Abstract
Within hours of intranasal challenge, mouse-adapted H1N1 A/Puerto Rico/8/34 (PR8) influenza genomic RNA is found in the olfactory bulb (OB) and OB pro-inflammatory cytokines are up-regulated. Severing the olfactory tract delays the acute phase response (APR) and the APR is attenuated by immunization.
Objectives
Determine if immunization affects OB-localization of influenza or the molecular brain mechanisms regulating the APR.
Methods
Male mice were immunized with PR8 influenza then OB viral RNA, APR, and influenza-related cytokine responses were determined after homologous viral challenge.
Results
Immunization did not prevent influenza OB viral invasion within 24 hour of viral challenge. However, it greatly attenuated OB viral RNA 6 days post-viral challenge and the APR including hypothermia and body weight loss responses. Within the OB, 24 hours after influenza challenge, prior immunization blocked virus-induced up-regulation of toll-like receptor 7 and interferon gamma (IFNγ) mRNAs. At this time, hypothalamic (HT) growth hormone releasing hormone receptor and tumor necrosis factor-alpha mRNAs were greatly enhanced in immunized but not in positive control mice. By 6 days post-viral challenge, OB and HT mRNAs returned towards baseline values. In lungs, mRNA up-regulations were greater than those in the brain and were maximized 6 days post-challenge. Lung IFNγ mRNA decreased at 24 hours but increased at 6 days post-challenge in the positive controls compared to negative controls. Immunization prevented the up-regulation of most of the flu-related mRNAs measured in lungs.
Conclusion
Collectively, these data suggest a role for OB and HT involvement in immunization protection against influenza infection.
Keywords: olfactory bulb, hypothalamus, cytokine, immunization, growth hormone releasing hormone receptor, acute phase response, interferon gamma, influenza
Introduction
Acute viral infections are responsible for much of the infectious illness in the world and they have a tremendous economic and social toll (1). Remarkably little is known about how such viruses produce systemic symptoms. Of the known respiratory viruses, influenza is probably the most dangerous; its death toll in 1918–19 has been estimated to exceed 40 million and the 1957 and 1968 pandemics may have killed up to 6 million. In non-pandemic years influenza routinely kills about 50,000 people in the U.S. and a half million people world-wide, primarily the very young and very old. Mouse-adapted human strains of influenza cause a lethal or sub-lethal pneumonitis (similar to influenza pneumonia in humans) in the mouse accompanied by early onset of severe systemic symptoms (e.g. weight loss, hypothermia, excess sleep) (2, 3). Influenza virus also induces a broad array of cytokines and other immune mediators in infected lung and brain (4, 5, 6). Influenza virus produces double-stranded (ds) RNA during replication that is recognized by pattern recognition receptors (PRRs) (7, 8, 9). PRRs in turn induce cytokines (10, 11, 12, 13). The strain of influenza virus used in this study, H1N1 A/Puerto Rico/8/34 (PR8) is considered incapable of invading the mouse brain, but it localizes to the olfactory bulb (OB) within hours of intranasal infection (5). Viral RNA and viral antigen expression in the OB are accompanied by cytokine induction within the OB as well as the hypothalamus (HT). Cytokines are well-characterized for their role in triggering the acute phase response (APR). Interestingly, if the olfactory tract is severed, the onset of the APR is delayed (14).
PR8 invades the OB 1–2 days before the onset of influenza APR symptoms in the mouse model used in our studies. Influenza is thought to enter the CNS by retrograde travel via the peripheral nervous system axons (e.g., the vagus nerve) and cranial nerves innervating the OB following intranasal inoculation (15, 16, 17). Consequently, the now widely used method of intranasal influenza immunization with live attenuated influenza virus (LAIV) provides a plausible route of neuro-invasion. However, in our mouse model it is unknown whether prior immunization blocks influenza virus OB localization; we report herein that it does not at 24 hours post viral challenge but greatly attenuates viral OB localization at 6 days post inoculation. Nevertheless, we demonstrate that immunization is associated with virus-induced changes in OB and HT mRNAs of multiple substances involved in the immune response suggesting a role for these brain areas in immunization protection against influenza virus.
Materials and Methods
General procedures
Virus purification and titration
A mouse-adapted strain of influenza, H1N1 A/Puerto Rico/8/34 H1N1 (PR8) (lot#3X010621) suspended in pyrogen-free allantoic fluid (Specific Pathogen-Free Avian Supply, North Franklin, CT) was purified by sucrose-gradient sedimentation by ultracentrifugation as previously described (18). The virus was diluted to a protein concentration of 200 µg/mL, aliquoted, frozen on dry ice, and stored at −80°C. A standard hemagglutination assay with two-fold dilutions of purified PR8 (10 µL) mixed with 50 µL chicken red blood cells and incubated at room temperature for 30 min yielded 1×107 virus particles/mL. Samples were tested for endotoxin levels by the Limulus lysate gel-clot assay (Biowhittaker, Walkersville, MD) and no endotoxin was detected. Additionally, the purified virus tested negative for mycoplasma and acholeplasma contamination by nested PCR (nPCR) (American Type Culture Collection, Manassas, VA) as previously described (19). In Madin-Darby canine kidney (MDCK) cells, viral titrations were assessed and expressed as median tissue culture infectious doses (TCID50) as previously described (18) . The starting titer of purified PR8 virus was 5× 104 TCID50/µL.
Mice
Male C57BL/6J mice (n = 117) were obtained from Jackson Laboratories (Bar Harbor, ME). Mice were housed in plastic filter-top cages and maintained at 23–24°C on a 12:12 hour light-dark cycle. All mice were provided food and water ad libitum. Mice were 12–14 weeks old at the start of experiments. All mouse procedures were approved by the Washington State University Animal Care and Use Committee and conformed to National Institutes of Health guidelines.
Virus administration
The concentrations of PR8 and heat-inactivated (HI) (100°C, 25 min) PR8 used for each experiment are described below. At light onset, mice were anesthetized briefly using 20% isoflurane/80% polyethylene glycol (PEG) anesthesia (19), then intranasally instilled with 25 µL of live PR8 or HI PR8 in Dulbecco’s phosphate buffered saline (DPBS) (Sigma-Aldrich) in each nostril, for a total inoculum of 50 µL.
Tissue collection
Mice were anesthetized with isoflurane and blood was collected via terminal cardiac puncture. Blood was placed in tubes containing ethylenediaminetetraacetic acid (EDTA), centrifuged at 1000 × g for 20 min and the plasma was saved. Lungs were collected, the brains were removed, and the OB and HT were dissected as previously described (19, 20). Tissue and plasma were flash-frozen in liquid nitrogen, and stored at −80°C until analyzed.
Experimental protocols
Determination of immunization dose
Five groups of mice (n = 4–5 per group) were intranasally inoculated with DPBS as control, or 50 TCID50, or 10 TCID50, 5 TCID50, or 1 TCID50 dilutions of PR8 to determine the minimal dosage of PR8 to induce alterations in the APR. Mice were weighed on the day of immunization and every 2 days at light onset for 20 days post-immunization. By day 8, the 50 TCID50, 10 TCID50 and 5 TCID50 dose groups showed weight loss (data not shown). The 5 TCID50 immunization dose was used because on day 8, but not on other post-challenge days, there was a detectable small weight reduction. The 1 TCID50 dose failed to induce any body temperature change. In subsequent studies, mice were hyper-immunized 5 weeks after the first immunization using the 5 TCID50 dose of live virus. The mice were then used for APR and molecular studies 5 weeks after the second immunization injection.
