Abstract
Purpose
Recent advances in technology now provide tools capable of tracking genome-wide expression changes occurring in progressive pathological processes. The present experiments were carried out to determine if acetylcholine receptor α 6 subunit (Chrna6) is a reliable retinal ganglion cell (RGC) marker in adult mouse eyes and if Chrna6 expression can be used to track progressive loss of RGCs, such as is observed in the DBA/2J glaucoma model.
Methods
Data sets derived from the BXD strains were used to extract gene expression signatures for RGCs. Pooled retinas from DBA/2J or C57BL/6J cases at 1–3 months, 12 months, and 16–17 months were prepared for gene-array and RT-PCR analysis. Globes were fixed in paraformaldehyde and sectioned for immunofluorescence with antibodies against Chrna6.
Results
Chrna6 has a cellular expression signature for RGCs with high correlation to Thy1 (r = 0.65), a recognized RGC marker. Immunofluorescence experiments confirm that in the young and adult mouse retina, Chrna6 is preferentially expressed by RGCs. We further show that C3H/HeJ retinas, which lack photoreceptors, also express Chrna6 in the RGC layer. Gene expression array analyses, confirmed by RT-PCR, show progressive loss of Chrna6 expression in retinas of the DBA/2J glaucomatous mouse retinas.
Conclusions
Quantitative trait locus analysis provides support for Chrna6 as a RGC marker. Chrna6 expression decreases with death of RGCs in glaucomatous DBA/2J mice and after optic nerve crush injury, further supporting Chrna6 as a reliable RGC marker. High expression of RGC Chrna6 in the absence of photoreceptors is suggestive that Chrna6 expression by RGCs is independent of photoreceptor-derived stimuli.
Keywords: Aging, Glaucoma, Neurodegenerative Diseases, Retina, Gene Network, QTL
INTRODUCTION
Neuronal nicotinic acetylcholine receptors (nAChRs) are ligand-gated cation channels dependent on the acetylcholine (ACh) ligand for function.1 At least twelve different genes are members of a heterogeneous family of pentameric nAChRs. The two main classes of nAChRs are subdivided based upon channel sensitivity to α-bungarotoxin (α-Bgtx) and epibatine (Epi). α7–α10 AChR subunits can form both homomeric and heteromeric receptors that are α-Bgtx sensitive, whereas α2–α6 and β2–β4 form α-Bgtx insensitive heteromeric receptors capable of binding Epi.2 In the CNS, functional variety in these pentameric receptors is achieved through combination of nine α and three ‘structural’ β subunits.3–6
Abundant expression of the acetylcholine receptor α 6 subunit (Chrna6) has been described in the substantia nigra, ventral tegmental area7–9 and retina.10,11 In the visual system, the retina, optic tract and its targets express high levels of Chrna6.10,12,13 Optic nerve (ON) Chrna6 expression density along with eye enucleation experimental data suggest Chrna6 expression in ON target areas is largely restricted to presynaptic terminals of RGC axons, where they may function to modulate glutamate release by RGCs to the brain.14–17 Although glutamate is the major neurotransmitter in the retina, nAChRs localized to RGC soma play an important functional role in visual system development.18 Light-independent spontaneous retinal activity, “retinal waves”, are initially cholinergic and are mediated by nAChRs before a functional switch occurs which makes the waves glutamatergic.19–21 Experimental disruption of retinal waves has highlighted its influence on both axonal and dendritic properties of RGC development,13,19,22 further defining the importance of nAChR expression by RGCs.
The present experiments were carried out to determine if Chrna6 is a reliable RGC marker in adult mouse eyes and if Chrna6 expression can be used to track progressive loss of RGCs, such as is observed in the glaucomatous DBA/2J mouse model of glaucoma. Using bioinformatic tools in GeneNetwork (genenetwork.org), we show that Chrna6 has a cellular expression signature for retinal ganglion cells with high correlation to Thy1 (r = 0.65), a recognized RGC marker. Immunofluorescence experiments confirm that in the young mouse retina, Chrna6 is preferentially expressed by RGCs. We further show that Chrna6 RGC expression is maintained through adulthood and that Chrna6 expression by RGCs is not dependent on photoreceptor derived stimuli. Gene chip analyses confirmed by RT-PCR show progressive loss of Chrna6 gene expression in the DBA/2J glaucoma mouse model.
METHODS
Animals
DBA/2J, C57BL/6J and C3H/HeJ animals were bred and handled according to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research. All animal procedures were approved by the University of Miami Institutional Animal Care and Use Committee.
Optic Nerve Crush Injury
Mice were anesthetized by intraperitoneal injection of ketamine (100 mg/kg Ketaset; Fort Dodge Animal Health, Fort Dodge, IA) and xylazine (10 mg/kg TranquiVed; Vedco, Inc., St. Joseph, MO). A limbal conjunctival peritomy was performed in the superior region and gently peeled back to allow access to the posterior region of the globe. The optic nerve was then exposed through a small window made between the surrounding muscle bundles and fatty tissue by gentle blunt dissection. Care was taken not to damage muscles or the vortex veins. At a site approximately 1 mm posterior to the globe, the optic nerve was clamped with a pair of Dumont no. 5 self-closing tweezers (Ted Pella Inc., Redding, CA) for 3–5 s. After this procedure, antibiotic ointment was applied to the surgical site and mice were observed. In the postoperative period, the mice exhibited normal eating and drinking behavior and were given Buprenorphine (0.1 mg/kg) subcutaneously, twice daily for two days following procedure for pain control.
