Abstract
In eukaryotes, genome organization can be observed on many levels and at different scales. This organization is important not only to reduce chromosome length but also for the proper execution of various biological processes. High-resolution mapping of spatial chromatin structure was made possible by the development of the chromosome conformation capture (3C) technique. 3C uses chemical cross-linking followed by proximity-based ligation of fragmented DNA to capture frequently interacting chromatin segments in cell populations. Several 3C-related methods capable of higher chromosome conformation mapping throughput were reported afterwards. These techniques include the 3C-carbon copy (5C) approach, which offers the advantage of being highly quantitative and reproducible. We provide here a reference protocol for the production of 5C libraries analyzed by next-generation sequencing or onto microarrays. A procedure used to verify that 3C library templates bear the high quality required to produce superior 5C libraries is also described. We believe that this comprehensive detailed protocol will help guide researchers in probing spatial genome organization and its role in various biological processes.
Keywords: Chromatin, transcription, epigenetics, genome organization, structure
1. Introduction
In eukaryotes, genomes are contained inside cell nuclei in the form of highly organized chromatin. The non-random nature of genome architecture is overall manifested by the confinement of chromosomes into distinct nuclear volumes that are known as “chromosome territories” [1]. This organization appears important to reduce the length of genomes in order to fit within small nuclear volumes [2] and for the proper execution of numerous biological processes such as transcription. Notwithstanding its relatively high plasticity, chromosome organization can be observed at different scales. For example, low-resolution studies indicate that gene-rich chromosomes tend to localize at the center of the nucleus, while those with less genes are most often found near the nuclear periphery. Similarly, genes that are adjacent to nuclear matrix attachment regions often display low transcription activities [3]. Co-regulated genomic domains have also been found to physically co-localize in the nucleus [4]. These observations together highlight an important relationship between three-dimensional genome structure and function.
The link between spatial chromatin organization and function can also be observed at a higher resolution. At the molecular level, chromatin structure appears dependent on both DNA sequence and associated proteins. Although predominantly composed of genomic DNA wrapped around nucleosomes, the primary 11-nm chromatin fiber associates with a very large repertoire of non-histone proteins. The composition of chromatin can dictate whether DNA is organized into the less compact and more transcriptionally permissive euchromatin, or into heterochromatin, which is denser and transcriptionally repressed. Chromatin composition can also affect how the genome folds through the formation of long-range interactions. Long-range chromatin contacts were shown to be important for transcription. For instance, it was found that distal transcription regulation could be mediated by physical contacts between control DNA elements and target genes. Many examples of distal enhancer-promoter contacts have been reported genome-wide and found to involve genes located on the same (cis) or on different chromosomes (trans) to the control element [5–10]. In addition to transcription factors and the basal transcription machinery, other non-histone chromatin binding proteins are known to mediate long-range DNA contacts. Such is the case for the CCCTC-binding factor (CTCF) and cohesin, which play critical roles in genome organization and gene expression [11–15].
There remain a tremendous number of unanswered questions relating to high-resolution genome organization that include the number of different contacts, what mediates them and how they are regulated [16]. Moreover, three-dimensional chromatin structure itself could represent an important transcription regulation mechanism [17]. These questions can now be addressed with a group of high-resolution spatial genome mapping methods. The techniques broadly derive from the chromosome conformation capture (3C) technology developed in 2002 by Dekker et al. [18], and include 4C [19–22], 5C [23], 6C [24], GCC [25], Hi-C [26], and ChIA-PET [7]. All 3C-related techniques use formaldehyde cross-linking of cell populations to capture physical interactions between chromatin segments. The DNA of cross-linked genomes is then fragmented either by restriction digest or sonication, and converted into unique ligation products by proximity-based ligation. DNA fragments interacting more frequently in vivo will therefore yield higher levels of ligation products in the cell population. Given that the average physical distance between DNA fragment pairs is inversely proportional to the interaction frequency of corresponding pair-wise ligation products, the average spatial organization of genomes can be inferred from any 3C-related technique. Amongst these techniques, 5C can be used to map the three-dimensional organization of a given locus quantitatively at high-resolution and by high-throughput (Fig. 1a). 5C is relatively economical and with complete support information can easily be implemented in any laboratory despite its sophistication. Reference protocols describing 5C library production [23, 27, 28] and analysis [29] were independently described previously. In this article, we present a detailed version of the protocol currently used to produce 5C libraries destined for next-generation sequencing (Fig. 1b). This protocol can also be used to generate 5C libraries for microarray analysis. We explain a procedure for correctly assessing the quality of 3C libraries prior to 5C library production and how to generate high quality 5C libraries. We believe that this article will help researchers generate superior 5C libraries and will facilitate data analysis and interpretation.
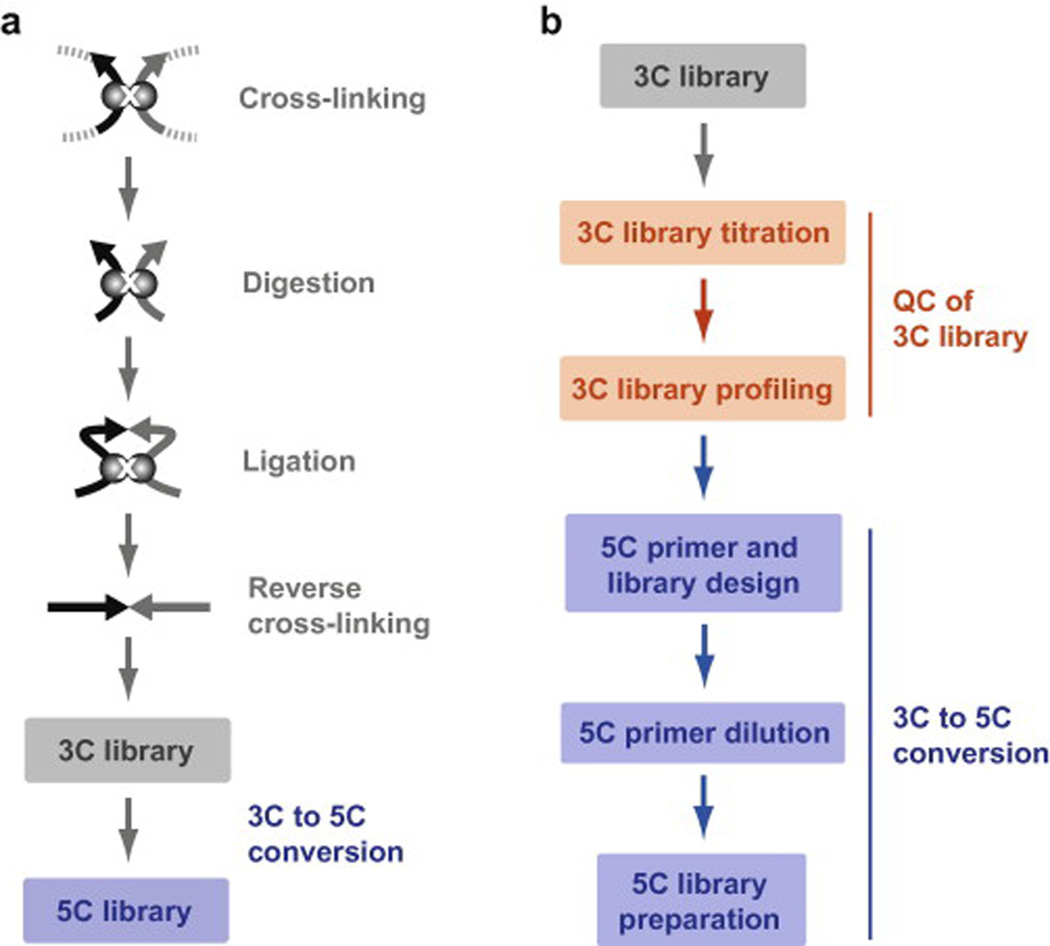
Fig. 1. Mapping chromosome conformation at high resolution with 5C technology.
(a) Schematic representation of the main experimental steps involved in 5C library production. 3C libraries are first prepared by capturing chromatin contacts in vivo with formaldehyde cross-linking, digestion with a restriction enzyme, inter-molecular ligation of cross-linked fragments with T4 DNA ligase, and reverse cross-linking. 5C libraries are next derived by annealing and ligating 5C primers onto predicted 3C junctions by ligation-mediated amplification (LMA; see text for details). (b) Flowchart of the 5C protocol steps described in this article. 3C library production has been described elsewhere and is presented in grey (see text for references).
2. 3C considerations
2.1 3C experimental design
The first step in performing 5C analysis is to prepare a 3C library. Selecting the right restriction enzyme to make the 3C library is a crucial decision because it will directly affect the type of experimental design that is most appropriate for the 5C analysis later on (discussed in section 4.2, below). Like 3C, 5C considers targeted genomic regions for analysis, and these should first be examined to identify the restriction enzyme that gives the best cutting pattern relative to the position of regulatory elements such as enhancers and transcription start sites. The enzyme should not bear significant star activity or be sensitive to DNA methylation. It should cut evenly and frequently within the region(s) of interest, and based on the resolution desired, a 4-cutter or 6-cutter may be selected to maximize the number of restriction fragments available for probing. Conversely, the regions studied should not contain significant repetitive sequence to facilitate 5C primer design and minimize the background.
2.2 Choosing the right concentration of formaldehyde
5C measures pair-wise 3C ligation products derived from proximity-based ligation of chemically cross-linked chromatin. When preparing 3C libraries, DNA fragments are cross-linked and generate 3C products in amounts that are more or less proportional to their average physical distance in cell populations. Hence, the concentration of cross-linking agent can have a significant impact on whether conformation can be resolved over short distances and if long-range contacts will rise above the background. Formaldehyde is mainly used as cross-linker because it forms cross-links between proteins and between DNA and protein, the cross-links are reversible and happen over short distances (<2 Å), hence favor proximal interactions. [m1] Although the mammalian 3C library cross-linking conditions correspond to those used during chromatin immunoprecipitation (ChIP) and thus might lend to a better integration of both types of datasets, a different type of cross-linker could be used or the concentration of formaldehyde could be titrated to achieve optimal detection of certain types of conformations.