Virus concentration necessary to enter OB
Twenty immunologically naïve mice (i.e. no previous exposure to PR8) (n = 5 per group) were intranasally inoculated with 50 TCID50, 2.5×103 TCID50, or 2.5×104 TCID50 of live PR8, or HI 2.5×104 TCID50 PR8 serving as a control to determine the minimal virus concentration necessary to enter the OB. Mice were sacrificed 24 hours post-virus challenge. OBs were analyzed by nested (n) PCR for PR8 minus and plus nucleoprotein (NP) RNA presence as previously described (5). Briefly, tissues were homogenized and RNA was extracted using Trizol reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. OB tissue cDNA was synthesized and then amplified using primers for the minus (genomic viral RNA) and plus strands (replication intermediates) of the PR8 NP gene. Primer sequences are shown in Table I. nPCR (NP) RNA products were visualized on Tris/Borate/EDTA (TBE) 1% agarose gels and (NP) RNA specific bands migrated as 450 base pair (bp) products.
Table I.
Primers used for RT-PCR.
| Gene of Interest | Sequence 5’–3’ | |
|---|---|---|
| NP-1 | plus strand | GGGAAGGATCCTAAGAAAAC |
| NP-2 | minus strand | TGCACTTTCCATCATCCTTA |
| NP-3 | plus strand | AATGATCGGAACTTCTGGAG |
| NP-4 | minus strand | CTTCGTCCCTTTGATGAAGC |
| Mx1 | sense | GAAGGCAAGGTCTTGGATG |
| antisense | GCTGACCTCTGCACTTGACT | |
| OAS-1a | sense | CTTTGATGTCCTGGGTCATGT |
| antisense | GCTCCGTGAAGCAGGTAGAG | |
| IFNγ | sense | CTGACCAATAAGAAACATTCAGAGCT |
| antisense | GAGTGTAGACATCTCCTCCCATCA | |
| IL1β | sense | CAACCAACACGTGATATTCTCCATG |
| antisense | GATCCACACTCTCCAGCTGCA | |
| TLR3 | sense | AGCATCAAAAGAAGCCGAAA |
| antisense | CTTGCTGAACTGCGTGATGT | |
| MDA-5 | sense | GCCTGGAACGTAGACGACAT |
| antisense | TCATCGAAGCAGCTGACACT | |
| RIG-1 | sense | AGAGAATTCGGCACCCAGAA |
| antisense | AGCTCTCGCTCGGTCTCATC | |
| GHRHR | sense | CTGGCCTTGACATCCGTGTAC |
| antisense | GTCGAGCTGGCAGAAGTTCAG | |
| TLR7 | sense | CTGGAGTTCAGAGGCAACCATT |
| antisense | TGTTATCACCGGCTCTCCATAGAA |
Core body temperatures and body weights post-viral challenge
Fourteen mice (n = 4–6 per group) were anesthetized with intraperitoneal (IP) injection of ketamine/xylazine (87/13 mg/kg) and surgically provided with small IP transponders (TAE-mitters, Mini Mitter, Bend, OR) and then placed on top of receiver plates in individual filter-top cages as previously described (14). After surgery, mice were allowed 2 weeks of recovery time prior to experimentation. Then, the immunologically naïve negative and positive control mice were intranasally inoculated with DPBS. The immunized mice received 5 TCID50 PR8 then a second immunization challenge 5 weeks later. Five weeks after hyper-inoculations or in separate groups of mice used for positive or negative control injections, baseline measurements were taken for 2 days. Core body temperature and locomotor activity data were collected in 6 min intervals, averaged over 2 hours, and processed with the VitalView analysis software (Mini Mitter). Mice were then challenged with either live PR8 (2.5×104 TCID50) (positive control and immunized mice) or the same dose of HI PR8 (negative control), at light onset and were then monitored for 12 days post-virus challenge to determine the effect of immunization on the APR. In addition, baseline body weight was taken at light onset and daily for 12 days post-virus challenge.
PR8 antibodies and molecular expressions in brain and lungs
Three groups of mice (n = 20 per group)(negative control, positive control and immunized mice) were treated as above but without the implantation of transponders, and tissues used for antibody and molecular determinations were collected 24 hours or 6 days post-virus challenge (10 mice/group sacrificed at each time point). However, an n = 5 was used for influenza virus detection in the OB 6 days post-virus challenge due to sample loss.
Anti-PR8 determination
A plaque assay based on Bachmann et al. (1999) was used to determine antibodies specific to PR8 (21). Briefly, plasma was treated at 56°C for 30 min to inhibit complement, diluted 1:20, and then incubated with 50 TCID50 PR8 for 1 hour. MDCK cell monolayers in 6-well tissue culture plates were inoculated with 200 µl of a virus/serum mixture, virus only, or no virus (i.e. saline) and incubated for 1 hour (37°C, 5% CO2) to allow cell attachment. A soft agar/media suspension was layered over the wells and allowed to solidify, and cells were incubated for 3 days (37°C with 5% CO2). The soft agar was then removed and the cells were stained with 1% crystal violet/70% methanol for 20 min, and then washed under water until clear. Plaques in the wells were counted in duplicate on wells under illumination (713 lux) by an investigator blinded to the treatments. Inter-rater reliability was tested by blindly counting the same randomly selected plates (r = 0.888, p = 0.018).
RNA determinations
OB, HT and lung tissues were taken 24 hours and 6 days post-infection challenge from each group and were processed and treated for determining the presence of minus and plus strand PR8 (NP) RNA using nPCR as described in (5). OB, HT, and lung tissue from each group were also analyzed for mRNA levels of murine genes known to be affected by influenza virus. Cyclophilin A (CycA), 2’,5’-oligoadenylate synthetase (OAS-1a), myxovirus resistance-1 (Mx1), interferon gamma (IFNγ), toll-like receptor 3 (TLR3), toll-like receptor 7 (TLR7), melanoma differentiation associated gene-5 (MDA-5), retinoic acid-inducible gene-1 (RIG-1), , tumor necrosis factor alpha (TNFα), interleukin-1 beta (IL1β), and GHRHR mRNA levels were measured by RT-PCR as previously described (22). The primers used are shown in Table I. RNA was extracted, and cDNA was prepared and analyzed as previously described (19, 20). CycA mRNA was used to compare expression of mRNAs and these data were analyzed by the delta threshold cycle (Ct) value method as previously described (22).
Statistics
Core body temperature and activity data were analyzed with univariate and mixed 2-way analysis of variance (ANOVA) repeated measures to determine significant time, treatment, and group differences. If main effects were found, the post-hoc analyses were used and those values are reported. Plaque formation numbers and mRNA data were analyzed between treatment groups using independent t-test analysis. Significance was set at p < 0.05.
Results
Virus concentration necessary to enter OB
Neither viral minus or plus strand (NP) RNA was present in the OB of mice inoculated with HI 2.5×104 TCID50 PR8 (Table II); this result is consistent with prior findings (5). After the lower live virus doses (50 TCID50 and 2.5×103 TCID50), viral NP RNA was also not detectable in the OB. However, virus minus and plus strand (NP) RNAs were detected in the OB of mice given the 2.5×104 TCID50 dose of PR8 (Table II). A 2.5×104 TCID50 viral dose was used in further experiments; this dose is about one hundredth the doses we used in prior studies (5, 19).
Table II.