RNA Isolation, Amplification and Labeling
Pooled whole retinas from age matched DBA/2J and control C57BL/6J mice were homogenized and total RNA was isolated using TRIZOL Reagent (15596-018; Invitrogen, Carlsbad, CA). The quantity of extracted RNA was measured by spectrophotometry (BioPhotometer 6131; Eppendorf, Hamburg, Germany). First strand cDNA was synthesized, fractionated and biotinylated for hybridization to Affymetrix GeneChip Mouse Genome 430 2.0 gene expression microarrays.
Microarray Data Analysis
Total RNA from six to eight pooled whole retinas of age matched DBA/2J or control C57BL/6J was isolated using TRIZOL Reagent (15596-018; Invitrogen, Carlsbad, CA) and Qiagen RNeasy column purification (Qiagen, Valencia, CA) according to manufacture’s instructions. Ten microgram samples of extracted RNA were analysed by using Affymetrix M430 2.0 microarrays. Target was prepared and hybridized using then vendor’s protocols. Total RNA (2 µg) was converted into cDNA using reverse transcriptase (Enzo Biosciences) and a modified oligo(dT) 24 primer that contains T7 promoter sequences (GenSet). After first strand synthesis, residual RNA was degraded by the addition of RNaseH and double-stranded cDNA was generated using DNA polymerase I and DNA ligase. The cDNA was then purified and concentrated using phenol:chloroform extraction followed by ethanol precipitation. The cDNA products were incubated with T7 RNA polymerase and biotinylated ribonucleotides using an in vitro transcription kit (Affymetrix). The resultant cRNA product was purified using a RNeasy column (Qiagen) and quantified by spectrophotometry.
cRNA target (20 µg) was incubated at 94°C for 35 min in fragmentation buffer (Tris, MgOAc, KOAc). The fragmented cRNA was diluted in hybridization buffer (MES, NaCl, EDTA, Tween 20, Herring Sperm DNA, Acetylated BSA) containing biotin-labeled OligoB2 and Eukaryotic Hybridization Controls (Affymetrix). The hybridization cocktail was denatured at 99°C for 5 min, incubated at 45°C for 5 min and then injected into a GeneChip cartridge. The GeneChip array was incubated at 42°C for at least 16 h in a rotating oven at 60 rpm. GeneChips were washed with a series of nonstringent (25°C) and stringent (50°C) solutions containing variable amounts of MES, Tween 20 and SSPE. The microarrays were then stained with streptavidin phycoerythrin and the fluorescent signal was amplified using a biotinylated antibody solution. Fluorescent images were converted to estimates of steady-state expression using the GeneChip software v 2.0 from Affymetrix. Array data were scaled to a median intensity setting of 500. Signal strength was computed using the Affymetrix Microarray Suite algorithm version 5.0 and GeneChips were scaled to a median intensity setting of 500.
Quantitative RT-PCR
Six to eight pooled whole retinas from age matched DBA/2J or control C57BL/6J mice were homogenized using a disposable Kontes Pellet Pestle with a cordless motor tissue grinder (Kimble Kontes, Vineland, NJ) on ice, and total RNA was isolated using TRIZOL Reagent (15596-018; Invitrogen, Carlsbad, CA). cDNA templates were synthesized from 2 µg of each RNA sample using Superscript III reverse transcription reagents according to the manufacturer’s protocol (#18080–085, Invitrogen, Carlsbad, CA). The following primers were designed with the aid of Primer323 and IDT design software (Integrated DNA Technologies, Coralville, IA): ARP, F 5’-CGACCTGGAAGTCCAACTAC-3’ and R 5’-ATCTGCTGCATCTGCTTG-3’; Chrna6, F 5’-AAAGGCAGTACAGGCTGTGAA-3’ and R 5’-CGCCGACAGCATTGTTATAC-3’. The qRT-PCR was completed (iCylcer iQ, Bio-Rad, Hercules, CA) using 100 ng cDNAs and iQ SYBR Green Supermix (#170–8882, Bio-Rad, Hercules, CA), in duplicate reactions. All primer sets underwent the same thermal cycling parameters: 95°C for 10 min, 40 cycles of 95°C for 15 s and 55°C for 1 min, 95°C for 1 min, 65°C for 2 min, and 80 cycles of 55°C for 10 s. SYBR green data was visualized using the iCylcer iQ v.3.1 software (Bio-Rad, Hercules, CA) and analyzed with Microsoft Excel (Microsoft Inc.). A standard curve for each primer set was assessed by serial diluting cDNA obtained from our control mice so that analyses are performed for 50, 25, 12.5, and 6.25 ng. The cycle number determined at the computer-selected threshold (CT) was plotted against the log cDNA concentration providing a linear curve. Using each corresponding standard curve, plotted values were obtained.