3. 3C experiment
The template 3C libraries used for 5C analysis are prepared the same way as those used during conventional 3C. Detailed protocols describing how to generate 3C libraries from mammalian cells have been described elsewhere and will not be covered here [27, 30, 31]. For reference, 3C library production can be summarized into five simple steps as follows:
Cross-linking of cells
Cells are first cross-linked by adding formaldehyde at a final concentration of 1% for 10 min in the culture media. The reaction is then stopped with glycine at a final concentration of 125 mM.
Cell lysis
Lysis is carried out by resuspending the cells in a hypotonic buffer containing 0.2% NP-40 in the presence of protease inhibitors, and using a dounce homogenizer. The lysed cells are then washed with a restriction buffer compatible with the enzyme used in the next step.
Solubilization and digestion of the chromatin
The fixed chromatin is solubilized by adding SDS and heated at 65°C for 10 min. Triton X-100 is then added to quench the excess SDS, which allows efficient digestion. The restriction enzyme is added and incubated on a rotating platform overnight at the recommended temperature. The digestion is stopped by adding SDS and incubating the sample for 30 min at 65°C.
Ligation of the chromatin under dilute conditions
The ligation is carried out at 16°C for 2 to 4 hours with T4 DNA ligase in the presence of ATP and DTT. The reaction is performed under ∼20 fold diluted condition in the presence of Triton X-100 to further quench the excess SDS.
Reverse cross-linking and purification of the 3C library
Treating with Proteinase K at 55°C [m2]reverses the cross-links, and the DNA is purified by standard phenol-chloroform extraction. The resulting DNA suspension represents the 3C library.
3.1 Quality control of 3C libraries
5C library preparation is directly affected by the quality of 3C libraries such that 3C libraries should first be tested before proceeding to 5C. In this section, we describe two procedures to systematically assess the grade of 3C templates. We describe how to titrate 3C libraries with primer pairs against a gene desert region, and to characterize interaction frequencies in a given locus. These two steps measure parameters that are important for the production of superior 5C libraries. While 3C library titration reveals the overall purity and concentration of the library, measuring a set of pair-wise interaction frequencies in a given locus profiles the library’s content in unique contacts.
3.1.1 Reagents
Cellular 3C library
Control 3C library
Deionized autoclaved water used in all solutions and dilutions
10X PCR buffer: 600 mM Tris-SO4, (pH 8.9), 180 mM (NH4)2SO4
50 mM MgSO4
25 mM dNTPs (Invitrogen, cat. no. 10297-117)
10 mg/ml salmon testes DNA (Sigma, cat. no. D7656-1ML)
1X Tris-EDTA (TE) buffer pH 8.0: 10 mM Tris-HCl (pH 8.0), 1 mM EDTA (pH 8.0)
80 µM 3C primer stocks reconstituted in 1X TE
Taq DNA polymerase (NEB, cat. no. M0273L) or AmpliTaq Gold® (Invitrogen, cat. no. 4398808)
Agarose (Wisent, cat. no. 800-015-CG)
10X Tris-borate-EDTA (TBE) buffer: 890 mM Tris Base, 890 mM Boric Acid, 20 mM EDTA
10 mg/ml ethidium bromide solution (Bio-Rad cat. no. 161-0433)
4X agarose gel loading solution: 10% Ficoll, 0.15% Xylene Cyanol.
3.1.2 3C library titration
3C libraries are usually titrated by semi-quantitative PCR with primer pairs against neighboring restriction fragments, and against distant fragments that are typically 60–100 kb away from each other (Fig. 2). PCR products are then resolved on agarose gels containing ethidium bromide (panel a) and quantified (panel b). Titrating 3C libraries before 5C library production is important for several reasons. First, it verifies that unique contacts are present in the library and that their intensities increase with increasing library volumes. A good 3C library will yield a single PCR product on agarose gel that will plateau at higher volumes and progressively disappear at greater dilutions (Fig. 2a). Titrations can sometimes display a biphasic pattern where PCR products initially decrease with increasing dilutions but transiently increase at greater dilutions. This type of profile is not desirable as it may indicate the presence of impurities that can interfere with 5C library production. The amplification of primer dimers at low library dilution and/or the absence of a smooth signal titration throughout the dilution range can also reflect the presence of impurities. We found it very helpful to use Amicon Ultra Centrifuge Filters (Milipore, cat# UFC5030BK) on the final steps of 3C libraries production [31]
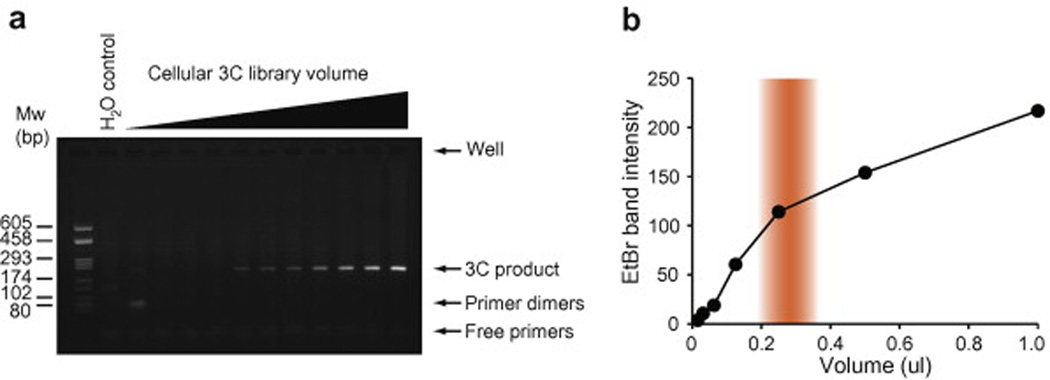
Fig. 2. Example of a cellular 3C library titration.
(a) Acceptable result of a cellular 3C library titration resolved on agarose gel and stained with ethidium bromide. This example shows the result of a 3C library analyzed by twofold serial dilution and amplified by endpoint PCR with GD34 and GD35 primers (Table 1). This primer pair recognizes neighboring fragments in a gene desert region (ENCODE region ENr313). Endpoint PCRs were resolved on a 1.5 % agarose gel containing 0.5 µg/ml ethidium bromide. (b) Gel quantification of the titration shown in (a). The volume of cellular 3C library stock is indicated on the x-axis. The intensity of PCR products stained with ethidium bromide is indicated on the y-axis. Ideal volume range is highlighted in orange.
We recommend titrating libraries with primers against a gene desert region because chromatin contacts do not typically change a lot with cellular state in these regions. Thus, using gene deserts can avoid introducing biases in PCR linear amplification range between samples that could influence the detection of differences in chromatin contacts in test loci. In human cells, we routinely titrate our 3C libraries by probing the ENCODE ENr313 gene desert region [32]. When titrating with primer pairs against neighboring fragments, the amount of 3C library selected for further chromatin contact profiling (see section 2.3.) should fall within the range preceding saturation and must be in the middle of the linear amplification range (Fig. 2b). 3C libraries yielding very little PCR products from neighboring gene desert fragments and failing to plateau at highest volumes may not be sufficiently concentrated for the production of 5C libraries (see section 3.3.). A set of validated gene desert primers designed for BglII 3C libraries is presented in Table 1[m3]. We describe below a procedure recommended for titrations performed with primer pairs against neighboring fragments in a gene desert region.
Table 1.
Validated BglII 3C library gene desert primers.
| Primer name | Primer sequence (5’-3’) | Distance from BglII restriction site (bp) |
|---|---|---|
| GD34 | GTGATGTTAAATGAAGGATTGGCAAACAG | 189 |
| GD35 | GGGGTACGCCTGATAGGAATATTACAAGG | 170 |
| GD36 | CAGGCTTTATTTCCATTCCTCTAATTGAGTTCCC |
151 |
| GD37 | GTATGACACAGAGAAAATGTGGTGCATTCACC |
266 |
| GD38 | CAATAGGACTCAATTGCTTTAAGTTCCTGGGG |
125 |
| GD39 | GCTGTGCATACCCTAAAGATTTGGTAAAGCC |
108 |
| GD40 | GCACATCACCTACAACCAACAAACAGGAAAC |
245 |
| GD41 | CTCTCTCTCTCACATACAAGATGGGCAGC | 171 |
| GD42 | CAACCTGCTCCTCAGTAGTGACTTTTGGG | 177 |
| GD43 | CATCCAAATGAAGCACCAATTCAGGAATGCAATC |
115 |
| GD44 | GCTGCCCATCGCAACCAGCTGATCTTCAAC |
309 |
| GD45 | CCAATTCCATTTAACAATGAGCATGCACTGAGC |
224 |
| GD46 | GTCTACAAATATGTATCTCCCTTTCAAGTCCAC |
140 |
| GD47 | CAGTGTCTTTCAGATTCTATCCTAGTCTTACTGG |
130 |
| GD48 | CATGAGTACACAAAAATGGCTGCTTGATGCC |
115 |
| GD49 | CTTCTTGTTGAACTGTAATTATAACAGAGCCACC | 104 |
| GD50 | CAGGAGGCCGAGTTATGCTACCCATTTGG | 213 |
| GD51 | CACCATATCTGGCCACTATAATAGATTTCTGACTC |
187 |
| GD52 | GATGAGAGAAAGGTGGTTCTATGAGAACATATGC |
101 |
| GD53 | CTTTGTGGAACAGCCATTGGCAAAAGTCC | 111 |
| GD54 | CATCGGGTAGGAATACCTCCCATACTACC | 174 |
| GD55 | CAGTCAAACTTAGATACGCAGAGGCAAGG | 175 |
| GD56 | GACTGTTTACTTAGCAGATTTGTGGTAGAATGG |
98 |
| GD57 | GCACACCTGTTTCAGACTAGGCTCCTCC | 238 |
| GD58 | CATCTCACTGCCATAAAGCACTTCAGAATG |
230 |
3.1.3 3C titration procedure
On ice, prepare twofold 3C library dilutions starting with approximately 250–500 ng of cellular library. Eight to ten dilutions should be sufficient. Include a water control to verify the absence of contamination in the PCR mix and to control for primer-dimer formation.