PR8 concentration necessary to enter the OB.
| Minus (Genomic) |
Plus (Replicating) | |
|---|---|---|
| Treatment Group | PR8 (NP) RNA | PR8 (NP) RNA |
| HI (2.5 × 104 TCID50) | 0/5 | 0/5 |
| 50 TCID50 | 0/5 | 0/5 |
| 2.5 × 103 TCID50 | 0/5 | 0/5 |
| 2.5 × 104 TCID50 | 4/5 | 3/5 |
Immunologically naïve mice were inoculated with various concentrations of PR8 via intranasal administration. 24 hours post-viral challenge data are shown. Mice challenged with HI (2.5 × 104 TCID50), 50 TCID50, and 2.5 × 103 TCID50 did not have detectable minus or plus strand NP (RNA) in the OB 24 hours post-viral challenge. However, mice given PR8 at 2.5 × 104 TCID50 had minus and plus strand (NP) RNA in the OB 24 hours post-challenge.
Acute phase responses to live and heat-inactivated virus
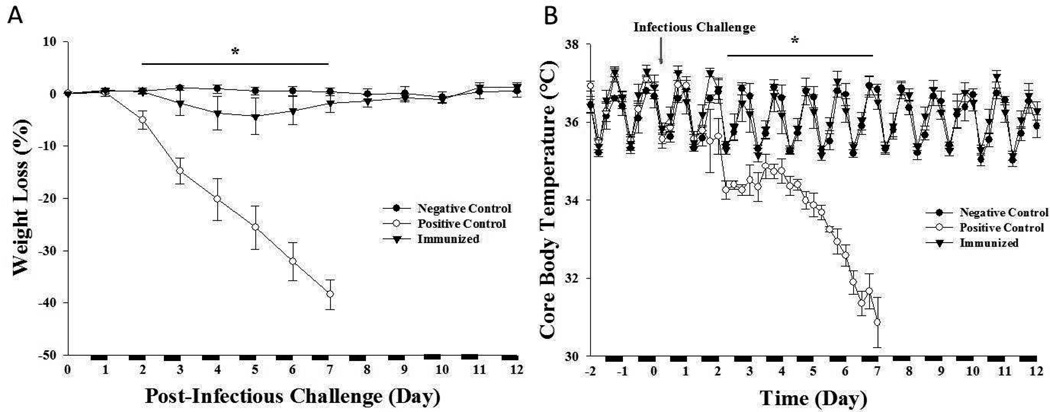
Immunologically naïve mice challenged with 2.5×104 TCID50 PR8 (positive controls) had significantly lower core body temperatures following virus challenge compared to baseline values [time: F (1, 13) = 36.034, p < 0.001; time×treatment group: F (2, 13) = 12.795, p = 0.001] (Figure 1B). This effect began 2 days post-viral challenge and persisted until day 7 when all positive control mice died. Core body temperatures of naïve mice challenged with HI 2.5×104 TCID50 PR8 (negative controls) were not significantly different from baseline values (Figure 1B). Mice immunized with live 5 TCID50 PR8 had similar core body temperatures as to those observed in the negative control mice. Body weights of positive control mice declined compared to the negative control and immunized mice [time: F (1, 13) = 57.361, p = 0.001; time×treatment group: F (2, 13) = 38.670, p < 0.001. These effects were evident by the second day after virus challenge and continued to decline until day 7 (Figure 1A). In contrast, the body weights of the negative controls and the virus-challenged immunized mice did not change significantly (Figure 1A).
Figure 1. Body weight and core body temperatures after PR8 viral challenge.
Body weight was not altered in negative control mice challenged with HI 2.5 × 104 TCID50 PR8 (A). Positive controls had a rapid loss in body weight (days 2–7) and died within 7 days of viral challenge. Immunized mice had a similar body weight response post-challenge (2.5 × 104 TCID50 PR8) as the negative control mice. Negative control and immunized mice showed normal cyclical temperature variation throughout the day and viral challenge did not alter core body temperature in these groups (B). Positive control mice had rapid loss of body temperature after the viral challenge. n = 4–6 per group. (*) = significant difference comparing positive controls to negative controls. On the x-axis, the black bar indicates dark cycle and white indicates light cycle (12 hour light:12 hour dark). Significance was set at p < 0.05.
Plaque assay for antibodies specific to PR8
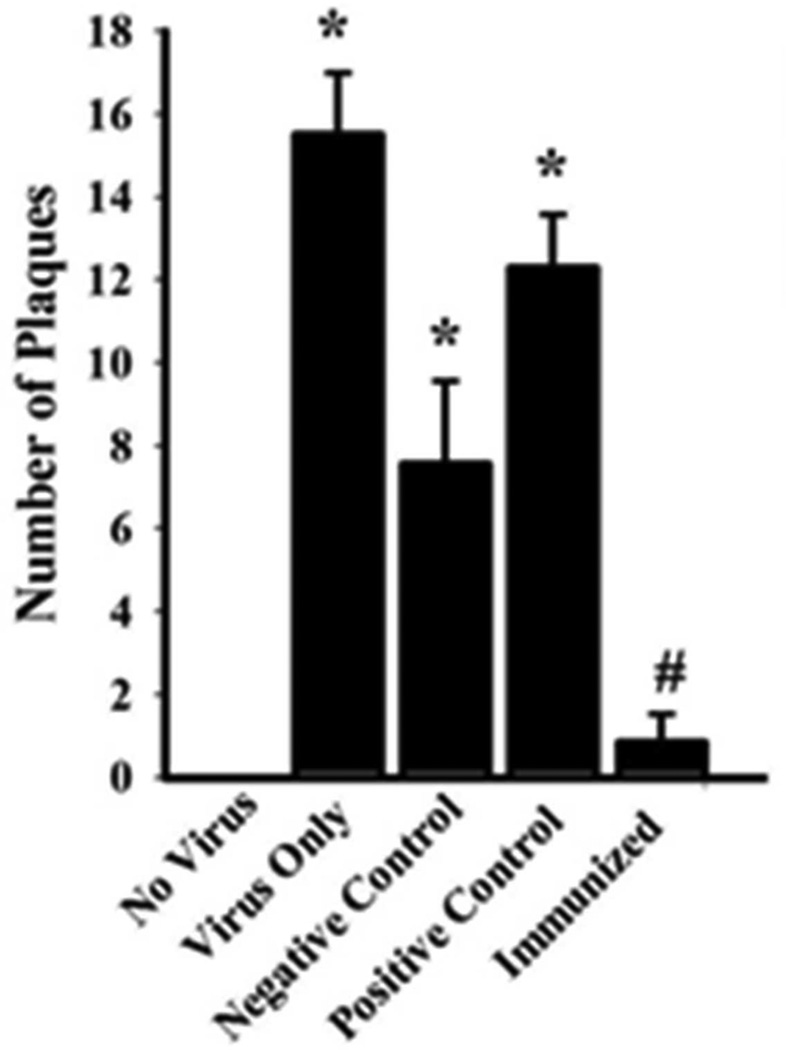
The plaque assay indicated that immunized mice had PR8-specific antibodies in their plasma (Figure 2). MDCK cells not exposed to virus had no plaques while MDCK cells exposed to virus had greater numbers of plaques [t (1, 2) = 10.333, p = 0.009]. Plasma from immunized mice prevented plaque formation in the MDCK cells compared to the positive control group [t (1, 2) = 8.785, p < 0.001]. The plasma from the negative control group also failed to prevent viral plaques in the MDCK cells.
Figure 2. Plaque Assay for Detection of Antibodies Specific to PR8.