Immunofluorescence and Microscopy
Primary antibodies were: rabbit anti-Chrna6 (1:500 dilution, ab65168, Abcam, Cambridge, MA), goat anti-Chrna6 (P13) (1:100 dilution, SC-27294, Santa Cruz Biotechnology, Santa Cruz, CA). Secondary antibodies were: Donkey anti-rabbit Alexa 647 (1:100 dilution, #31573, Molecular Probes, Eugene, OR) and Donkey anti-goat DyLight 649 (1:100 dilution, #705-496-147, Jackson ImmunoResearch Laboratories, West Grove, PA). Globes were fixed using 4% paraformaldehyde (Cat. #15714-S; Electron Microscopy Sciences, Hatfield, PA) in 1× phosphate buffered saline (PBS; 127 mM NaCl, 2.7 mM KCl, 10 mM phosphate) buffer at 25°C. Following fixation, globes were embedded in paraffin, sectioned and mounted on glass slides. Sections were de-parafinized in xylene, sequentially rinsed with an ethanol gradient wash, and treated at 95°C with Trilogy (CMX833-C; Cell Marque, Rocklin, CA) antigen retrieval reagent. Tissue sections were subsequently blocked with Rodent Block M (RBM96; BioCare Medical, Concord, CA) for 30 min to reduce nonspecific binding. Slides were incubated overnight at 4°C in PBS containing primary antibodies, washed with PBS, incubated in antibody buffer containing secondary antibodies overnight at 4°C, washed with PBS, and mounted onto glass coverslips using DAPI Vectashield (Vector, Burlingame, CA). Confocal microscopy images were obtained with a Leica TCP SP5 spectral confocal microscope (Leica, Exton, PA) (63×, water-immersion, 1.2 NA objective).
RESULTS
Expression Covariation Data for a C57BL/6J × DBA/2J Cross (the BXDs) Demonstrates Chrna6 Expression has a RGC Specific Profile
In recent work we and others24 have exploited massive open-access gene expression data sets to define expression signatures for specific cell populations in the eye. The Chrna6 1450426_at probe set covaries well with both Thy1 (r = 0.65, n = 72 BXD strains using probe set 1423135_at) and with Gap43 (r = 0.58 using 1423537_at). Using the combination of these three transcripts to define a joint gene expression we find the first principal component explains ~75% of the variance in expression of these transcripts and loads most heavily on Chrna6 (r = 0.88) and Thy1 (r = 0.85). Furthermore, using this joint RGC signature or eigengene to generate a list of other expression covariates we find Stmn2 (1423280_at), Sept3 (1439833), Nrip3 (1448954_at), and Fam19a5 (1419490_at), Hpcla4 (1433987_at), Kif5a (1434670_at), Prkcb (1460419_at), Rab33a (1417529_at), and Cplx1 (1417746_at) among the top 10 transcripts of this RGC signature. Several of these genes are known to be specifically expressed in RGCs, including Stmn225 and Cplx1.26
Chrna6 mRNA Expression in Retina Decreases as DBA/2J Retinas Progressively Lose Retinal Ganglion Cells
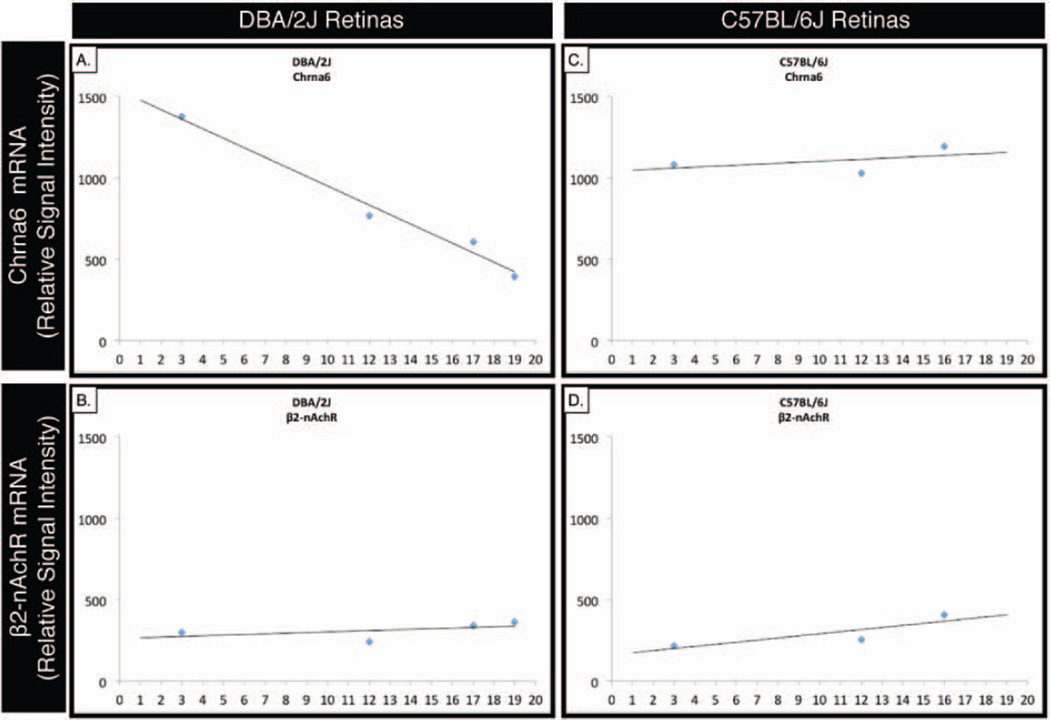
Expression levels of Chrna6 were assessed by GeneChip analysis of DBA/2J and age-matched C57BL/6J control mouse retinas. The GeneChip expression levels of Chrna6 mRNA (Figure 1A) progressively decrease as DBA/2J mice lose retinal ganglion cells with increasing age. A similar decrease in gene expression was not observed in the pentameric receptor binding partner, β2-nAChR (Figure 1B). Control C57BL/6J gene expression demonstrated relatively unchanged expression levels of Chrna6 (Figure 1C) or β2-nAChR (Figure 1D) in age-matched retinas. Gene expression levels were determined and analyzed with the Affymetrix Microarray Suite 5.0 analysis program. In order to account for the built-in redundancy in the gene chip design, available probes for each analysed gene with a p value >0.05 were averaged and plotted relative to age. A best-fit line portrays the changing trend in expression level over time.