-
On ice, assemble the PCR reactions in individual tubes or a 96-well plate, according to Table 2.
* All reagents should be chilled before mixing. The Taq DNA polymerase may be added directly to the master mix when using an enzyme modified to automate the Hot Start technique such as AmpliTaq Gold. Otherwise, we recommend adding the Taq DNA polymerase at the very end of the experimental setup to minimize primer dimer formation. This can be achieved by first dispensing a master mix lacking the water and Taq, followed by the 3C template, and finally the Taq dilution.
Amplify 3C products according to the PCR program outlined in Table 3.
Add 8 µl of 4X agarose gel loading solution and mix by pipetting.
-
Resolve 15 µl of each PCR sample on a 1.5% Agarose gel containing ethidium bromide (0.5 µg/ml). Include a molecular weight ladder to verify the expected size of PCR product.
* Expected PCR product sizes are calculated by adding the distance of each primer from their respective restriction site (Table 1).
Catalog titration result with a gel documentation system. Verify that only one PCR product of the correct size is linearly amplified throughout the dilution series. There should be no primer dimers and PCR signals should progressively decrease with increasing dilution as illustrated in Fig. 2a.
Quantify PCR products in each lane and plot signal intensities against the amount (?) of[m4] 3C template used in corresponding reaction as shown in Fig. 2b.
Select a volume (amount) of 3C library in the linear amplification range as shown in Fig. 2b.
Table 2.
3C PCR reaction composition.
| Stock component | Volume (µl) per rxn* | Final per rxn |
|---|---|---|
| 10X PCR buffer | 2.5 | 1× |
| 50 mM MgSO4 | 2.0 | 4.0 mM |
| 25 mM dNTPs | 0.2 | 0.2 mM |
| 100 ng/µl STD** | 1.5 | 150 ng |
| 5 µM 3C Primer A | 2.0 | 0.4 µM |
| 5 µM 3C Primer B | 2.0 | 0.4 µM |
| Taq DNA polymerase 5 U/µl | 0.2 | 1 U |
| H2O | 10.6 | |
| 3C library dilution | 4.0 |
Refers to 25 µl reactions
STD; salmon testis DNA stock diluted in water. Adding STD in the reaction mixture can help reduce the background but is optional.
Table 3.
3C PCR reaction conditions.
| Cycle number | Denature | Anneal | Extend | Cool |
|---|---|---|---|---|
| 1 | 94 °C | |||
| 5 min | ||||
| 2–35 | 94 °C | 65 °C | 72 °C | |
| 30 s | 30 s | 30 s | ||
| 36 | 94 °C | 65 °C | 72 °C | |
| 30 s | 30 s | 8 min | ||
| 37 | 10 °C | |||
| infinity |
3.1.4 3C library content profiling [m5]
Once 3C libraries are found to titrate well and amounts have been selected, the next step in 3C library quality control involves measuring interaction frequencies in a given locus. This process verifies the presence of expected chromatin contact patterns that are reflective of proper cross-linking, digestion, and ligation during 3C library preparation. 3C library profiling also assesses technical and biological reproducibility, and verifies that library volumes defined from titrations are neither too high nor too low before proceeding to 5C. As 5C data produced later should recapitulate results obtained from 3C library profiling, these data can also be compared to assess the quality of 3C to 5C library conversion.
We recommend profiling the interaction frequency of random collisions in a gene desert region or if known, measuring long-range interactions in a locus of interest (Fig. 3). Gene deserts are regions spanning ∼0.5–1 Mb of genomic DNA where no genes are annotated or predicted. The rationale for choosing a gene desert region lies on the premise that the absence of genes reduces the probability of regulated long-range interactions. We have successfully used the pilot ENCODE region ENr313 of human genome as gene desert region [13, 33]. Regardless of whether random collisions or long-range contacts of a locus of interest are used, interaction frequencies should vary with distance, with interaction frequency decreasing with increasing genomic distance (panel a), and looping contacts from an “anchored” site in the genome transiently displaying greater interaction frequencies (panel b). Because conventional 3C analysis is performed “point-by-point” with pair-wise primer combinations, control 3C libraries must be generated to normalize differences in primer pair efficiencies. Control 3C libraries are usually produced from BAC clones featuring the corresponding test genomic regions and has been described in detail elsewhere [28]. Since no cross-linking is involved, BAC DNA is used to make a library of randomly ligated restriction fragments in the absence of any DNA-protein or protein-protein interactions. A randomly ligated library should contain all possible restriction fragment combination irrespective of distance between them and hence any primer pair tested for 3C titration should theoretically result in equal amplification unless there is a bias in primer efficiency, which can be corrected using this control [27, 30]. We describe below how to measure the frequency of random collisions in a gene desert locus.
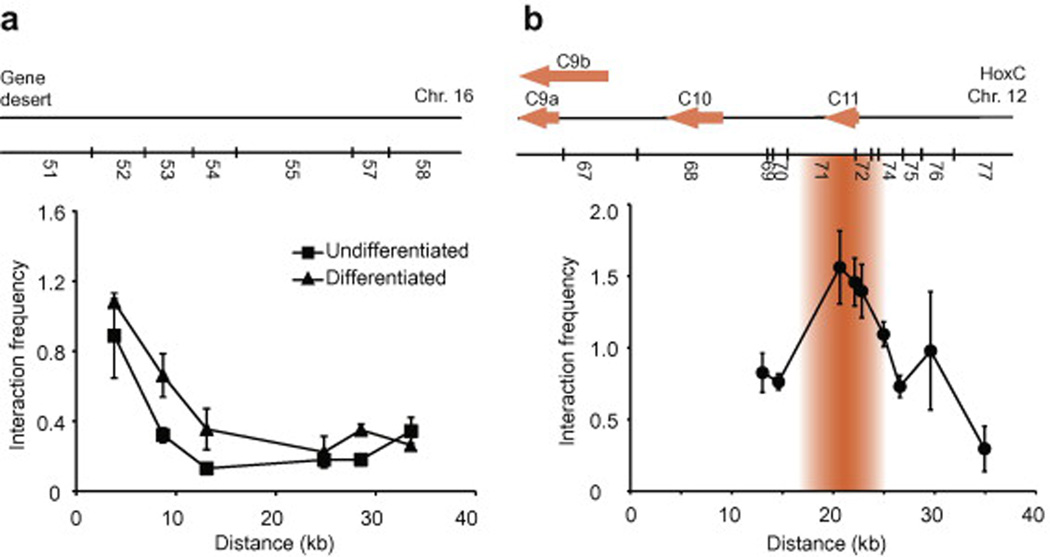
Fig. 3. Examples of 3C library content profiling.
(a) Profiling 3C libraries using random collisions. This example shows a “fixed point” analysis of two 3C libraries in a gene desert region. 3C libraries were from a differentiation time-course of NT2/D1 cells with retinoic acid as described previously [13]. “Fixed point” analysis was performed by combining the GD51 primer pair-wise with oligos GD52 to GD58 recognizing downstream BglII restriction fragments (Table 1). Interaction frequencies measured in the ENr313 gene desert region derive from random collisions, which decrease with increasing distance as this region is devoid of looping contacts. Error bars show the standard deviation (SD). (b) Profiling 3C libraries using known long-range interactions in a given locus. This example shows a “fixed point” analysis of the human HoxC cluster 5’ end in undifferentiated NT2/D1 cells as described previously [13]. Interaction frequencies between a region containing the HoxC9 gene promoter and downstream cluster genes confirm the presence of a looping contact with HoxC11 in this cell line. Error bars show the standard error of the mean (SEM). The x-axis indicates the linear genomic distance between interacting chromatin fragments. Interaction frequencies are indicated on the y-axis. Linear representations of the corresponding genomic regions and predicted BglII restriction patterns are shown above each graph.
3.1.5 3C profiling procedure [m6]
-
On ice, assemble the PCR reactions in individual tubes or a 96-well plate according to Table 2. 3C library volumes defined from the titrations should be adjusted to 4 µl with water. Prepare at least duplicates of 3 PCRs for each primer pair for both cellular and control 3C libraries
* Chill all reagents before mixing. Note that when measuring random collisions from an “anchored” site, only one primer remains constant in the master mix. The variable primer should be added independently from the master mix and the template.
Amplify 3C products according to the PCR program outline in Table 3.
Add 8 µl of 4X agarose gel loading solution to each sample and mix by pipetting.
-
Resolve 15 µl of each PCR sample and a molecular weight ladder on a 1.5% agarose gel containing 0.5X TBE and ethidium bromide (0.5 µg/ml). For each primer pair, load cellular and control technical triplicates side by side on the gel to avoid noise from differences in ethidium bromide staining along the gel.
* Always verify expected PCR product sizes by adding the distance of each primer from their respective restriction site.
Catalog titration result with a gel documentation system. Verify that only one PCR product of the correct size is amplified from each primer pair and that there are no primer dimers. Exclude all samples exhibiting these problems and repeat PCR triplicates when necessary.
Quantify PCR products individually and subtract the background. For every pair-wise interaction, divide each cellular signal by each control library signal, and calculate the average ratio to obtain corresponding interaction frequencies and standard deviations.
-
Plot interaction frequencies relative to genomic distance as shown in Fig. 3a.
* The genomic distance between the 3’ end restriction site of the “anchored” site and the 3’ end restriction site of the variable fragment is usually considered as the distance between pair-wise interactions.