Plaque numbers for each treatment group are presented as means ± SE (n = 10 per group). No plaques were formed on the MDCK monolayer when no virus was applied. PR8 induced plaque formation if only virus was applied (i.e. no plasma). Significantly fewer plaques were formed when plasma from immunized mice was applied to the MDCK monolayer compared to when plasma from the other treatment groups was used, indicating the presence of PR8-specific antibodies in the immunized mice. Similar numbers of plaques were found in negative and positive control groups suggesting that plasma from both groups lacked anti-PR8 antibodies. (*) = significant when compared to MDCK monolayers that had no virus applied. (#) = significance when compared to positive control mice. Significance was set at p < 0.05.
Viral NP minus and plus RNA occurrence in the OB and lungs
Neither minus or plus viral NP RNA was found in the OBs or lungs of the negative control mice 24 hours after viral challenge (Table III). In contrast, in the positive control mice, the minus NP RNA, but not the plus NP RNA, was present in the OBs of 8 of the 10 animals tested. In lungs, all the immunized and positive control mice had both minus and plus viral NP RNA 24 hours post-viral challenge. In immunized mice, minus NP RNA, but not plus strands, was found in the OB within 24 hours of challenge. In the lungs of immunized mice, most of the mice had both minus and plus NP RNA strands 24 hours post-viral challenge. By day 6 post-viral challenge, the occurrence of NP RNA was similar as that found after 24 hours with the exception of the immunized mice. On day 6, only minus NP RNA was detected in only one mouse, and none of the mice had the plus strand, in the OB suggesting that immunization greatly attenuated OB viral localization. In lungs 6 days after challenge, only a few of the immunized mice possessed viral NP RNA strands (Table III).
Table III.
Minus and plus strand PR8 RNA presence in the OB and lungs 24 hours and 6 days post-viral challenge.
| OB | Lung | |||
|---|---|---|---|---|
| 24 hours | Minus | Plus | Minus | Plus |
| (Genomic) | (Replicating) | (Genomic) | (Replicating) | |
| Treatment Group | PR8 (NP) RNA | PR8 (NP) RNA | PR8 (NP) RNA | PR8 (NP) RNA |
| Negative control | 0/9 | 0/9 | 0/10 | 0/10 |
| Positive control | 8/10 | 0/10 | 10/10 | 10/10 |
| Immunized | 7/10 | 0/10 | 9/10 | 6/10 |
| OB | Lung | |||
| Day 6 | Minus | Plus | Minus | Plus |
| (Genomic) | (Replicating) | (Genomic) | (Replicating) | |
| Treatment Group | PR8 (NP) RNA | PR8 (NP) RNA | PR8 (NP) RNA | PR8 (NP) RNA |
| Negative control | 0/5 | 0/5 | 0/8 | 0/8 |
| Positive control | 4/5 | 3/5 | 10/10 | 10/10 |
| Immunized | 1/5 | 0/5 | 3/10 | 1/10 |
mRNA expressions in the OB, HT, and lung 24 hours post-viral challenge
OB
The changes in OB mRNA levels 24 hours post-viral challenge were all below 5-fold except for IFNγ (Figure 3). TLR7 mRNA levels were greater in positive control mice compared to negative control mice [t (1, 16) = 3.641, p = 0.003]. TLR7 mRNA levels in immunized mice were similar to the negative control group and lower than the positive control group [t (1, 17) = 3.295, p = 0.004]. Overall, immunization had little effect on altering TLR3, MDA-5, and RIG-1 mRNAs in the OB. Thus, TLR3 mRNA levels were greater in the positive control and immunized groups vs. the negative control group [t (1, 16) = 2.257, p = 0.038; t (1, 16) = 3.368, p = 0.004, respectively] although in immunized mice they were not significantly different from the positive control mice. Immunized mice had greater MDA-5 and RIG-1 mRNA levels compared to negative controls [t (1, 16) = 2.265, p = 0.038; t (1, 16) = 3.685, p = 0.002, respectively]. However, MDA-5 and RIG-1 mRNA levels of the immunized group were not significantly different from the positive control group.
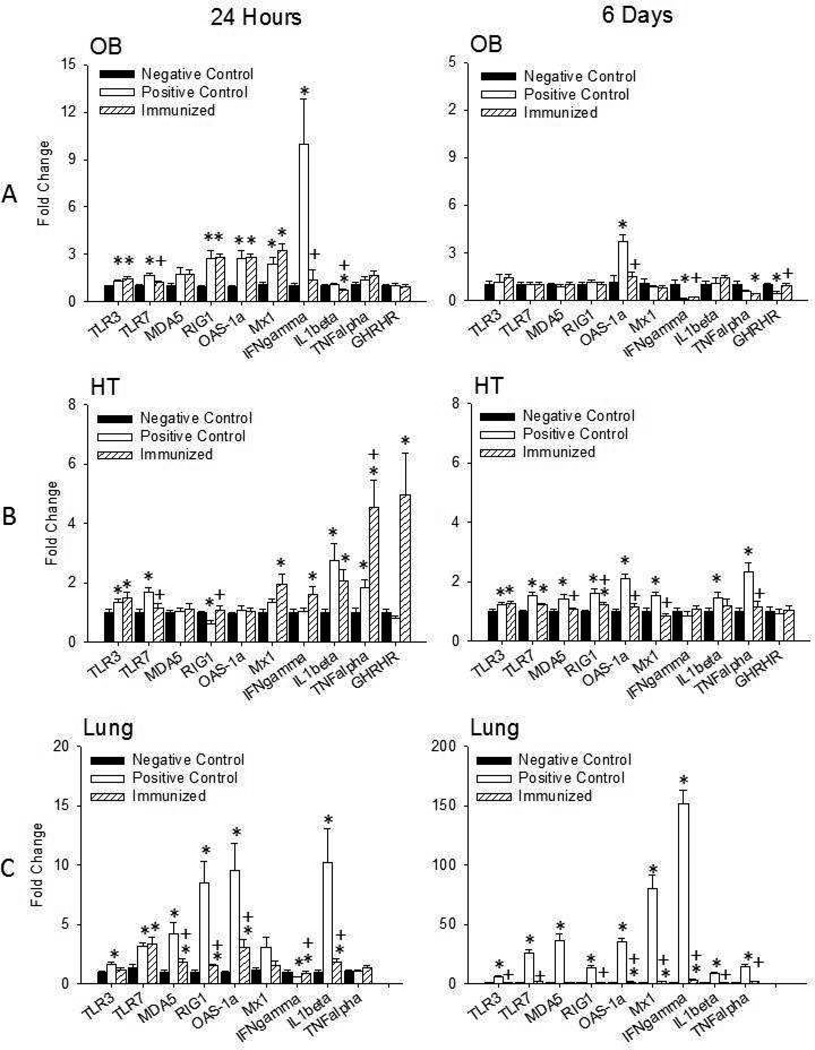
Figure 3. Virus-related cytokines in the olfactory bulb, hypothalamus, and lung 24 hours and 6 days post-viral challenge.
Influenza virus induced small enhancements of most cytokine mRNAs in the olfactory bulb (OB) (A) of the positive control mice 24 hours post-viral challenge. Immunization had little effect on altering mRNA levels in the OB after viral challenge with the exception of IFNγ and TLR7 mRNAs. In the hypothalamus (HT) (B), the influenza-induced enhancement of TLR7 was attenuated with immunization, although Mx1, TNFα, and GHRHR mRNA levels were enhanced. In the lung (C), immunization inhibited PR8-induced elevations in most of the mRNAs measured; this effect was more pronounced 6 days post-viral challenge. (*) = significant difference compared to negative controls. (+) = significant difference compared to positive controls. Significance was set at p < 0.05.