FIGURE 1.
Chrna6 mRNA expression by GeneChip analysis is inversely proportional to retinal ganglion cell loss in DBA/2J glaucomatous progression. Gene expression levels were measured from 4–6 pooled retinas using Affymetrix whole genome chip arrays (Mouse Genome 430 2.0 Array). Expression levels of Chrna6 RNA in the retina progressively decrease as the DBA/2J mice age (A) and lose retinal ganglion cells. In contrast, expression levels of Chrna6 in C57BL/6J retinas (C) and pentameric receptor binding partner, β2-nAChR (B, D), do not change with increasing age. Expression levels of all Affymetrix probes available for each gene with a p value <0.05 were averaged and subsequently plotted on the graph with a best fit line highlighting the trend of expression over time.
Real Time PCR Confirms Decreased Chrna6 mRNA Expression as DBA/2J Retinas Progressively Lose Retinal Ganglion Cells
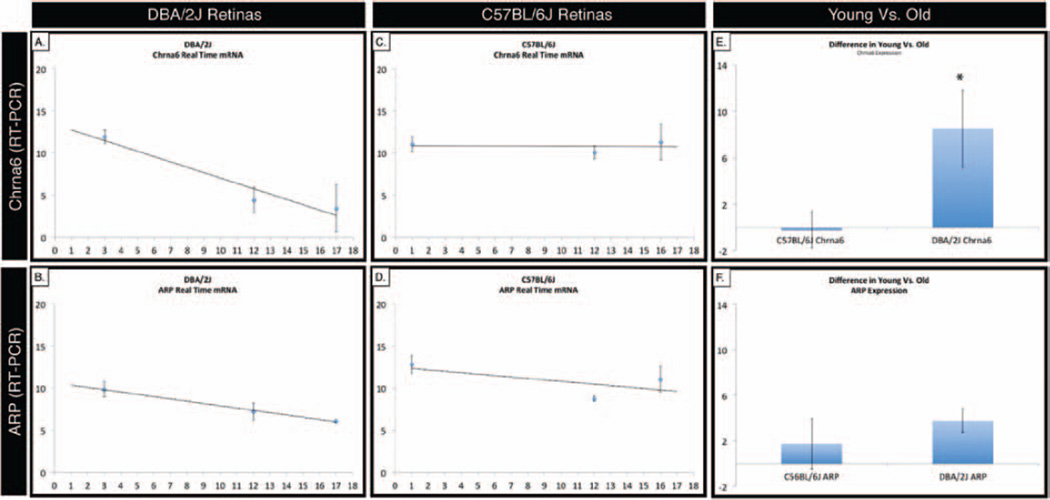
In agreement with gene chip expression data, semi-quantitative real-time RT-PCR measurements show a decrease in Chrna6 expression (Figure 2A) as DBA/2J retinas age, whereas age-matched control C57BL/6J Chrna6 expression levels remain largely unchanged (Figure 2C). In both DBA/2J and C57BL/6J retinas, the ubiquitous acidic ribosomal protein (ARP) loading control shows a similar decrease in expression (Figure 2B and 2D) as animals increase in age. A t-test comparison of the age dependent decrease in mRNA expression levels between DBA/2J and C57BL/6J retinas shows a statistically significant change (p < 0.001) in Chrna6 (Figure 2E), while no significant difference is observed for control ARP (Figure 2F). cDNA was prepared from pooled retina from either DBA/2J or C57BL/6J mice of selected age groups, and real-time PCR was performed using primers which span an intron, assessing only processed mature cDNA (using SYBR green as the quantification dye). Error bars represent standard deviation. A best-fit line portrays the changing trend in expression level over time.
FIGURE 2.
Correlation of decrease Chrna6 mRNA expression with retinal ganglion cell loss is confirmed by real time PCR analysis. Real-time PCR was performed using primers that spanned an intron, measuring only mature cDNA. In agreement with gene chip expression data, RT-PCR analysis shows a decrease in Chrna6 levels (A) as DBA/2J retinas age, whereas age-matched control C57BL/6J Chrna6 signal levels remain largely unchanged (C). The ubiquitous acidic ribosomal protein (ARP) loading control shows a similar decrease in expression (B, D) as both DBA/2J and C57BL/6J retinas increase in age. The age related decrease in Chrna6 mRNA expression levels (E) in DBA/2J relative to the C57BL/6J retinas is statistically significant (p < 0.001, student’s t-test). There is no significant age related decrease in ARP loading control mRNA expression levels (F) in DBA/2J relative to the C57BL/6J retinas. Error bars represent standard deviation. A best-fit line portrays the changing trend in expression level over time.