When test regions are compared in two or more libraries, gene desert “anchored” profiles can be used to estimate a normalization factor to correct the strength of interaction frequencies in test regions. This is done by calculating the averaged log(ratio) from each corresponding pair-wise interactions. Normalization factors represent the inverse of this average.
4. 5C considerations
3C libraries contain new ligation products in addition to other genomic DNA that was either not digested or digested but not ligated. There are several types of ligation products in 3C libraries. These include self-circularized and so-called “head-to-head” (H-H), “tail-to-tail” (T-T), “head-to-tail” (H–T), and “tail-to-head” (T-H) products, with “head” and “tail” referring to the restriction sites at the 5’ and 3’ ends of DNA fragments, respectively. [m7]During 3C, primers are usually designed unidirectionally along chromosomes and simply mixed pair-wise to measure either H-H or T-T products [18, 27, 30]. [m8]This experimental design avoids detecting mixtures of uncut and partially re-ligated DNA fragments, or the highly abundant self-ligated products. Since H-H and T-T products derive from proximity-based ligation in quantities that are proportional to their average physical distance, their relative abundance can be used to infer three-dimensional chromatin organization.
5C measures the same H-H or T-T products as 3C but can detect a very large number of different interactions simultaneously rather than point-by-point (Fig. 4). [m9]During 5C, the junctions of H-H or T-T 3C products are quantitatively detected with 5C primers using a modified ligation-mediated amplification (LMA) approach, which can be conducted at a very high level of multiplexing. LMA is performed in two simple steps that include annealing 5C primer pairs on the same DNA strand at predicted 3C junctions followed by ligation of annealed primers with an NAD-dependent DNA ligase. Because this class of enzyme only ligates nicks in dsDNA, the perfectly paired forward and reverse 5C primers annealed onto 3C templates imitates a “nick” and hence can be ligated efficiently. The relative abundance of selected 3C ligations products can thus be quantitatively converted into a collection of pair-wise ligated 5C primers. This “carbon-copy” of selected 3C junctions represents the 5C library, which is then amplified with universal primers dedicated for either microarray hybridization or high-throughput DNA sequencing. As 5C primer annealing efficiency onto 3C junctions is not significantly different within multiplexed primer pools, a control 3C library is only needed when microarray hybridization the selected probing method because differences in hybridization efficiency must be corrected [27, 28]. Below, we explain how to design 5C experiments, and provide an updated protocol to generate high-quality 5C libraries.
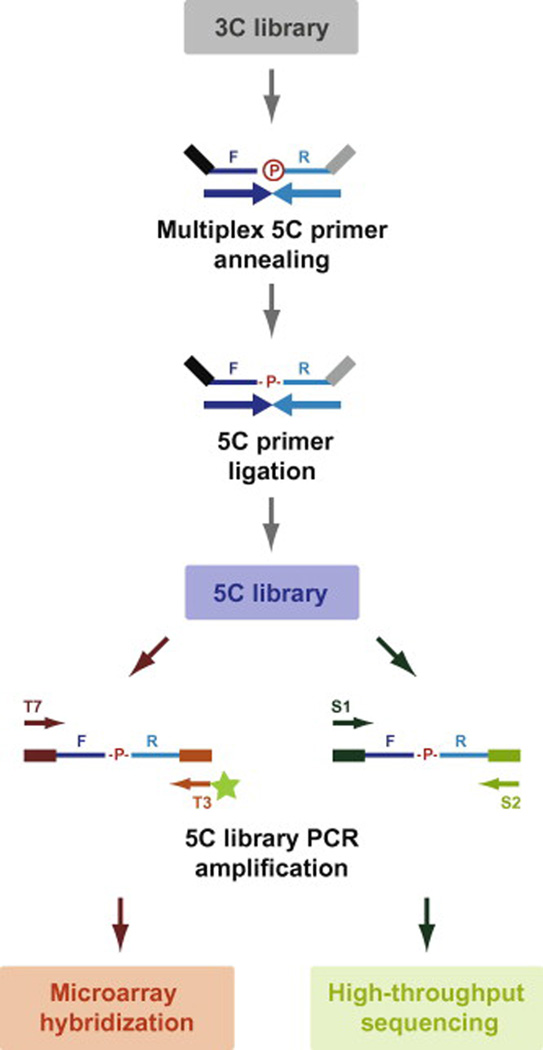
Fig. 4. 3C to 5C library conversion.
5C libraries are produced by annealing 5C primers at predicted 3C junctions in a multiplex setting followed by specific ligation of annealed primers with an NAD-dependent DNA ligase. The universal tails of 5C primers are illustrated as black and grey lines and are used to amplify libraries in a single PCR step. Universal tail and corresponding one-step PCR primer sequences depend on whether microarray hybridization or high-throughput sequencing is selected for analysis. Microarray forward and reverse tail sequences are T7: 5’-TAATACGACTCACTATAGCC-3’ and T3c: 5’-TCCCTTTAGTGAGGGTTAATA-3’ respectively. Reverse universal PCR primers used for microarray analysis must be fluorescently labeled at their 5’ ends as highlighted by the green star. High-throughput sequencing forward and reverse tail sequences are S1: 5’-CCTCTCTATGGGCAGTCGGTGAT-3’ and S2c: 5’- AGAGAATGAGGAACCCGGGGCAG-3’ respectively.
4.1 Regions of interest
5C can be used to measure pair-wise interaction frequencies at several genomic regions simultaneously. When multiple loci are interrogated, we recommend probing regions that are similar in size and no larger than a few Mb. Whenever possible, we also recommend that regions be probed with 5C primer schemes that are similar in complexity to even out representation in sequencing datasets. Combining primer schemes that yield a comparable expected number of different 5C ligation products will promote equal representation of all regions considered in the sequencing data as described below.
4.2 5C experimental design
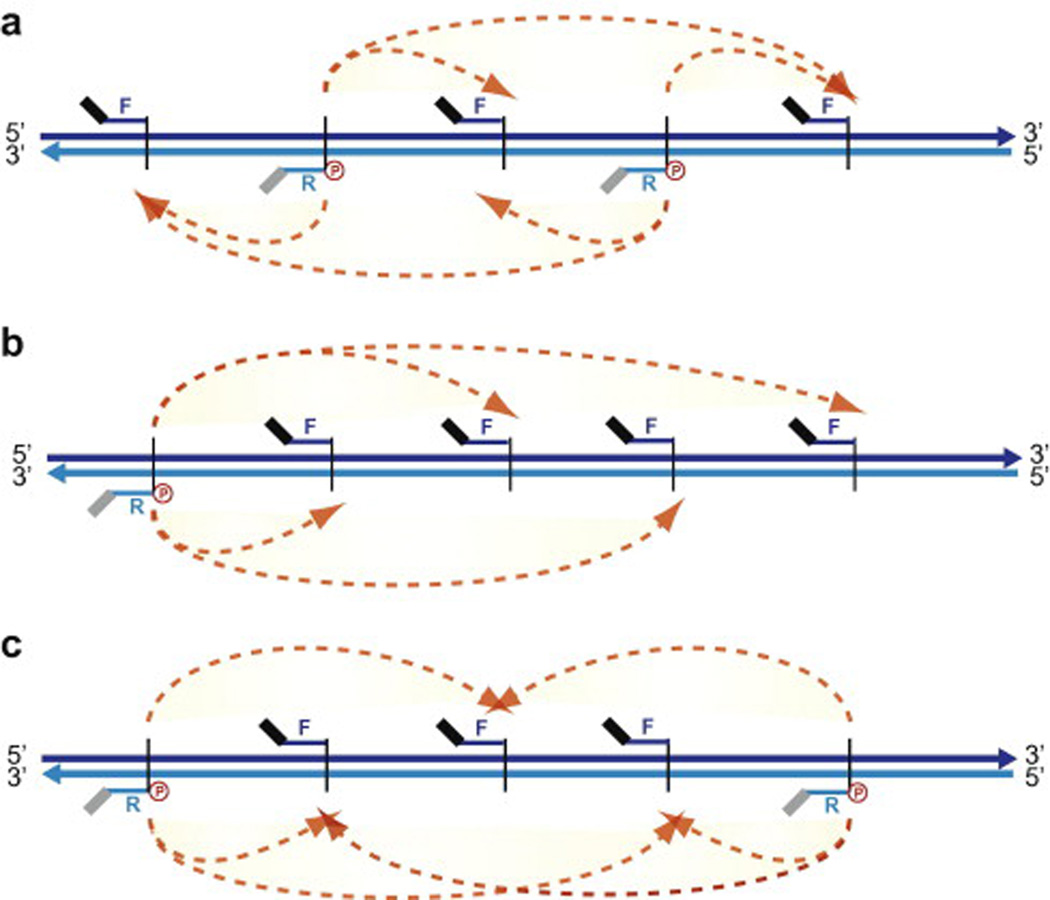
In addition to tailoring the 3C library design to the type of 5C question, the 5C primer design scheme can be adapted to accommodate different kinds of questions, ranging from long-range looping contacts to general chromatin architecture. There are several important general considerations when designing a 5C library. First, contacts can only be detected between fragments when one fragment is represented by a forward 5C primer and the other by a reverse primer. This is because primers must anneal onto the same strand of H-H or T-T 3C junctions. Second, because the unique sequences of forward and reverse 5C primers are exactly anti-sense to each other, any given restriction fragment can only be probed by either a reverse or forward 5C primer (Fig.5). Thus the maximum number of interaction frequencies measured per 5C library is reached when consecutive forward and reverse primers are mixed. Combining different 5C library designs for higher coverage can potentially circumvent this inherent 5C limitation [29]. This option is particularly important when mapping compaction profiles to estimate average chromatin fiber structures. When measuring long-range interactions from a given fixed genomic region however, we recommend representing the “anchored” fragment by a reverse 5C primer and the remaining surrounding domain with either forward primers, or a mixture of both. There are essentially three types of 5C design schemes as illustrated in Fig. 5 and outlined below.
Fig. 5. Types of 5C experimental design.