The largest mRNA effect in the OB 24 hours post-viral challenge occurred with IFNγ. IFNγ mRNA levels were increased by about 10 fold in positive control mice compared to negative control mice [t (1, 17) = 3.291, p = 0.004]. IFNγ mRNA levels in immunized mice were significantly less than positive controls [t (1, 18) = 3.255, p = 0.004]. The IFNγ mRNAs were not significantly different in immunized mice vs. negative controls. In the OB 24 hours post challenge, IL1β, and TNFα mRNA levels were similar in all 3 groups. The immunized group exhibited slightly lower IL1β mRNA levels compared to the positive control group [t (1, 18) = 2.148, p = 0.046], but the positive control group was higher than negative control group [t (1, 15) = 2.927, p = 0.015]. OAS-1a mRNA and Mx1 mRNA responses were less in magnitude but were greater in the positive control group compared to the negative control group [t (1, 16) = 3.886, p = 0.004; t (1, 15) = 3.807, p = 0.002, respectively]. The immunized group also had greater OAS-1a and Mx1 mRNA levels compared to negative controls [t (1, 15) = 7.143, p < 0.001; t (1, 17) = 5.072p < 0.001, respectively], although OAS-1a and Mx1 mRNA levels were similar to the positive control group. GHRHR mRNA levels were not significantly different between any treatment groups.
HT
Changes in HT TLR3, TLR7, RIG1, Mx1, and IFNγ mRNAs were all small, 2 fold or less, 24 hours post-challenge. In immunized mice, HT mRNA levels of TLR3, IFNγ and Mx1 were significantly higher than the negative control group [t (1, 17) = 2.612, p = 0.02; t (1, 17) = 2.739, p = 0.02; t (1, 18) = 3.017, p < 0.01, respectively]. Further, TLR3 and TLR7 mRNA expressions were up-regulated and RIG1 mRNA was down-regulated in the positive control vs. negative group [t (1, 18) = 2.414, p = 0.027; t (1, 17) 3.758, p < 0.001; t (1, 18) = 3.334, p = 0.005, respectively] and immunization blocked the up-regulation of TRL7 mRNA and the down-regulation of RIG1 mRNA (Figure 3). HT MDA-5 and OAS-1a mRNAs 24 hours post-viral challenge were similar between groups. In contrast, HT IL1β and TNFα mRNA levels were greater in positive controls vs. negative controls [t (1, 18) = 3.365, p = 0.003; t (1, 18) = 2.845, p = 0.011, respectively]. Further, IL1β and TNFα mRNA levels were greater in immunized mice compared to negative control [t (1, 17) = 2.874, p = 0.011; t (1, 18) = 4.396, p < 0.001, respectively]. Immunized mice had similar IL1β mRNA but greater TNFα mRNA levels as positive control mice [t (1, 18) = 3.241, p = 0.005]. The largest effect of immunization occurred with GHRHR mRNA; the immunized group had greater GHRHR mRNA levels than the positive and negative control groups [t (1, 15) = 2.302, p = 0.036; t (1, 15) = 2.780, p = 0.032, respectively].
Lungs
Within 24 hours of virus challenge, lung levels of TLR3, TLR7, MDA5, RIG-1, OAS-1a and IL1β mRNAs were up-regulated in positive controls relative to negative controls [t (1, 12) = 2.525, p = 0.027; t (1, 13) = 2.866, p = 0.013; t (1, 12) = 2.875, p = 0.014; t (1, 13) = 3.896, p = 0.002; t (1, 13) = 3.405, p = 0.005; t (1, 12) = 2.560, p = 0.025, respectively]. The increases in RIG1, OAS-1a and IL1β mRNAs were substantial being 7–9 fold greater (Figure 3). Immunization blocked the up-regulations of MDA5, RIG1, OAS-1a and IL1β mRNAs [t (1, 13) = 2.363, p <0.05; t (1, 13) = 3.882, p = 0.006; t (1, 14) = 2.652, p = 0.029; t (1, 12) = 2.851, p = 0.025, respectively]. In contrast, Mx1 and TNFα mRNA did not differ between groups 24 hours after viral challenge. Lung IFNγ mRNA was significantly reduced in the positive control mice compared to the other two groups [t (1, 14) = 2.803, p = 0.023; t (1, 13) = 2.646, p = 0.033, respectively].
mRNA expressions in the OB, HT and Lungs 6 days post-viral challenge
OB
Similarly to Day 1, overall mRNA levels in the OB 6 days after viral challenge are limited to less than 5-fold changes compared to baseline (Figure 3). The most prominent change were the greater levels of OAS-1a mRNA in positive control mice compared to negative controls [t (1, 6) = 3.176, p = 0.006]. Immunized mice also had lower levels of OAS-1a mRNA compared to positive control mice [t (1, 9) = 4.278, p = 0.008]. TNFα mRNA levels in immunized mice were lower levels than that occurring in the negative control group [t (1, 12) = 2.709, p = 0.03]. OB IFNγ mRNA levels were very low in both the immunized mice compared to the negative and positive controls of mice sacrificed 6 days after viral challenge [t (1, 12) = 2.287, p = 0.041; t (1, 8) = 2.483, p = 0.038]. In contrast, GHRHR mRNA levels were reduced in the positive control group compared to both negative control and immunization groups [t (1, 9) = 4.184, p = 0.014; t (1, 8) = 3.543, p = 0.03, respectively].
HT
The changes in hypothalamic mRNAs 6 days after viral challenge were all under 2 fold (Figure 3). Nevertheless, some of the differences were significant, TLR3, TLR7, MDA5, RIG1, OAS-1a, Mx1, IL1β, and TNFα mRNAs were all higher in the positive controls than the negative control mice [t (1, 16) = 2.561, p = 0.021; t (1, 18) = 3.802, p = 0.001; t (1, 18) = 2.845, p = 0.011; t (1, 17) = 3.750, p = 0.002; t (1, 17) = 7.217, p < 0.001; t (1, 16) = 3.807, p = 0.002; t (1, 18) = 2.844, p = 0.012; t (1, 18) = 4.528, p < 0.001, respectively]. Further, immunization blocked the up-regulations of MDA5, RIG1, OAS-1a, Mx1 and TNFα mRNAs [t (1, 16) = 2.389, p <0.05; t (1, 16) = 2.349, p = 0.037; t (1, 17) = 5.562, p < 0.001; t (1, 16) = 5.045, p < 0.001; t (1, 17) = 3.54, p < 0.001, respectively]. On the 6 day post-viral challenge, HT IFNγ and GHRHR mRNA did not change between groups.