Retina Chrna6 Protein Expression is Enriched in the Retinal Ganglion Cell Layer
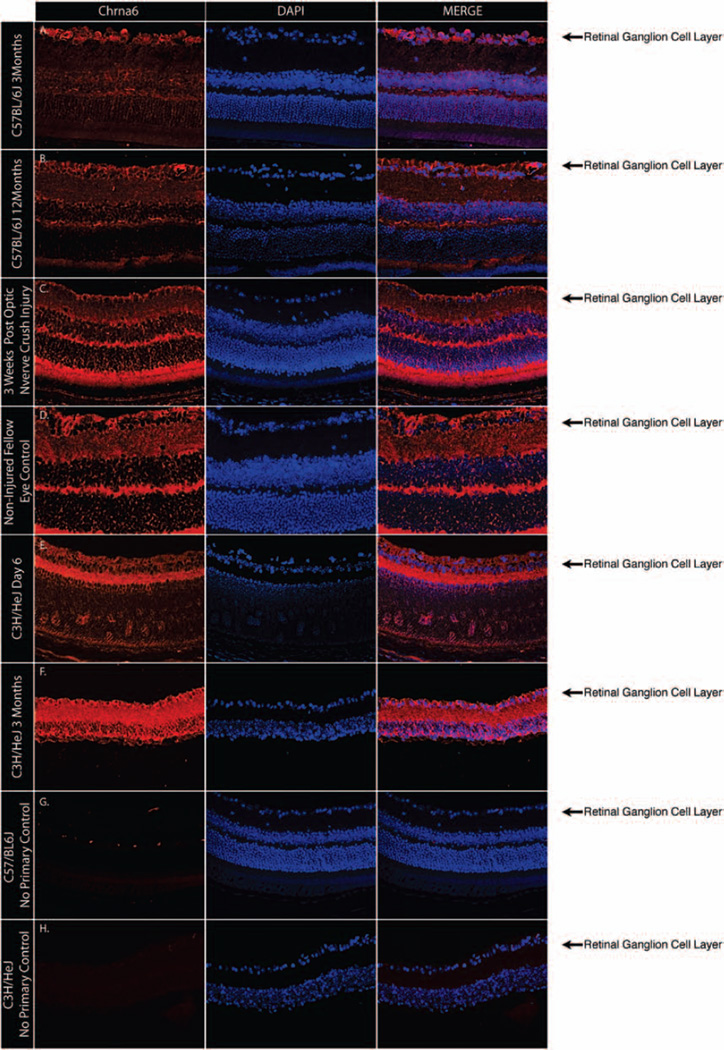
Chrna6 expression at the protein level was assessed in whole retinas of young and adult C57BL/6J mice. Immunostaining demonstrated Chrna6 expression most prominently in the RGC layer of both young and old C57BL/6J mice (Figure 3A and 3B). Chrna6 expression in optic nerve crush injured retinas is decreased when compared to noninjured control fellow eyes (Figure 3C and 3D). Early postnatal C3H/HeJ (Figure 3E) and adult C3H/HeJ (Figure 3F) whose retinas lack a photoreceptor outer nuclear layer, show high Chrna6 expression levels in the RGC layer and inner plexiform layer. All shown images are representative results from experiments repeated in at least three separate times with at least two different retinas.
FIGURE 3.
Chrna6 protein is present within the retinal ganglion cell layer of mouse retinas. Representative globe cross-sections immunostained with Chrna6 antibody (red) show Chrna6 expression in the retinal inner plexiform layer and an enriched level of expression in the RGC layer of both young and old C57BL/6J mice (A, B) and a decrease in the number of retinal ganglion cells expressing Chrna6 is observed 3 weeks following optic nerve crush injury (C, D). Immunostaining of C3H/HeJ retinas show similar Chrna6 protein expression levels in RGCs and the inner plexiform layer of early postnatal (E) and outer nuclear layer lacking adult (F) retinas. Nuclear DAPI staining (blue) and merged blue and red channels are also shown. Control sections lacking primary antibody (G, H) represent a minimal level of auto-fluorescence in tissue samples.
DISCUSSION
The identification of molecular markers to track disease progression is becoming increasingly important and a major focus of clinical research. Little is known about the molecular identity of RGCs and whether different molecularly defined RGC-subtypes respond differently to disease-related stimuli.25 Recent advances in technology provide scientists with tools capable of tracking genome-wide expression changes that occur in progressive pathological processes, such as an optic neuropathy like glaucoma. Although a major cause of irreversible blindness worldwide, glaucoma pathogenesis is poorly understood for most types of glaucoma. Animal models of glaucoma, if they closely mimic human glaucoma, are important tools for dissecting the molecular pathogenesis of RGC death under glaucomatous stimuli, thus providing insight and targets relevant to patient clinical care. Identification and confirmation of novel RGC markers using glaucoma animal models provides a first step toward identifying subtype-specific populations of RGCs highly susceptible to disease.
In this work we identified Chrna6 as an effective marker with which to track RGC loss in glaucoma animal models of disease. Using data sets and tools that are part of the GeneNetwork system, we were able to define robust expression covariance signatures for RGCs and confirmed membership of Chrna6 within the RGC cell type of the retina using new array data sets and RT-PCR tracking through a progressive RGC loss mouse line. Chrna6 can be added as reliable biomarker for RGCs and RGC loss secondary to glaucoma. It is important to note that in addition to providing evidence for Chrna6 expression as a reliable part of the RGC genetic signature, it is possible that like other well-recognized RGC markers such as Thy-1, Chrna6 expression levels may also serve as a marker for RGC stress.27,28 To further validate that Chrna6 expression as a RGC marker we demonstrated that protein levels are high in the RGC layer of the retina in young and adult mice.
