umbarger MA, Toro E, Wright MA, Porreca GJ, Baù D, Hong SH, Fero MJ, Zhu LJ, Marti-Renom MA, McAdams HH, Shapiro L, Dekker J, Church GM. Mol Cell. 2011;44:252–64. doi: 10.1016/j.molcel.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, Gribnau J, Barillot E, Blüthgen N, Dekker J, Heard E. Nature. 2012;485:381–5. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanyal A, Lajoie BR, Jain G, Dekker J. Nature. 2012;489:109–13. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Science. 2009;326:289–93. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. Nature. 2010;465:363–7. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanizawa H, Iwasaki O, Tanaka A, Capizzi JR, Wickramasinghe P, Lee M, Fu Z, Noma K. Nucleic Acids Res. 2010;38:8164–77. doi: 10.1093/nar/gkq955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Cell. 2012;148:458–72. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 35.Horike S, Cai S, Miyano M, Cheng JF, Kohwi-Shigematsu T. Nat Genet. 2005;37:31–40. doi: 10.1038/ng1491. [DOI] [PubMed] [Google Scholar]
- 36.Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, Orlov YL, Velkov S, Ho A, Mei PH, Chew EG, Huang PY, Welboren WJ, Han Y, Ooi HS, Ariyaratne PN, Vega VB, Luo Y, Tan PY, Choy PY, Wansa KD, Zhao B, Lim KS, Leow SC, Yow JS, Joseph R, Li H, Desai KV, Thomsen JS, Lee YK, Karuturi RK, Herve T, Bourque G, Stunnenberg HG, Ruan X, Cacheux-Rataboul V, Sung WK, Liu ET, Wei CL, Cheung E, Ruan Y. Nature. 2009;462:58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Handoko L, Xu H, Li G, Ngan CY, Chew E, Schnapp M, Lee CW, Ye C, Ping JL, Mulawadi F, Wong E, Sheng J, Zhang Y, Poh T, Chan CS, Kunarso G, Shahab A, Bourque G, Cacheux-Rataboul V, Sung WK, Ruan Y, Wei CL. Nat Genet. 2011;43:630–8. doi: 10.1038/ng.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li G, Ruan X, Auerbach RK, Sandhu KS, Zheng M, Wang P, Poh HM, Goh Y, Lim J, Zhang J, Sim HS, Peh SQ, Mulawadi FH, Ong CT, Orlov YL, Hong S, Zhang Z, Landt S, Raha D, Euskirchen G, Wei CL, Ge W, Wang H, Davis C, Fisher-Aylor KI, Mortazavi A, Gerstein M, Gingeras T, Wold B, Sun Y, Fullwood MJ, Cheung E, Liu E, Sung WK, Snyder M, Ruan Y. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, Shafer A, Neri F, Lee K, Kutyavin T, Stehling-Sun S, Johnson AK, Canfield TK, Giste E, Diegel M, Bates D, Hansen RS, Neph S, Sabo PJ, Heimfeld S, Raubitschek A, Ziegler S, Cotsapas C, Sotoodehnia N, Glass I, Sunyaev SR, Kaul R, Stamatoyannopoulos JA. Science. 2012;337:1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dekker J. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 41.Simonis M, Kooren J, de Laat W. Nat Methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- 42.Published in this issue, DOI 10.1016/j.ymeth.2012.07.022
- 43.Published in this issue, DOI 10.1016/j.ymeth.2012.06.010
- 44.Published in this issue, DOI 10.1016/j.ymeth.2012.06.017
- 45.Published in this issue, DOI 10.1016/j.ymeth.2012.04.009
- 46.Published in this issue, DOI 10.1016/j.ymeth.2012.04.003
- 47.Published in this issue, DOI 10.1016/j.ymeth.2012.06.019
- 48.Published in this issue, DOI
- 49.Published in this issue, DOI 10.1016/j.ymeth.2012.05.001
- 50.Published in this issue, DOI 10.1016/j.ymeth.2012.06.018
- 51.Published in this issue, DOI 10.1016/j.ymeth.2012.08.009
- 52.Published in this issue, DOI 10.1016/j.ymeth.2012.04.004
- 53.Published in this issue, DOI 10.1016/j.ymeth.2012.04.010