Lungs
The largest changes in mRNA expressions found in this study were evident in the lungs of positive control mice 6 days after challenge. Every mRNA determined was significantly greater, about 10 fold or higher, in the positive controls mice than in the negative control mice [TLR3, TLR7, MDA5, RIG1, OAS-1a, Mx1, IFNγ, IL1β, and TNFα, [t (1, 17) = 8.168, p < 0.001; t (1, 17) = 6.363, p < 0.001; t (1, 17) = 6.274, p < 0.001; t (1, 17) = 7.424, p < 0.001; t (1, 18) = 9.799, p < 0.001; t (1, 17) = 6.782, p < 0.001; t (1, 17) = 12.986, p < 0.001; t (1, 18) = 8.875, p < 0.001; t (1, 18) = 8.642, p < 0.001 respectively]. Further, immunization blocked these upregulations [t (1, 16) = 7.288, p < 0.001; t (1, 16) = 5.699, p < 0.001; t (1, 16) = 5.860, p < 0.001; t (1, 16) = 6.702, p < 0.001; t (1, 16) = 8.527, p < 0.001; t (1, 16) = 6.301, p < 0.001; t (1, 16) = 11.988, p < 0.001; t (1, 17) = 7.803, p < 0.001; t (1, 16) = 7.140, p < 0.001 respectively]. In the lungs 6 days after viral challenge, OAS-1a, Mx1, IFNγ, and TNFα mRNAs were increased in the immunized group mice relative to the negative control mice [t (1, 17) = 2.444, p < 0.001; t (1, 16) = 2.699, p = 0.02; t (1, 16) = 2.909, p =0.022; t (1, 17) = 4.057, p < 0.001 respectively] (Figure 3).
Discussion
Our major finding is that immunization of mice failed to prevent homologous influenza virus from invading the OB within 24 hours of intranasal viral challenge as determined by the presence of viral genomic (minus) RNA although it almost completely blocked OB viral presence 6 days post inoculation. The occurrence of virus in the OB 24 hours post inoculation is consistent with prior findings of influenza invading the OB within just a few hours of viral challenge (5), although we now show that virus is found in the OB after much lower virus doses. In addition, current results indicate that virus is present in the OB of positive control mice 6 days after inoculation. Although this might suggest viral replication within the OB, it seems more probable that the virus continuously replicates over the course of several days within the nasal epithelium and then is transported to the OB. Immunization attenuated this process because both minus and plus viral RNA detection was substantially lower in immunized mice than in the positive controls 6 days after viral challenge (Table III). Immunization also prevented the hypothermia and weight loss associated with the APR to influenza and influenza-induced mortality as previously shown by others (23, 24). Further, as discussed below, immunization had profound effects on the expression of several mRNAs within brain and lungs. Other viruses also localize to the OB. For instance, a closely related virus to H1N1—the highly pathogenic H5N1 influenza (i.e., H5N1 avian influenza), enters the CNS of mice around 7 days after intranasal inoculation (25). Parainfluenza virus also penetrates the OB of mice and its viral RNA is present in olfactory glomeruli after intranasal inoculation (26). Whether OB localization of viral RNA or DNA modifies immunization outcomes remains unknown. Regardless, current results suggest that immunization does not affect viral transport to the OB within 24 hours of viral challenge. However, immunization attenuates OB virus localization over the multiple day time course of the infection.
IFNγ mRNA is substantially up-regulated in the OB within 24 hours of viral challenge and this response is almost completely attenuated in immunized mice. These results clearly indicate that immunization affects OB production of molecules known for their involvement in host defenses against viruses (27). Further, they suggest that OB IFNγ is involved in the initiation of the morbidity associated with the APR but not maintenance of the APR since the OB- IFNγ mRNA response is absent by 6 days post-inoculation.
Influenza viron particles contain negative sense single stranded (ss) RNA as their genomic RNA and positive sense strands are synthesized during replication, forming viral double-stranded (ds) RNA that is recognized by the PRRs such as TLR3, MDA-5, and RIG-1. These actions lead to pro-inflammatory cytokine production and activation (28). Mice lacking TLR3 have attenuated APR sickness behavior responses to PR8 influenza infection indicating a role of PRRs in influenza pathogenesis (29). Immunization did not block influenza-induced OB up-regulation of TLR3, MDA-5, and RIG-1 mRNAs, suggesting these ds RNA receptors retain a role in host-defense after immunization. In contrast, the attenuation of virally-induced up-regulated OB TLR7 mRNA with immunization might have altered viral pathogenicity including viral invasion into the lungs, lung cytokines, and thus the APR. Thus immunization-induced inhibition of OB TLR7 would attenuate the recognition of the ss RNA influenza RNA and thereby limit the production of down-stream events such as IL1β and IFNγ production. In addition, TLR7 influences the production of influenza-specific antibodies (30). Further, after influenza vaccination, TLR7−/− mice fail to gain protective immunization (31). Finally, the immunization-alteration of OB TLR7, IFNγ and IL1β mRNA levels 24 hours post viral challenge is consistent with the prior demonstration that TLR7 agonists induce local production of pro-inflammatory cytokines such as TNFα and IL1β within brain (32).
The IFN-inducible genes OAS-1a and Mx1 have antiviral functions (33) , and the Mx1 gene antiviral activity is specific for orthomyxoviruses, including influenza (27, 34). However, immunization did not alter viral-induced OB expression of these genes compared to positive controls despite the large up-regulation of OB IFNγ mRNA suggesting that the OAS-1a and Mx-1 genes in the OB are not involved in the induction of the APR.
In the lungs, immunization attenuated enhanced IL1β and OAS-1a mRNAs 24 hours after infectious challenge and these effects may have contributed to the immunization-associated attenuated APR. By 6 days post-viral challenge lung IFNγ, IL1β and TNFα mRNA levels are elevated in the positive controls relative to the immunized mice. Further, systemic injection of IFNγ, IL1β or TNFα induce several facets of the APR including reduced locomotor activity, body temperature changes, and enhanced brain cytokine levels suggesting that these, and perhaps other pro-inflammatory cytokines of lung origin, maintain the influenza-induced APR (35, 36, 37).
Another notable finding reported herein is the very large up-regulation of HT GHRHR mRNA 24 hours after influenza challenge in immunized mice. It seems likely that the GHRHR plays some type of protective role since mice that lack functional GHRHRs, lit/lit mice, have higher morbidity and mortality rates upon influenza challenge (38). An independent study, using quantitative genetic analysis, also implicated the GHRHR as playing a role in the influenza-induced APR (3). The GHRHR is expressed in both the HT and pituitary; since growth hormone does not rescue the lit/lit mice from influenza challenge, we hypothesized that HT GHRHR is involved in host responses to influenza. The differences between positive controls and immunized mice is not likely due to the presence of virus because Alt et al (2003) showed that 24 hours after influenza challenge (38), using a higher dose than that used in the present study, there were no replicating viruses in the brain and lung viral titers were similar in WT and lit/lit mice. Naïve WT mice, but not lit/lit mice, have enhanced HT IL1β, TNFα and GHRHR mRNA levels 38 hours after influenza challenge confirming the HT involvement of these substances as part of the molecular APR (39). The GHRHR is up-regulated by IL1β (40), but HT IL1β mRNA was increased in both positive control mice and immunized mice, thus, this does not seem to be the cause of increased GHRHR mRNA observed only in immunized mice. The actions of IL1β on either pituitary release of growth hormone or on sleep are blocked by anti-GHRH antibodies suggesting that GHRH and its receptor are down-stream from IL1β (41); whether this sequence of events is also involved in immunization and occurs in the HT remains unknown. Regardless of such speculation, current results suggest that hypothalamic GHRHR may play a fundamental role in the protection provided by immunization.
Immunization inhibits viral replication and the APR and is mediated, in part, by antibody-specific B cells and attenuated pro-inflammatory cytokine responses (42). Activation of antibody secreting cells (ASCs) in the lungs, particularly of memory B cells secreting large amounts of IgG and IgA antibodies specific to the particular strain of influenza virus, plays a significant role in reducing viral titers in subsequent infections (43). Our results demonstrate that immunization attenuated the influenza-induced APR, reduced mortality, enhanced PR8-specific antibodies, and attenuated lung pro-inflammatory cytokines are consistent with prior findings (3, 14, 44). The occurrence of PR8-specific antibodies in immunized mice and lack of APR after the influenza immune challenge with the same virus indicates that these aspects of adaptive immunity can modulate innate immune functions regulating the APR.