(a) Alternating scheme. Alternating forward and reverse 5C primers represent each consecutive fragment. (b) Anchored scheme. A reverse 5C primer represents a given fragment while forward 5C primers represent the rest. (c) Mixed scheme. This experimental design features both fixed anchored and alternating schemes.
Alternating scheme
In this scheme, forward and reverse 5C primers are selected alternatively on consecutive restriction fragments. This scheme is ideal when probing the three-dimensional organization of domains or to investigate the existence of long-range looping interactions in an unbiased manner such as when no prior information on the regulatory elements is available.
Anchored scheme
This scheme is more targeted and can be used to examine either the interaction pattern of a specific class of regulatory DNA element (e.g. DHS site, TSS, enhancer, insulator) with other elements or with the surrounding genomic region. For example, the anchored scheme could be used to measure long-range looping interactions between the promoters of genes and their enhancers by representing one type of element with forward 5C primers and the other with reverse oligos. In this case, a priori knowledge of the position of elements, which is increasingly available for many species, is necessary [34–37].
Mixed scheme
The mixed scheme includes elements from both alternating and anchored designs to examine chromatin architecture. When this design is used, anchored sites are usually incorporated first in the pattern. The remaining restriction fragments are then each represented by alternating forward and reverse 5C primers, while trying to keep the representation of each type of 5C primer equal, and the overall design complexity comparable to the other regions when more than one domain is examined.
4.3 5C primer design
As 5C products derive from the ligation of perfectly and contiguously annealed primer pairs onto the same strand of H-H or T-T 3C library junctions, each 5C product therefore consists of one forward and one reverse 5C primer as described above and previously [23, 27]. Only reverse 5C primers are phosphorylated at their 5’ ends to facilitate ligation between 5′-P of the reverse primer and 3′-OH of the forward primer. Forward and reverse 5C primers comprise two parts; one universal sequence and a unique sequence corresponding to the sense or antisense strand of restriction fragments. Universal tail sequences differ between forward and reverse primers and serve to PCR-amplify 5C libraries. For this reason, tail sequences are located at the 5’ end of forward primers and 3’ end of reverse 5C primers (Figs. 4 and 5). Since H-H and T-T are thought to exist in equimolar ratios, we usually measure only one of these configurations and design primers with unique sequences corresponding to the 3’ end of restriction fragments.
The optimum threshold for size, melting temperature (Tm), BLAST score, and GC content for 5C primers are similar to that of any other PCR primer. Oligos not adequately fulfilling these criteria should be excluded from the design. 5C primers should not form extensive secondary structure or be complementary to each other. Similarly, primers corresponding to very short (<500 bp when a six-cutter restriction enzyme used for a 3C library production ) or very large (>15 kb) restriction fragments should be excluded. We recommend centering all 5C primers at a specific Tm (usually around 55°C by default it is 65 deg C for my5C for the entire primer including universal tail), and to adjust primer lengths with non-specific sequence at the 5' and 3' ends of the forward and reverse primers, respectively.
At least two web-based programs are now available to design 5C primers from a given genomic region around commonly used restriction enzyme sites. The first program is called “5CPrimer” and was developed to generate 5C libraries probed by microarray hybridization [29, 33, 38]. This program predicts forward and reverse primer sequences based on melting temperature and can also set a maximum synthesis cycle number to match 5C primer lengths to microarray feature lengths as previously described [23, 29, 33]. The .txt file generated by this program can be opened directly in excel making batch primer orders very convenient.
The second program available for 5C primer design can be found on the my5C platform [39]. A full suite of web-based 5C tools was developed for the design, analysis and data visualization of 5C data using my5C. These tools allow users to design a 5C experiment for any given locus and species just by pasting the DNA sequence of interest. The web tool will then automatically design the 5C primers using chosen primer layout and filtering processes to create a 5C primer pool, which are optimally designed taking into considerations all the parameters. Once primers are designed and subsequently experiments are done, a full spectrum of analysis, integration and visualizations tools are available for the data analysis. Both sites are highly intuitive although design through my5C offers the advantage of excluding low complexity and non-specific primers systematically through blasting and repeat masker. This feature is particularly important considering that low complexity primers tend to yield high noise levels.
4.3.1 Using different 5C tails
Universal sequences are usually approximately 20 nt in length but can vary in length and composition. Similarly, the unique regions can also vary but are usually between 25 to 30 nt long. For these reasons, the average length of 5C library products is typically never less than 100 bp and no more than 150 bp. Universal 5C primer tails can have any sequence as long as this sequence is not found in the corresponding 3C library genome. We have previously used the T7 and complementary T3 sequences successfully to produce 5C libraries probed on microarrays or high-throughput DNA sequencing [23, 33]. Note that reverse universal PCR primers used to amplify libraries hybridized onto microarrays must be fluorescently labeled at their 5’ ends. Other sequences have also been used during high-throughput DNA sequencing (Fig. 4 and [40])(put here reference for “emulsion” sequence Bau, Sanyal and Lajoie et al NSMB 2011, Sanyal and Lajoie et al ENCODE Nature Sept 2012 in press). [m10]
For the downstream analysis of 5C libraries with short read next-generation sequencing (NGS), each 5C primer can be uniquely barcoded using an n-mer (usually 4–6 nucleotides) sequence that is added between universal tail and the specific complementary sequence. The sequence of the “universal” tails gets sequenced in every NGS reads by default, which limits the sequencing of unique bases to <15. Hence, these barcodes can be used effectively to assign the sequencing reads to the corresponding primers and ultimately map to the genomic coordinates. Alternatively, the sequence of the sequencing primers themselves (Illumina or SOLiD) can be used as “universal” tail in 5C primers to get sequencing read with sufficient coverage of unique bases and effectively assign that read to the corresponding primer during mapping. In the future, adaptation of multiplexed sequencing using unique identifiers or “index” may facilitate sequencing of multiple 5C libraries (tagged with unique identifiers) in a single sequencing run and also reduce the cost.
Once 5C primers and libraries are designed, the generation of high quality 5C libraries is relatively quick and simple. A key step in 5C library production is the dilution of 5C primers, which can yield very high non-specific background when improperly conducted. We describe below how to prepare diluted primer pools and produce 5C libraries from validated 3C templates.
5. 5C experiment
5.1 Titrating 5C primers and 3C libraries
When a small number of 5C primers are used, the individual 5C primers can be ordered in a lyophilized form at a reduced cost. If so, the 5C primer stocks must first be resuspended into 1x TE. We usually reconstitute primers at 80 µM in 1X TE and heat stocks 15 minutes at 65°C prior use. To avoid contamination of stocks, we suggest aliquoting all stocks and storing them at −80°C.
When the same primer set is used often or if a large number of primers are required for the analysis, we recommend first pooling the 5C primers. Primer pools will simplify and accelerate 5C analysis and reduce the variability between experiments. Large 5C primer sets can be ordered already resuspended in 1X TE at a given concentration (typically between 50 to 100 µM) in a 96 well plate format. Pools can be prepared directly from stock aliquots by mixing an equal volume of each primer into an eppendorf tube. We recommend pooling forward and reverse primers separately to reduce the incidence of template-independent 5C products. When analyzing multiple regions, we also suggest keeping the forward and reverse primer pools of each region separate such that regions may be analyzed independently if desired without having to make new pools. At a cost, the 5C primers can be ordered already 5' end-phosphorylated. Alternatively, reverse 5C primer pools can be phosphorylated with Polynucleotide kinase using the standard procedure. Large primer pools can take a long time to generate, and we suggest aliquoting and storing them at −20°C.
Forward and reverse 5C primers should be kept separate until immediately before use[m11], diluted and mixed in equimolar concentrations before proceeding with the titration. In general, for human and mouse, we found that pools where individual 5C primer concentrations range between 0.5 to 1 nM [m12](We typically use 0.5–1fmol of individual primer in each 5C annealing reaction or 1uL of 0.5–1 nM per 5C annealing reaction (we use 20uL annealing reaction volume) hence it comes to 25–50 pM per 5C primer per 5C annealing reaction) in the reactions work best. Similarly, we find that on average a volume of 3C library containing 100,000 to 200,000 genome copy equivalents works well. [m13]When several cell lines are analyzed with the same 5C primer pool, we recommend that primer titration be conducted for each cell type since the same region can be present in different copy numbers. Also, the efficiency of 5C library production can differ between samples because of differences in cross-linking efficiency or purity. When more than one region is characterized, we recommend creating 5C libraries separately, and combining them before analysis to avoid differences due to copy number variations.
5.2 5C primer pool titration
The optimal 5C primer pool concentration will depend on the number of 5C primers used and the factors mentioned above. We generally titrate primer pools over a range 5- 500 pM of each 5C primer per 5C annealing reaction as per above calculations). Below is a brief example of a titration scheme for a pool of 100 primer stocks (50 forward and 50 reverse) each at 100 µM.
Mix 5 µl of each forward or reverse 5C primer stock in separate eppendorf tubes.
-
Pipet up and down and briefly spin down at maximum speed at 4°C. The concentration of 5C primers in each pool is now 2 µM.
*Tubes and primers should be ice-cold before mixing to minimize background. Mix on ice.
-
Dilute each 5C primer pool individually 10-fold in water by mixing 5 µl of each primer pool with 45 µl of water. Mix by pipetting up and down and briefly spin down each dilution at maximum speed at 4°C. The primer pools are now each at 200 nM.
*Tubes, water and primers should be ice-cold before diluting to minimize background. Dilute on ice.
On ice, mix 25 µl of each forward and reverse primer pool for a final individual 5C primer concentration of 100 nM.