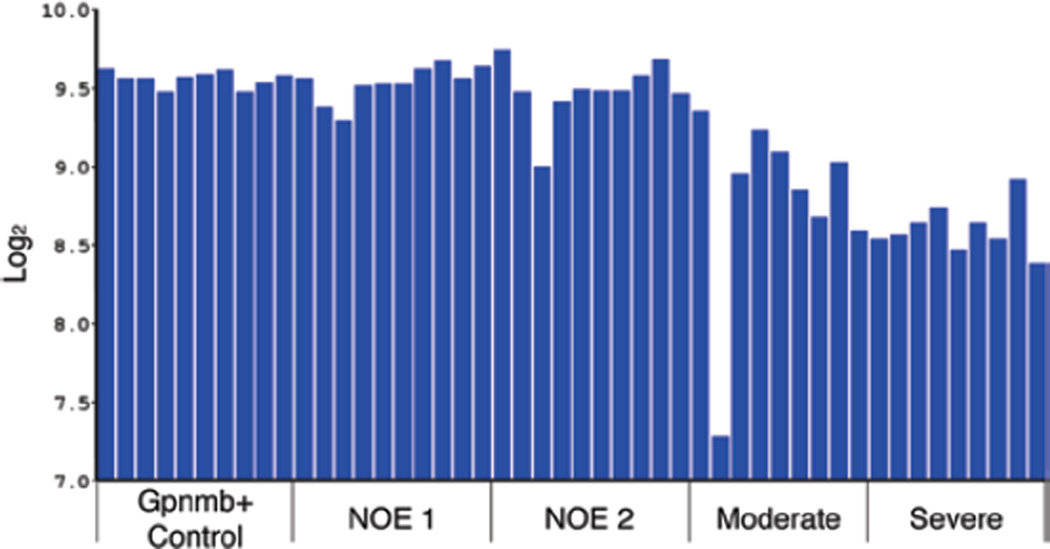
As an independent test for the ability of Chrna6 levels to monitor the loss of retinal ganglion cells, we performed a meta-analysis of the data presented by Howell and colleagues29 regarding changes in gene expression in the DBA/2J model of glaucoma. In this study, the authors related changes in the transcriptome of retinas from DBA/2J mice directly to the severity of the glaucoma (as defined by determining the degree of degeneration in the optic nerve). Analyzing microarray data using GeneNetwork, we plotted Chrna6 expression levels from all of the individual animals and identified severity groups (Figure 4). The data clearly demonstrates down-regulation of Chrna6 as the severity of glaucoma progresses within this cohort of DBA/2J mice. Thus, Chrna6 accurately reflects the loss of RGC in a naturally occurring model of glaucoma.
FIGURE 4.
The level of Chrn6 expression in a meta-analysis using data from Howell et al. (2011) is shown for each individual DBA/2J mouse in the Howell et al. dataset. The scale to the left is expression from the microarray studies expressed in log base 2 with the mean expression of mRNA across the microarray set to 8. The mice were classified as WT Gpnmb controls (Gpnmb+ Controls), no detectable glaucoma 1 (NOE 1) no detectable glaucoma 2 (NOE2), moderate glaucoma (Moderate) and severe glaucoma (Severe). Notice the decrease in expression of Chrna6 in the retina as the severity of glaucoma increases.
The C3H/HeJ rd/rd mouse is an inbred mouse strain displaying a spontaneous retinal degeneration phenotype, and is widely regarded as a model for retinitis pigmentosa. The initial phenotypic findings of rod and cone degeneration in C3H/HeJ mice begin around postnatal day 8, resulting in virtually no photoreceptors by four weeks of age.30,31 Apoptosis of photoreceptors also results in the lack of photoreceptor driven glutamatergic input into RGCs. Although the loss of photoreceptors in the rd/rd mouse results in progressive dendritic atrophy of first order bipolar neurons, no appreciable changes to RGC dendritic architecture or viability are observed.32 This implies that neuronal functional integrity may be dependent on cell-cell contacts and proper chemical stimulation between neighboring cells of the retina. Through the elimination of the outer nuclear layer in C3H/HeJ retinas, we demonstrate that Chrna6 expression by RGCs is not dependent on photoreceptor light-driven signaling.
One powerful approach to define cell-specific markers and signatures is to identify genes that have well correlated expression patterns across highly diverse inbred strains. Based on highly accurate transmission electron microscopic analysis, the RGC population varies from 50,000 to 70,000 cells across strains of mice, with very few outliers.33,34 This modest variation lines up well with relatively modest but still detectible differences in mRNAs for several RGC markers.24 As we have confirmed here using a glaucoma model with more extreme effects on RGC number, Chrna6 is a significant expression marker identified for retinal ganglion cells. Expression of Chrna6 mRNA or protein can be used as a proxy of RGC loss in a mouse model of pigment dispersion glaucoma.
DBA/2J mice develop age-dependent progressive ocular abnormalities including elevated IOP, iris atrophy and secondary angle closure.35–37 Anterior chamber pathology in DBA/2J mice typically has its onset at approximately 6 months of age, followed by elevation of IOP between 8–9 months, and severe ON damage and RGC loss by 12 months of age in the majority of animals.35–37 We have also recently reported on glutamate-induced excitotoxicity as a possible contributing factor for DBA/2J glaucomatous pathology.38 Our finding that Chrna6 is a reliable RGC marker helps account for the heterogeneous and geographically asymmetric characteristics of RGC death in age-dependent animal models of glaucoma. As DBA/2J retinas progressively lose RGCs, levels of Chrna6 expression from pooled retinas also decrease as demonstrated by GeneChip expression analysis and confirmatory RT-PCR.
A high level of Chrna6 expression has previously been described in the retina.10,11 Brain target nuclei for the ON also contain high levels of Chrna6.10,12,13 Chrna6 present in ON target areas are largely restricted to presynaptic ends of RGC axons, where they may function to modulate glutamate release onto the brain.14–17 The ON tract has been described to have the highest density of Chrna6 protein in the CNS.14 These findings contribute to the hypothesis that RGCs express large levels of Chrna6 for delivery to ON targets, supporting our finding of Chrna6 as a reliable marker for RGCs in glaucoma progression.