The present study of influenza immunization using intranasal-administered live virus is relevant to intranasally administrated human vaccines. The recently approved seasonal LAIV FluMist® (MedImmune, Inc., Gaithersburg, MD, USA) provided protection to pandemic 2009 H1N1 and secondary bacterial infections primarily through CD4+ T cells (24). Further, mice intranasally immunized with various LAIV preparations have reduced lung viral titers that are dependent on viral replication efficiency associated with the local induction of cytokines (45). That a close proximity between the intranasal inoculation site and the exposed OB neuron projections exists in the nasal cavity is a concern because of the ready access of the virus into the brain (46). In humans, intranasal administration of the virus may be associated with viral localization to the OB as occurs in mice (this study). Conversely, the effectiveness of LAIV may be dependent upon OB molecular events induced by the intranasal application of virus.
In conclusion, we show that intranasal immunization does not prevent viral invasion into the OB, but attenuates production of viral plus RNA and the viral up-regulated IFNγ mRNA that occurs within 24 hours of challenge. Further, immunization is associated with enhanced hypothalamic IFNγ, TNFα and GHRHR mRNAs and the attenuation of facets of the APR regulated by the brain.
Acknowledgements
This work was supported by the National Institutes of Health, Grant # HD036520 to JMK. We thank Dr. Christopher J. Davis for his review of the manuscript and help with data analyses.
Footnotes
(a) Papers published in periodicals: Sun J, Koto H, Chung KF: Interaction of ozone and allergen challenges on bronchial responsiveness and inflammation in sensitised guinea pigs. Int Arch Allergy Immunol 1997;112:191–195.
References
- 1.Oxford JS. Influenza A pandemics of the 20th century with special reference to 1918: virology, pathology and epidemiology. Rev Med Virol. 2000;10:119–133. doi: 10.1002/(sici)1099-1654(200003/04)10:2<119::aid-rmv272>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 2.Fang J, Sanborn CK, Renegar KB, Majde JA, Krueger JM. Influenza viral infections enhance sleep in mice. Proc Soc Exp Biol Med. 1995;210:242–252. doi: 10.3181/00379727-210-43945. [DOI] [PubMed] [Google Scholar]
- 3.Toth LA, Williams R. A quantitative genetic analysis of slow-wave sleep in influenza-infected CXB recombinant inbred mice. Behav Genet. 1999;29:339–348. doi: 10.1023/a:1021661901196. [DOI] [PubMed] [Google Scholar]
- 4.Conn CA, McClellan JL, Maassab HF, Smitka CW, Majde JA, Kluger MJ. Cytokines and the acute phase response to influenza virus in mice. Am J Physiol. 1995;268:R78–R84. doi: 10.1152/ajpregu.1995.268.1.R78. [DOI] [PubMed] [Google Scholar]
- 5.Majde JA, Bohnet SG, Ellis GA, Churchill L, Leyva-Grado V, Wu M, Szentirmai É, Rehman A, Krueger JM. Detection of a mouse-adapted human influenza virus in the olfactory bulb of mice within hours after intranasal infection. J NeuroVirol. 2007;13:399–409. doi: 10.1080/13550280701427069. [DOI] [PubMed] [Google Scholar]
- 6.Van Reeth K. Cytokines in the pathogenesis of influenza. Vet Microbiol. 2000;74:109–116. doi: 10.1016/s0378-1135(00)00171-1. [DOI] [PubMed] [Google Scholar]
- 7.Majde JA. Viral double-stranded RNA, cytokines and the flu. J Interferon Cytokine Res. 2000;20:259–272. doi: 10.1089/107999000312397. [DOI] [PubMed] [Google Scholar]
- 8.Majde JA, Brown RK, Jones MW, Dieffenbach CW, Maitra N, Krueger JM, Cady AB, Smitka CW, Maassab HF. Detection of toxic viral-associated double-stranded RNA (dsRNA) in influenza-infected lung. Microb Pathogen. 1991;10:105–115. doi: 10.1016/0882-4010(91)90071-h. [DOI] [PubMed] [Google Scholar]
- 9.Majde JA, Guha-Thakurta N, Chen Z, Bredow S, Krueger JM. Spontaneous release of stable viral double-stranded RNA into the extracellular medium by influenza virus-infected MDCK epithelial cells: Implications for the viral acute phase response. Arch Virol. 1998;143:2371–2380. doi: 10.1007/s007050050467. [DOI] [PubMed] [Google Scholar]
- 10.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303:1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 11.Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 12.Matsumoto M, Kikkawa S, Kohase M, Miyake K, Seya T. Establishment of a monoclonal antibody against human Toll-like receptor 3 that blocks double-stranded RNA-mediated signaling. Biochem Biophys Res Commun. 2002;293:1364–1369. doi: 10.1016/S0006-291X(02)00380-7. [DOI] [PubMed] [Google Scholar]
- 13.Scumpia PO, Kelly KM, Reeves WH, Stevens BR. Double-stranded RNA signals antiviral and inflammatory programs and dysfunctional glutamate transport in TLR3-expressing astrocytes. Glia. 2005;52:153–162. doi: 10.1002/glia.20234. [DOI] [PubMed] [Google Scholar]
- 14.Leyva-Grado VH, Churchill L, Harding J, Krueger JM. The olfactory nerve has a role in the body temperature and brain cytokine responses to influenza virus. Brain Behav Immun. 2010;24:281–288. doi: 10.1016/j.bbi.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwasaki T, Itamura S, Nishimura H, Sato Y, Tashiro M, Hashikawa T, Kurata T. Productive infection in the murine central nervous system with avian influenza virus A (H5N1) after intranasal inoculation. Acta Neuropathol. 2004;108:485–492. doi: 10.1007/s00401-004-0909-0. [DOI] [PubMed] [Google Scholar]
- 16.Matsuda K, Park CH, Sunden Y, Kimura T, Ochiai K, Kida H, Umemura T. Vagus nerve is one route of transneural invasion for intranasally inoculated infuenza A virus in mice. Vet Pathol. 2004;41:101–107. doi: 10.1354/vp.41-2-101. [DOI] [PubMed] [Google Scholar]
- 17.Shinya K, Shimada A, Ito T, Otsuki K, Morita T, Tanaka H, Takada A, Kida H, Umemura T. Avian influenza virus intranasally inoculated infects the central nervous system of mice through the general visceral afferent nerve. Arch Virol. 2000;145:187–195. doi: 10.1007/s007050050016. [DOI] [PubMed] [Google Scholar]
- 18.Chen L, Duricka D, Nelson S, Mukherjee S, Bohnet S, Taishi P, Majde JA, Krueger JM. Influenza virus-induced sleep responses in mice with targeted disruptions in neuronal or inducible nitric oxide synthases. J Appl Physiol. 2004;97:17–28. doi: 10.1152/japplphysiol.01355.2003. [DOI] [PubMed] [Google Scholar]