-
Prepare two-fold serial dilutions of this primer pool over a range of 100 nM to 0.625 nM. 1 µl of each dilution will be used to titrate primers (over a range of .0625 to 10 nM final concentrations) [m14]as described in section 5.5.[m15]
For me it’s always easier to think in femto moles of each individual primer in annealing reaction. So I put in titration 5, 2.5, 1, 0.5 and 0.1 fmol of each primer. 1fmol /20mkl = 0.05nM or 50pM
5.3 Diluting 5C pool stocks
Once an optimal 5C primer pool concentration has been identified, frozen 5C primer pools should be diluted in water and mixed before each experiment. Below, we provide an example of a dilution scheme for mixing ten primer pools (each at 2 µM final 5C primer concentration) to an optimal final concentration of 1 nM in the annealing reaction.
-
On the day of the experiment, dilute an aliquot of each 5C primer pool stock (2 µM) individually ten fold in water. Mix by pipetting up and down and briefly spin down each dilution at maximum speed at 4°C. Primer stocks are now each at 200 nM.
* Tubes, water and primers should be ice-cold before preparing dilutions to minimize background. Prepare dilutions on ice.
-
Immediately before use combine 10 µl of each primer pool dilution into an eppendorf tube. Add 100 µl of ice-cold water for a final 5C primer pool concentration of 10 nM.
* Unused primer dilutions in water must be discarded on the same day and should never be stored for later use. Never freeze-thaw primers stored in water as their activities may decrease.
-
For each set of reactions, prepare a 5C primer master mix by combining 1.0 µl of 5C primers pools with 1 µl of cold 10X NEB Buffer 4 (10X annealing buffer). 2 µl of each 5C primer pool master mix will be used in each 10 µl 5C annealing reaction for a final concentration of 1 nM.
* Prepare on ice immediately before use.
5.4 3C template titration
In general, approximately 600 ng of cellular 3C library is used as template for 5C library production. This amount of cellular library is approximately ten times the amount of library used during conventional 3C analysis, and corresponds to approximately 100,000 human genome equivalents if we consider 6 billion bp for a diploid human genome, and an average bp mass of 660 Da. Since the quantities mentioned above can vary based on the complexity, level of cross-linking, and purity of 3C libraries, and on the predicted 5C library complexity, we recommend first performing 3C template titrations to test new experimental designs. This step simply involves using a range of 3C library amounts and verifies that the 5C libraries produced are both quantitative and sufficient for further analysis. We suggest titrating 3C libraries two-fold over a range of 50,000–800,000 haploid genome equivalents as a starting point.
5.5 Making a 5C library
5.5.1 Reagents
Cellular 3C library
Control 3C library (required only when probing by microarray hybridization)
Deionized autoclaved water used in all solutions and dilutions
Forward 5C primer pool stocks resuspended in 1X TE (desalted)
Reverse 5C primer pool stocks resuspended in 1X TE (desalted and phosphorylated)
80 µM universal tail PCR primer stocks (reverse microarray PCR primers must be fluorescently labeled at their 5’ ends)
10 mg/mlNEB Buffer 4
10 mg/ml salmon testes DA (Sigma, cat. no. D7656-1ML)
Taq DNA ligase and buffer (NEB, cat. no. M0208S)
10X PCR buffer: 600 mM Tris-SO4, (pH 8.9), 180 mM (NH4)2SO4
50 mM MgSO4
25 mM dNTPs (Invitrogen, cat. no. 10297-117)
Taq DNA polymerase (NEB, cat. no. M0273L) or AmpliTaq Gold® (Invitrogen, cat. no. 4398808)
QIAquick gel extraction kit (Qiagen, cat. no. 28704) or MinElute PCR purification kit (Qiagen, cat. no. 28004) if libraries are analyzed with microarrays.
Agarose (Wisent, cat. no. 800-015-CG)
10X Tris-borate-EDTA (TBE) buffer: 890 mM Tris Base, 890 mM Boric Acid, 20 mM EDTA
10 mg/ml ethidium bromide solution (Bio-Rad cat. no. 161–0433)
4X agarose gel loading solution: 10% Ficoll, 0.15% Xylene Cyanol.
5.5.2 Procedure
The 5C library preparation protocol begins with mixing 3C library templates with salmon testes DNA to increase the specificity of LMA product formation. 5C primers are then added to the mixture and left to hybridize overnight. Annealed primers are ligated the following morning, and resulting 5C libraries are PCR-amplified for microarray or next-generation DNA sequencing. Amplified 5C sequencing libraries are finally purified from agarose gels to remove unincorporated 5C and PCR primers, and primer dimers. 5C libraries analyzed onto microarrays can either be purified on gel or from solution providing no primer dimers are present. [m16]The production of 5C libraries should always be conducted with a set of controls that include a “no ligase”, “no 5C primer”, “no template” 5C control, and a PCR “water control”. These controls verify the absence of contamination and PCR artifacts.
Annealing
-
On ice in 0.5 ml PCR tubes, mix 3C templates, water, and salmon testis DNA to a final DNA amount of 1.5 ug and a volume of 8 µl. Include the controls as outlined in Table 4.
* Always pre-chill all reagents and tubes before mixing.
Mix by pipetting and centrifuge briefly at maximum speed at 4°C.
Add 2 µl of 5C primer master mix (section 5.3) to each tube. Mix by pipetting and centrifuge briefly at maximum speed at 4°C.
Incubate samples 5 minutes at 95°C to denature 3C libraries and 5C primers.
-
Incubate overnight (16 hours) at 48°C to anneal 5C primers at 3C ligation junctions.
Note that the annealing temperature depends on the Tm selected for designing 5C primers. After the denaturation step, slowly decrease the temperature (slow ramp) in the PCR machine (0.1°C per second to required annealing temp). Slow cooling will promote efficient annealing of the 5C primers.
Table 4.
Example of a controlled 5C annealing reaction*.
| Ligation number |
Sample name |
** Volume of 3C library (µl) |
Volume of wate (µl) |
Salmon testes DNA (1 µg/µl) (µl) |
5C primer master mix (µl) |
|---|---|---|---|---|---|
| 1 | *** Cellular 5C | 2.5 | 4.6 | 0.9 | 2.0 |
| 2 | Cellular 5C “no ligase” |
2.5 | 4.6 | 0.9 | 2.0 |
| 3 | Control “no template” |
0 | 7.1 | 0.9 | 2.0 |
| 4 | Cellular 5C “no 5C primers” |
2.5 | 6.6 | 0.9 | 0 |
Total annealing volume is 10 µl. We do annealing in 20uL volume in 1X NEBuffer 4, NEB
Based on a 3C library concentration of 240 ng/ul and an optimal amount of 600 ng
More than one cellular 5C library will be required for analysis with either DNA sequencing or onto microarrays. This number should be adjusted based onto the 5C library complexity and desired sequencing depth. Consult Table 5 to select a number of reactions.
Ligation
-
6
The next day add 20 µl of ligation master mix containing 1X Taq DNA ligase buffer and 0.25 µl (10U) of Taq DNA ligase (40U/µl) to each tube except for the “no ligase” control. Mix by pipetting and centrifuge briefly at maximum speed at 4°C. To minimize non-specific product formation, keep the samples at the annealing temperature in the PCR machine.
* Ligation master mix should be prepared immediately before use and left at room temperature. 20 µl of 1X Taq DNA ligase buffer without Taq DNA ligase should be added to the “No ligase” controls.
-
7
Incubate samples at 48°C (or other temperature if applicable) for 1 hour to ligate annealed primers.
-
8
Incubate samples at 75°C for 10 minutes to end ligation reactions. Ligation products represent 5C libraries.
Amplification
We generally amplify 5C libraries with as few cycles as possible to minimize PCR amplification biases. To identify the cycle number corresponding to the lower end of the linear amplification range, we recommend first testing a range of PCR cycle numbers. The protocol used for regular PCR library amplification outlined in steps 9 to 13 can be used for this process.
-
9
On ice, assemble the PCR reactions in individual tubes or in a 96-well plate according to Table 5.
We recommend preparing at least six PCR reactions for each 5C library when analyzing onto microarrays and at least ten when using next-generation sequencing. Corresponding libraries can then be pooled before purification. We also recommend adding a PCR water control (no 5C reaction) to verify the absence of contaminants.
-
10
Amplify 5C products according to the PCR program outlined in Table 6.
Please mention that 5C library intended for deep sequencing by Illumina should be amplified using 5’ phosphorylated universal primers during LMA step to ensure PE adapter ligation during preparation of 5C library for deep sequencing, therefore one can buy HPLC purified 5’-P universal primers and use it directly.
-
11
Pool corresponding 5C PCR reactions into single tubes and transfer 5 µl of each pooled sample to a second set of tubes.
-
12
Add 2 µl of 4X agarose gel loading buffer and 3 µl of water to each 5 µl PCR sample and mix by pipetting.