ACKNOWLEDGEMENTS
We would like to thank Nicklaus P. Atria, Frederick A. Kweh and Amit K. Patel for their contribution to this manuscript.
Declaration of interest: RKL was supported by NIH NEI grant EY016775. The Bascom Palmer Eye Institute is supported by an unrestricted research grant from Research to Prevent Blindness and NIH center grant EY014801. SKB is supported by NIH NEI grant EY016112 and a Research to Prevent Blindness Career Award. EEG is supported by NEI grant RO1EY017841 and an unrestricted grant from Research to Prevent Blindness to the Hamilton Eye Institute, University of Tennessee.
REFERENCES
- 1.Corringer PJ, Bertrand S, Galzi JL, et al. Molecular basis of the charge selectivity of nicotinic acetylcholine receptor and related ligand-gated ion channels. Novartis Found Symp. 1999;225:215–224. doi: 10.1002/9780470515716.ch14. discussion 224. [DOI] [PubMed] [Google Scholar]
- 2.Sargent PB. The diversity of neuronal nicotinic acetylcholine receptors. Annu Rev Neurosci. 1993;16:403–443. doi: 10.1146/annurev.ne.16.030193.002155. [DOI] [PubMed] [Google Scholar]
- 3.Gotti C, Moretti M, Zanardi A, et al. Heterogeneity and selective targeting of neuronal nicotinic acetylcholine receptor (nAChR) subtypes expressed on retinal afferents of the superior colliculus and lateral geniculate nucleus: identification of a new native nAChR subtype alpha3beta2(alpha5 or beta3) enriched in retinocollicular afferents. Mol Pharmacol. 2005;68:1162–1171. doi: 10.1124/mol.105.015925. [DOI] [PubMed] [Google Scholar]
- 4.Hogg RC, Bertrand D. Regulating the regulators: the role of nicotinic acetylcholine receptors in human epilepsy. Drug News Perspect. 2003;16:261–266. doi: 10.1358/dnp.2003.16.5.829313. [DOI] [PubMed] [Google Scholar]
- 5.Millar NS, Harkness PC. Assembly and trafficking of nicotinic acetylcholine receptors (Review) Mol Membr Biol. 2008;25:279–292. doi: 10.1080/09687680802035675. [DOI] [PubMed] [Google Scholar]
- 6.Millar NS, Gotti C. Diversity of vertebrate nicotinic acetylcholine receptors. Neuropharmacology. 2009;56:237–246. doi: 10.1016/j.neuropharm.2008.07.041. [DOI] [PubMed] [Google Scholar]
- 7.Azam L, Winzer-Serhan UH, Chen Y, et al. Expression of neuronal nicotinic acetylcholine receptor subunit mRNAs within midbrain dopamine neurons. J Comp Neurol. 2002;444:260–274. doi: 10.1002/cne.10138. [DOI] [PubMed] [Google Scholar]
- 8.Klink R, de Kerchove d’Exaerde A, Zoli M, et al. Molecular and physiological diversity of nicotinic acetylcholine receptors in the midbrain dopaminergic nuclei. J Neurosci. 2001;21:1452–1463. doi: 10.1523/JNEUROSCI.21-05-01452.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Le Novère N, Zoli M, Changeux JP. Neuronal nicotinic receptor alpha 6 subunit mRNA is selectively concentrated in catecholaminergic nuclei of the rat brain. Eur J Neurosci. 1996;8:2428–2439. doi: 10.1111/j.1460-9568.1996.tb01206.x. [DOI] [PubMed] [Google Scholar]
- 10.Fucile S, Matter JM, Erkman L, et al. The neuronal alpha6 subunit forms functional heteromeric acetylcholine receptors in human transfected cells. Eur J Neurosci. 1998;10:172–178. doi: 10.1046/j.1460-9568.1998.00001.x. [DOI] [PubMed] [Google Scholar]
- 11.Yang K, Hu J, Lucero L, et al. Distinctive nicotinic acetylcholine receptor functional phenotypes of rat ventral tegmental area dopaminergic neurons. J Physiol (Lond) 2009;587:345–361. doi: 10.1113/jphysiol.2008.162743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Champtiaux N, Han ZY, Bessis A, et al. Distribution and pharmacology of alpha 6-containing nicotinic acetylcholine receptors analyzed with mutant mice. J Neurosci. 2002;22:1208–1217. doi: 10.1523/JNEUROSCI.22-04-01208.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feller MB. The role of nAChR-mediated spontaneous retinal activity in visual system development. J Neurobiol. 2002;53:556–567. doi: 10.1002/neu.10140. [DOI] [PubMed] [Google Scholar]
- 14.Cox BC, Marritt AM, Perry DC, et al. Transport of multiple nicotinic acetylcholine receptors in the rat optic nerve: high densities of receptors containing alpha6 and beta3 subunits. J Neurochem. 2008;105:1924–1938. doi: 10.1111/j.1471-4159.2008.05282.x. [DOI] [PubMed] [Google Scholar]
- 15.Gotti C, Moretti M, Clementi F, et al. Expression of nigrostriatal alpha 6-containing nicotinic acetylcholine receptors is selectively reduced, but not eliminated, by beta 3 subunit gene deletion. Mol Pharmacol. 2005;67:2007–2015. doi: 10.1124/mol.105.011940. [DOI] [PubMed] [Google Scholar]
- 16.Marritt AM, Cox BC, Yasuda RP, et al. Nicotinic cholinergic receptors in the rat retina: simple and mixed heteromeric subtypes. Mol Pharmacol. 2005;68:1656–1668. doi: 10.1124/mol.105.012369. [DOI] [PubMed] [Google Scholar]