- 19.Hodgson NR, Bohnet SG, Majde JA, Krueger JM. Influenza virus pathophysiology and brain invasion in mice with functional and dysfunctional Mx1 genes. Brain Behav Immun. 2012;26:83–89. doi: 10.1016/j.bbi.2011.07.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zielinski MR, Taishi P, Clinton JM, Krueger JM. 5'-Ectonucleotidase-knockout mice lack non-REM sleep responses to sleep deprivation. Eur J Neurosci. 2012;35:1789–1798. doi: 10.1111/j.1460-9568.2012.08112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bachmann MF, Ecabert B, Kopf M. Influenza virus: A novel method to assess viral and neutralizing antibody tiers in vitro. J Immunol Meth. 1999;225:105–111. doi: 10.1016/s0022-1759(99)00034-4. [DOI] [PubMed] [Google Scholar]
- 22.Taishi P, Churchill L, De A, Obal F, Jr, Krueger JM. Cytokine mRNA induction by interleukin-1β or tumor necrosis factor α in vitro and in vivo. Brain Res. 2008;1226:89–98. doi: 10.1016/j.brainres.2008.05.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ambrose CS, Levin MJ, Belshe RB. The relative efficacy of trivalent live attenuated and inactivated influenza vaccines in children and adults. Influenza Other Respi Viruses. 2011;5:67–75. doi: 10.1111/j.1750-2659.2010.00183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun K, Ye J, Perez DR, Metzger DW. Seasonal FluMist Vaccination Induces Cross-Reactive T Cell Immunity against H1N1 (2009) Influenza and Secondary Bacterial Infections. J Immunol. 2011;186:987–993. doi: 10.4049/jimmunol.1002664. [DOI] [PubMed] [Google Scholar]
- 25.Jang H, Boltz D, Sturm-Ramirez K, Shepherd KR, Jiao Y, Webster R, Smeyne RJ. Highly pathogenic H5N1 influenza virus can enter CNS. Proc Natl Acad Sci USA. 2009;106:14063–14068. doi: 10.1073/pnas.0900096106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mori I, Komatsu T, Takeuchi K, Nakakuki K, Sudo M, Kimura Y. Parainfluenza virus type 1 infects olfactory neurons and establishes long-term persistence in the nerve tissue. J Gen Virol. 1995;76:1251–1254. doi: 10.1099/0022-1317-76-5-1251. [DOI] [PubMed] [Google Scholar]
- 27.Malmgaard L. Induction and regulation of IFNs during viral infections. J Interferon Cytokine Res. 2004;24:439–454. doi: 10.1089/1079990041689665. [DOI] [PubMed] [Google Scholar]
- 28.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 29.Majde JA, Kapas L, Bohnet SG, De A, Krueger JM. Attenuation of the influenza virus sickness behavior in mice deficient with Toll-like receptor 3. Brain Behav Immun. 2010;24:306–315. doi: 10.1016/j.bbi.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heer AK, Shamshiev A, Donda A, Uematsu S, Akira S, Kopf M, Marsland BJ. TLR signaling fine-tunes anti-influenza B cell responses without regulating effector T cell responses. J Immunol. 2007;178:2182–2191. doi: 10.4049/jimmunol.178.4.2182. [DOI] [PubMed] [Google Scholar]
- 31.Koyama S, Ishii KJ, Kumar H, Tanimoto T, Coban C, Uematsu S, Kawai T, Akira S. Differential role of TLR- and RLR-signaling in the immune responses to influenza A virus infection and vaccination. J Immunol. 2007;179:4711–4720. doi: 10.4049/jimmunol.179.7.4711. [DOI] [PubMed] [Google Scholar]
- 32.Butchi NB, Pourciau S, Du M, Morgan TW, Peterson KE. Analysis of the neuroinflammatory response to TLR7 stimulation in the brain: comparison of multiple TLR7 and/or TLR8 agonists. J Immunol. 2008;180:7604–7612. doi: 10.4049/jimmunol.180.11.7604. [DOI] [PubMed] [Google Scholar]
- 33.Eskildsen S, Justesen J, Shierup MH, Hartmann R. Characterization of the 2“-5-”oligoadenylate synthetase ubiquitin-like family. Nucleic Acids Res. 2003;31:3166–3173. doi: 10.1093/nar/gkg427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haller O, Arnheiter H, Lindenmann J, Gresser I. Host gene influences sensitivity to interferon action selectively for influenza virus. Nature. 1980;283:660–662. doi: 10.1038/283660a0. [DOI] [PubMed] [Google Scholar]
- 35.Dantzer R, Kelley KW. Twenty years of research on cytokine-induced sickness behavior. Brain Behav Immun. 2007;21:153–160. doi: 10.1016/j.bbi.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krueger JM, Majde JA. In: Immunity: The wiley guide to sleep medicine: a concise reference and review. Lee-Chiong, editor. New York: John Wiley & Sons, Inc; 2009. pp. 223–226. [Google Scholar]
- 37.Majde JA, Krueger JM. Links between the innate immune system and sleep. J Allergy Clin Immunol. 2005;116:1188–1198. doi: 10.1016/j.jaci.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 38.Alt JA, Obal F, Jr, Traynor TR, Gardi J, Majde JA, Krueger JM. Alterations in EEG activity and sleep after influenza viral infection in GHRH receptor-deficient mice. J Appl Physiol. 2003;95:460–468. doi: 10.1152/japplphysiol.01190.2002. [DOI] [PubMed] [Google Scholar]
- 39.Alt JA, Bohnet S, Taishi P, Duricka D, Obal F, Jr, Traynor T, Majde JA, Krueger JM. Influenza virus-induced glucocorticoid and hypothalamic and lung cytokine mRNA responses in dwarf lit/lit mice. Brain Behav Immun. 2007;21:60–67. doi: 10.1016/j.bbi.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 40.Taishi P, De A, Alt J, Gardi J, Obal F, Jr, Krueger JM. Interleukin-1beta stimulates growth hormone-releasing hormone receptor mRNA expression in the rat hypothalamus in vitro and in vivo. J Neuroendocrinol. 2004;16:113–118. doi: 10.1111/j.0953-8194.2004.01138.x. [DOI] [PubMed] [Google Scholar]
- 41.Obál F, Jr, Fang J, Payne LC, Krueger JM. Growth-hormone-releasing hormone mediates the sleep-promoting activity of interleukin-1 in rats. Neuroendocrinology. 1995;61:559–565. doi: 10.1159/000126880. [DOI] [PubMed] [Google Scholar]
- 42.Waffarn EE, Baumgarth N. Protective B cell responses to flu--no fluke! J Immunol. 2011;186:3823–3829. doi: 10.4049/jimmunol.1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joo HM, He Y, Sangster MY. Broad dispersion and lung localization of virus-specific memory B cells induced by influenza pneumonia. Proc Natl Acad Sci USA. 2008;105:3485–3490. doi: 10.1073/pnas.0800003105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kapas L, Bohnet SG, Traynor TR, Majde JA, Szentirmai E, Magrath P, Taishi P, Krueger JM. Spontaneous and influenza virus-induced sleep are altered in TNF-double-receptor deficient mice. J Appl Physiol. 2008;105:1187–1198. doi: 10.1152/japplphysiol.90388.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lau YF, Santos C, Torres-Velez FJ, Subbarao K. The magnitude of local immunity in the lungs of mice induced by live attenuated influenza vaccines is determined by local viral replication and induction of cytokines. J Viro. 2010;85:76–85. doi: 10.1128/JVI.01564-10. l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clements ML, Betts RF, Murphy BR. Advantage of live attenuated cold-adapted influenza A virus over inactivated vaccine for A/Washington/80 (H3N2) wild-type virus infection. Lancet. 1984;1:705–708. doi: 10.1016/s0140-6736(84)92222-0. [DOI] [PubMed] [Google Scholar]