-
13
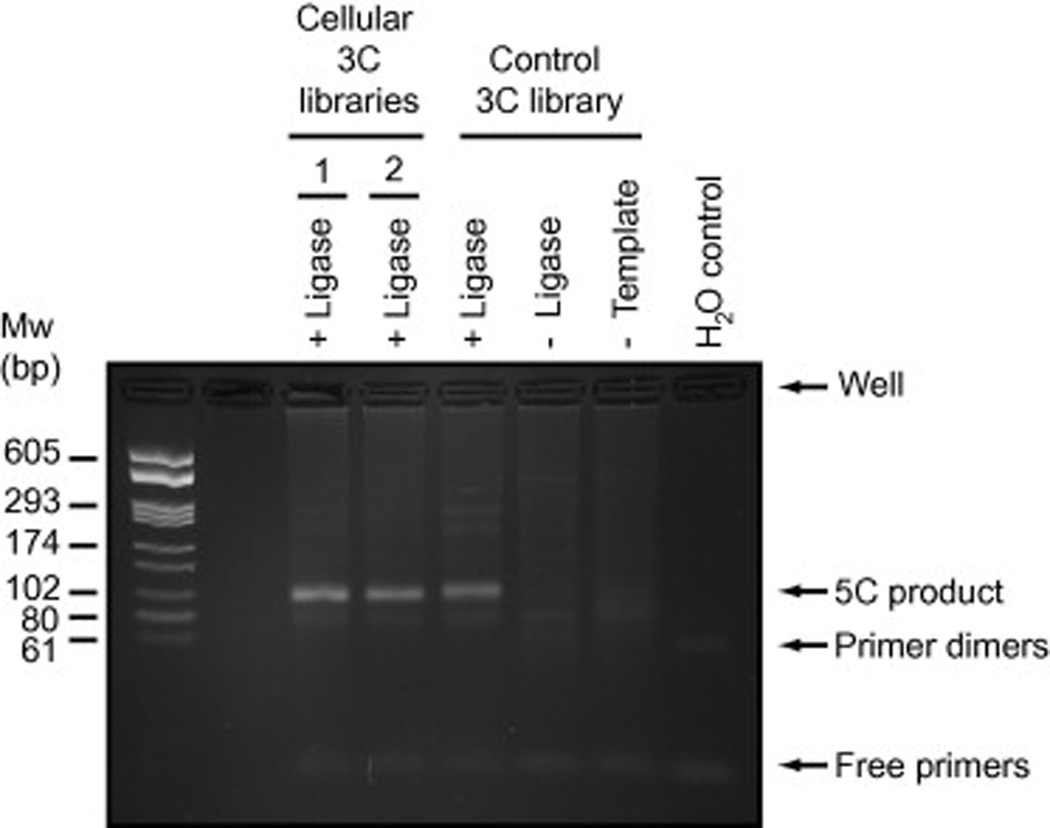
Resolve samples and a molecular weight ladder on a 2.5% agarose gel containing 0.5X TBE and 0.5 µg/ml ethidium bromide. An example of a controlled and successful 5C library production is shown in Fig. 6.
-
14a
If 5C library production is as expected (Fig. 6) and destined for microarray analysis, purify remaining pooled 5C libraries on MinElute columns as described by the manufacturer. Elute each sample into 15 µl of the elution buffer provided. These libraries are ready for microarray hybridization and can be quantified as described in steps 15 and 16.
-
14b
If 5C libraries are intended for sequencing, the remaining pooled 5C libraries must be resolved on agarose gel, and the expected PCR band extracted with the QIAquick gel extraction kit as described in the next section.
-
15
Resolve 2 µl of each purified sample along with a molecular weight ladder on a 2.5% agarose gel to verify the size, purity and concentration of 5C library products.
-
16
Catalog 5C library results with a gel documentation system. Quantify 5C products individually on gel and subtract the background. We usually use approximately 100 ng of amplified 5C library containing less than 1000 different contacts to hybridize microarrays.
Table 5.
5C PCR reaction composition.
| Stock component | Volume (µl) per rxn* | Final per rxn |
|---|---|---|
| 10X PCR buffer | 2.5 | 1X |
| 50 mM MgSO4 | 2.0 | 4.0 mM |
| 25 mM dNTPs | 0.2 | 0.2 mM |
| 100 ng/µl STD** | 1.5 | 150 ng |
| 5 µM universal forward primer 1 | 2.0 | 0.4 µM |
| 5 µM universal reverse primer 2 *** | 2.0 | 0.4 µM |
| **** Taq DNA polymerase 5 U/µl | 0.2 | 1 U |
| H2O | 10.6 | |
| 5C library | 6.0 |
Refers to 25 µl reactions
STD; salmon testis DNA stock diluted in water
Universal reverse primer should be fluorescently labeled when 5C libraries are hybridized onto microarrays.
Use AmpliTaq Gold when preparing 5C libraries for next-generation sequencing.
Table 6.
5C library PCR amplification conditions.
| Cycle number | Denature | Anneal* | Extend | Cool |
|---|---|---|---|---|
| 1 | 95 °C | |||
| 5 min | ||||
| 2 to 25–30** | 95 °C | 48 °C | 72 °C | |
| 30 s | 30 s | 30 s | ||
| 26 | 94 °C | 48 °C | 72 °C | |
| 30 s | 30 s | 8 min | ||
| 27 | 6 °C | |||
| infinity |
Annealing temperature corresponds to T7 and T3 universal primers and can vary for other universal primer pairs.
Use as few cycles as possible to minimize PCR amplification biases.
Fig. 6. Controlled 5C library production.
This example shows the results for the generation of 5C libraries from two cellular states and one control 3C library. “No ligase” and “ No template” controls for at least one library should be included in the initial 5C library production to verify the absence of contamination and the specificity of 5C product formation. 5C libraries were amplified with T7 and Cy3-labeled T3c primers and resolved on a 2.5 % agarose gel containing 0.5 µg/ml ethidium bromide. The “No 5C primer” control is not shown here.
Purification of 5C libraries on agarose gels
Run 5C libraries on long (20 cm) preparative 2% agarose gels for 3 h at 90 V and at 4°C.
Excise desired band from the gel and cut it into tiny pieces with a razor blade.
-
Dissolve gel in 3 volumes of QC buffer at *room temperature (RT) with frequent mixing (QIAquick gel extraction kit). Proceed to the next step immediately after the gel has dissolved completely.
*Do not melt gel at 50°C in QC buffer as this decreases the representation of A/T-rich sequences as reported previously [41].
Add library solutions to two MinElute columns and spin 10 sec at 6000 rpm in a microcentrifuge at RT. Discard flow through. Repeat if necessary.
Spin columns 30 sec at maximum speed to completely remove excess liquid.
Wash columns twice with 600 µl PE buffer. Spin column for 10 sec at 6000 rpm in a microcentrifuge at RT. Discard flow through.
Transfer columns to new collection tubes and spin for 2 min at maximum speed.
Transfer each column to a set of new tubes. Remove any visible leftover PE buffer from inside of the column: there is usually a small drop on the rim above the membrane. PE buffer contains ethanol such that any remaining amount of buffer will greatly affect elution of the DNA from the column.
Spin the column at maximum speed for 1 min, this time turning the column 180° towards the center of the centrifuge.
Elute 5C libraries from columns by adding 15 µl of hot (65°C) EB buffer in the middle of the column. Incubate for 3 min. Spin down at 6000 rpm for 1 min.
Repeat step 10 one more time. Run 0.5 µl of the purified library on the gel and quantify the amount of DNA in the sample.
5.6 Preparing a 5C library for deep sequencing
Using next-generation sequencing as readout for 5C libraries is the most adequate approach when the expected number of interactions is high. For instance, the number of potential contacts can easily exceed millions when characterizing entire chromosomes or large physical networks of DNA elements. In contrast to microarrays where contacts must be featured on the chip to be detected, sequencing measures any interactions present in 5C libraries. Sequencing also has a much larger detection range and a very high throughput with over hundreds of sequencing read per sample for technologies such as Illumina or SOLiD. The very high throughput of sequencing lends to multiplexing 5C libraries if prepared with proper barcoding, thereby reducing cost even as compared to microarrays in some cases.
We previously used the Illumina GAIIX paired end sequencing platform in a 36 bp format to probe 5C libraries. Preparing libraries for this type of sequencing takes 2 days, but several pause points exist along the protocol. Gel purified 5C libraries are first treated with Taq DNA polymerase in the presence of dATP to add a single A base at the 3' ends. The A-labeled 5C libraries are then ligated to Illumina Paired End (PE) adapters and purified on agarose gel to select for the correct ligation product. As the PE adapter size is 33bp, all ligation products will have similar sizes and we recommend running long preparative gels to separate primer dimers from actual 5C products. The purified 5C libraries are then amplified by PCR with PE primer 1.0 and 2.0 at a minimal PCR cycle number to maintain linearity (never go beyond 18 cycle as recommended by Illumina). We strongly recommend using an enzyme such as Pfu Ultra DNA polymerase for this step because it performs best towards GC-rich Illumina PE primers. Amplified 5C libraries are finally purified on agarose gel.
5.6.1 Reagents
QIAquick gel extraction kit (Qiagen, cat. no. 28704)
Taq DNA polymerase (NEB, cat. no. M0273S) with 10X Taq buffer containing MgCl2
10X T4 DNA ligase buffer (Invitrogen, cat. no. B0202S)
T4 DNA ligase (Invitrogen, cat. no. 15224) with 5X T4 DNA ligase buffer
QIAquick PCR purification kit (Qiagen, cat. no. 28104)
Pfu Ultra II Fusion DNA polymerase (Stratagene, cat. no. 600670) with 10X Pfu Ultra buffer
200 µM dNTPs and 200 µM dATP
TOPO TA cloning Kit (pCR2.1 TOPO) (Invitrogen, cat. no. K450002)
-
Illumina PE Adapter Oligo Mix (Illumina, San Diego, CA)
PE adapter sequences
5' P- GATCGGAAGAGCGGTTCAGCAGGAATGCCGAG
5' ACACTCTTTCCCTACACGACGCTCTTCCGATCT
-
PE primer 1.0 and PE primer 2.0 (Illumina, San Diego, CA)
PE PCR primer sequences
5'AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT
5'CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAACCGCTCTTCCGATCT
5.6.2 Procedure
Ligation of PE adapters to 5C libraries
To add A-tails at the 3' ends of 5C products, mix at least 2µg of purified on the gel[m17] 5C library (up to 35 µl), 4.2 µl 10X Taq standard buffer, 1.4 µl Taq polymerase, and 1.4 µl 10 mM dATP in an eppendorf tube. Add water for a final volume of 42 µl. Incubate the reaction at 72°C for 1 h.
To ligate PE adapters to the 5C library, add 6 µl of 10X T4 DNA ligase buffer, 5 Units (5 µl) of T4 DNA ligase, 7 µl of Illumina PE adapter Oligo Mix, 1µl of 100 mM ATP(1mM ATP final concentration), and incubate at RT for 100 min followed by a 25-minute incubation at 65°C.
Although it is recommended that the gel elution step be performed immediately, the reaction can be cooled down slowly to RT and kept at −20°C. To gel purify, load the entire ligation reaction on the 2% agarose preparative gel and run at 230 V for a minimum of 90 minutes at 4°C. The library usually looks like a ladder on the gel. Excise the expected band (size of 5C library + 66bp adapter sequences). Purify DNA on gel as described above. The adapter-modified 5C library should be stored at −20°C.