- 17.Swanson LW, Simmons DM, Whiting PJ, et al. Immunohistochemical localization of neuronal nicotinic receptors in the rodent central nervous system. J Neurosci. 1987;7:3334–3342. doi: 10.1523/JNEUROSCI.07-10-03334.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossi FM, Pizzorusso T, Porciatti V, et al. Requirement of the nicotinic acetylcholine receptor beta 2 subunit for the anatomical and functional development of the visual system. Proc Natl Acad Sci USA. 2001;98:6453–6458. doi: 10.1073/pnas.101120998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bansal A, Singer JH, Hwang BJ, et al. Mice lacking specific nicotinic acetylcholine receptor subunits exhibit dramatically altered spontaneous activity patterns and reveal a limited role for retinal waves in forming ON and OFF circuits in the inner retina. J Neurosci. 2000;20:7672–7681. doi: 10.1523/JNEUROSCI.20-20-07672.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feller MB, Wellis DP, Stellwagen D, et al. Requirement for cholinergic synaptic transmission in the propagation of spontaneous retinal waves. Science. 1996;272:1182–1187. doi: 10.1126/science.272.5265.1182. [DOI] [PubMed] [Google Scholar]
- 21.Zhou ZJ, Zhao D. Coordinated transitions in neurotransmitter systems for the initiation and propagation of spontaneous retinal waves. J Neurosci. 2000;20:6570–6577. doi: 10.1523/JNEUROSCI.20-17-06570.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong RO. Retinal waves and visual system development. Annu Rev Neurosci. 1999;22:29–47. doi: 10.1146/annurev.neuro.22.1.29. [DOI] [PubMed] [Google Scholar]
- 23.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 24.Geisert EE, Lu L, Freeman-Anderson NE, et al. Gene expression in the mouse eye: an online resource for genetics using 103 strains of mice. Mol Vis. 2009;15:1730–1763. [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanov D, Dvoriantchikova G, Barakat DJ, et al. Differential gene expression profiling of large and small retinal ganglion cells. J Neurosci Methods. 2008;174:10–17. doi: 10.1016/j.jneumeth.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reim K, Wegmeyer H, Brandstätter JH, et al. Structurally and functionally unique complexins at retinal ribbon synapses. J Cell Biol. 2005;169:669–680. doi: 10.1083/jcb.200502115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlamp CL, Johnson EC, Li Y, et al. Changes in Thy1 gene expression associated with damaged retinal ganglion cells. Mol Vis. 2001;7:192–201. [PubMed] [Google Scholar]
- 28.Huang W, Fileta J, Guo Y, et al. Downregulation of Thy1 in retinal ganglion cells in experimental glaucoma. Curr Eye Res. 2006;31:265–271. doi: 10.1080/02713680500545671. [DOI] [PubMed] [Google Scholar]
- 29.Howell GR, Walton DO, King BL, et al. Datgan, a reusable software system for facile interrogation and visualization of complex transcription profiling data. BMC Genomics. 2011;12:429. doi: 10.1186/1471-2164-12-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carter-Dawson LD, LaVail MM, Sidman RL. Differential effect of the rd mutation on rods and cones in the mouse retina. Invest Ophthalmol Vis Sci. 1978;17:489–498. [PubMed] [Google Scholar]
- 31.Jiménez AJ, García-Fernández JM, González B, Foster RG. The spatio-temporal pattern of photoreceptor degeneration in the aged rd/rd mouse retina. Cell Tissue Res. 1996;284:193–202. doi: 10.1007/s004410050579. [DOI] [PubMed] [Google Scholar]
- 32.Strettoi E, Porciatti V, Falsini B, et al. Morphological and functional abnormalities in the inner retina of the rd/rd mouse. J Neurosci. 2002;22:5492–5504. doi: 10.1523/JNEUROSCI.22-13-05492.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strom RC, Williams RW. Cell production and cell death in the generation of variation in neuron number. J Neurosci. 1998;18:9948–9953. doi: 10.1523/JNEUROSCI.18-23-09948.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams RW, Strom RC, Goldowitz D. Natural variation in neuron number in mice is linked to a major quantitative trait locus on Chr 11. J Neurosci. 1998;18:138–146. doi: 10.1523/JNEUROSCI.18-01-00138.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.John SW, Smith RS, Savinova OV, et al. Essential iris atrophy, pigment dispersion, and glaucoma in DBA/2J mice. Invest Ophthalmol Vis Sci. 1998;39:951–962. [PubMed] [Google Scholar]
- 36.Libby RT, Anderson MG, Pang IH, et al. Inherited glaucoma in DBA/2J mice: pertinent disease features for studying the neurodegeneration. Vis Neurosci. 2005;22:637–648. doi: 10.1017/S0952523805225130. [DOI] [PubMed] [Google Scholar]
- 37.Schlamp CL, Li Y, Dietz JA, et al. Progressive ganglion cell loss and optic nerve degeneration in DBA/2J mice is variable and asymmetric. BMC Neurosci. 2006;7:66. doi: 10.1186/1471-2202-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munguba GC, Camp AS, Risco M, et al. Vesicular glutamate transporter 3 in age-dependent optic neuropathy. Mol Vis. 2011;17:413–419. [PMC free article] [PubMed] [Google Scholar]