Quantify the linkered 5C library on agarose gel by running 10% of each gel-purified 5C library on a 2% agarose/0.5X TBE gel containing 0.5 µg/ml ethidium bromide along with a molecular weight marker of known concentration.
Amplify linkered 5C library by PCR as indicated in Table 7.
Amplify 5C products according to the PCR program shown in Table 8.
Pool PCR reactions and immediately gel purify as described above. Note that the size of the amplified library will increase by approximately 60 bp after amplification. If gel purification must be postponed, purify libraries on Qiagen columns to remove the DNA polymerase.
Quantify 5C libraries on agarose gel or by Nanodrop.
Store the amplified and linkered 5C library at −20°C. This library is ready for Paired End sequencing with an Illumina Genome Analyzer GAIIX sequencer.
Table 7.
5C linkered library PCR reaction conditions.
| Reaction component | Volume (µl) per rxn |
|---|---|
| Linkered 5C library | variable (∼10 ng) |
| PE primer 1.0 | 0.7 |
| PE primer 2.0 | 0.7 |
| 25 mM dNTP | 0.4 |
| 10X Pfu Ultra buffer | 5.0 |
| Pfu Ultra II Fusion DNA polymerase | 1.0 |
| H2O | to 50 µl final |
Table 8.
5C linkered library PCR amplification conditions.
| Cycle number | Denature | Anneal* | Extend | Cool |
|---|---|---|---|---|
| 1 | 98 °C | |||
| 30s | ||||
| 2–17 | 98 °C | 65 °C | 72 °C | |
| 10 s | 30 s | 30 s | ||
| 18 | 72 °C | |||
| 5 min | ||||
| 19 | RT* |
After the PCR is completed, keep the reactions at RT and D as soon as possible. Do not store reactions at 4°C for an extended time period as DNA polymerase remains active and can produce primer dimers.
Quality control of an adapter-modified 5C library
At least the first time a library is produced, we recommend cloning part of it and sequencing a few clones to verify that the DNA band extracted from the agarose gel contains genuine 5C ligation products and not primer dimers or other artifacts. This can be achieved by cloning approximately 10% of sequencing 5C libraries by TOPO cloning into Zero Blunt vectors (Invitrogen), and sequencing 20 to 50 clones by the regular Sanger method.
6. Conclusion
Job can you write a short conclusion? I put down a few ideas below but write whatever you prefer.
The 3C technologies measure chromatin contacts in cell populations. How does this affect what we can detect and can conclude about what is detected?
How does 5C relate to other 3C–derived approaches? What can 5C do that other methods cannot? What does it not do that others can? What does it do that others can?
-
What kind of questions can we address with 5C, and how will this protocol help in answering these questions?
We hope that this updated protocol will assist researchers in …
Acknowledgments
We thank members of our laboratories for helpful discussions and critical reading of this article. This work was funded by a grant from the Canadian Institutes of Health Research (CIHR MOP-86716) to J. Dostie, and by grants from the NIH (HG003143) and the Keck Foundation to J. Dekker. M.A.F. was supported by a fellowship from the Canadian Cancer Society Research Institute (CCSRI). J. Dostie is a CIHR New Investigator and FRSQ Research Scholar (Fonds de la Recherche en Santé du Québec).
Role of the funding source
The authors declare no involvement of the funding sources in study design, in data collection, analysis, or interpretation, in the writing of this article or in the decision to submit the paper for publication.
Contributor Information
Maria A. Ferraiuolo, Email: maria.ferraiuolo@mail.mcgill.ca.
Amartya Sanyal, Email: Amartya.Sanyal@umassmed.edu.
Natalia Naumova, Email: Natalia.Naumova@umassmed.edu.
Job Dekker, Email: Job.Dekker@umassmed.edu.
Josée Dostie, Email: josee.dostie@mcgill.ca.
References
- 1.Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2(3):a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fussner E, Ching RW, Bazett-Jones DP. Living without 30nm chromatin fibers. Trends Biochem Sci. 36(1):1–6. doi: 10.1016/j.tibs.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Towbin BD, Meister P, Gasser SM. The nuclear envelope--a scaffold for silencing? Curr Opin Genet Dev. 2009;19(2):180–186. doi: 10.1016/j.gde.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 4.Pombo A, Branco MR. Functional organisation of the genome during interphase. Curr Opin Genet Dev. 2007;17(5):451–455. doi: 10.1016/j.gde.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 5.Vakoc CR, et al. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17(3):453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 6.Spilianakis CG, et al. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435(7042):637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 7.Fullwood MJ, et al. An oestrogen-receptor-[agr]-bound human chromatin interactome. Nature. 2009;462(7269):58–64. doi: 10.1038/nature08497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yochum GS, et al. A beta-catenin/TCF-coordinated chromatin loop at MYC integrates 5'rsquo; and 3'rsquo; Wnt responsive enhancers. Proc Natl Acad Sci U S A. 2009;107(1):145–150. doi: 10.1073/pnas.0912294107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon H, Boss JM. PU.1 binds to a distal regulatory element that is necessary for B cell-specific expression of CIITA. J Immunol. 2010;184(9):5018–5028. doi: 10.4049/jimmunol.1000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kagey MH, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467(7314):430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137(7):1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merkenschlager M. Cohesin: a global player in chromosome biology with local ties to gene regulation. Curr Opin Genet Dev. 2010;20(5):555–561. doi: 10.1016/j.gde.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Ferraiuolo MA, et al. The three-dimensional architecture of Hox cluster silencing. Nucleic Acids Res. 2010;38(21):7472–7484. doi: 10.1093/nar/gkq644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hadjur S, et al. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature. 2009;460(7253):410–413. doi: 10.1038/nature08079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt D, et al. A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 2010;20(5):578–588. doi: 10.1101/gr.100479.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang XQD, Crutchley JL, Dostie J. Shaping the Genome with Non-coding RNA. Current Genomics. 2011;12(5):307–321. doi: 10.2174/138920211796429772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crutchley JL, et al. Chromatin conformation signatures: ideal human disease biomarkers? Biomark Med. 2010;4(4):611–629. doi: 10.2217/bmm.10.68. [DOI] [PubMed] [Google Scholar]
- 18.Dekker J, et al. Capturing Chromosome Conformation. Science. 2002;295(5558):1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 19.Simonis M, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat Genet. 2006;38(11):1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38(11):1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 21.Duan Z, et al. A three-dimensional model of the yeast genome. Nature. 465(7296):363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Würtele H, Chartrand P. Genome-wide scanning of HoxB1-associated loci in mouse ES cells using an open-ended Chromosome Conformation Capture methodology. Chromosome Research. 2006;14(5):477–495. doi: 10.1007/s10577-006-1075-0. [DOI] [PubMed] [Google Scholar]
- 23.Dostie J, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Research. 2006;16(10):1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tiwari VK, et al. A novel 6C assay uncovers Polycomb-mediated higher order chromatin conformations. Genome Res. 2008;18(7):1171–1179. doi: 10.1101/gr.073452.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rodley CD, et al. Global identification of yeast chromosome interactions using Genome conformation capture. Fungal Genet Biol. 2009;46(11):879–886. doi: 10.1016/j.fgb.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 26.Lieberman-Aiden E, et al. Comprehensive Mapping of Long-Range Interactions Reveals Folding Principles of the Human Genome. Science. 2009;326(5950):289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dostie J, Dekker J. Mapping networks of physical interactions between genomic elements using 5C technology. Nat Protoc. 2007;2(4):988–1002. doi: 10.1038/nprot.2007.116. [DOI] [PubMed] [Google Scholar]
- 28.Dostie J, Zhan Y, Dekker J. Chromosome conformation capture carbon copy technology. Curr Protoc Mol Biol. 2007;Chapter 21:14. doi: 10.1002/0471142727.mb2114s80. p. Unit 21. [DOI] [PubMed] [Google Scholar]
- 29.Fraser J, et al. Computing chromosome conformation. Methods Mol Biol. 2010;674:251–268. doi: 10.1007/978-1-60761-854-6_16. [DOI] [PubMed] [Google Scholar]
- 30.Miele A, et al. In: Mapping chromatin interactions by Chromosome Conformation Capture (3C), in Current Protocols in Molecular Biology. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Hoboken, N.J.: John Wiley & Sons; 2006. pp. 11–20. p. 21.11.1–21. [DOI] [PubMed] [Google Scholar]
- 31.Naumova N, et al. Analysis of long-range chromatin interactions using Chromosome Conformation Capture. Methods. 2012 doi: 10.1016/j.ymeth.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 447(7146):2007. 799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fraser J, et al. Chromatin conformation signatures of cellular differentiation. Genome Biol. 2009;10(4):R37. doi: 10.1186/gb-2009-10-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.ENCODE-consortium, Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447(7146):799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Elnitski LL, et al. The ENCODEdb portal: simplified access to ENCODE Consortium data. Genome Res. 2007;17(6):954–959. doi: 10.1101/gr.5582207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stamatoyannopoulos JA, et al. An encyclopedia of mouse DNA elements (Mouse ENCODE) Genome Biol. 2012;13(8):418. doi: 10.1186/gb-2012-13-8-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenbloom KR, et al. ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res. 2012;40(Database issue):D912–D917. doi: 10.1093/nar/gkr1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. http://Dostielab.biochem.mcgill.ca.
- 39. http://msy5c.umassmed.edu/welcome/welcome.php. Available from: http://genome.ucsc.edu/
- 40.Bau D, et al. The three-dimensional folding of the alpha-globin gene domain reveals formation of chromatin globules. Nat Struct Mol Biol. 18(1):107–114. doi: 10.1038/nsmb.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quail MA, et al. A large genome center'rsquo;s improvements to the Illumina sequencing system. Nat Methods. 2008;5(12):1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]