Abstract
Heart failure (HF) is a common cause of morbidity and mortality in congenital heart disease (CHD), with rising prevalence due to improved interventional and surgical treatment options and outcomes. The interplay between genetic factors and acquired postnatal factors in CHD might play a major role in the progression to HF. In this review, we propose three putative routes which lead to HF in CHD: rare monogenic entities that cause both CHD and HF, severe CHD lesions in which acquired hemodynamic effects of CHD or surgery result in HF, and most commonly a combined effect of complex genetics in overlapping pathways and acquired stressors caused by the primary lesion. Novel sequencing technologies have allowed a better understanding of the genomic architecture of HF and CHD with multiple overlapping pathways and hypothesized mechanisms through which pathway deficiencies that led to CHD can increase the risk of progression to HF. Through next-generation sequencing technologies and drug screening using patient-specific induced pluripotent stem cells, the coming years hold a tremendous promise of better understanding this confluence and yielding effective therapeutics.
Keywords: Heart Failure, Congenital Heart Disease, Genomics, Adult
Introduction
Congenital heart disease encompasses all malformations of the heart that occur in utero and exist at birth and is estimated to affect at least 8.1 per thousand live births.1 The prevalence of complex and hemodynamically significant lesions, which result in early deaths if not addressed via surgical or percutaneous methods, is 2.3 per thousand infants.2 The past three decades have witnessed marked progress in pediatric cardiology and cardiac surgery, allowing patients to survive to adulthood. As a result, the prevalence of an adult population with CHD (ACHD) outnumbers the pediatric population with CHD in the USA.2,3 An analysis of 71,686 patients with CHD between 1987 and 2005, revealed that infant CHD mortality has decreased dramatically, but with a shifting mortality burden towards adulthood.4 Heart failure (HF) is the major problem in ACHD, as nearly one quarter develops heart failure at 30 years.5 The events inciting HF in ACHD patients are different than in other forms of HF, however there are limited data available to guide their management. Consensus guidelines for diagnosis and treatment of HF are often used for HF ACHD patients because adequate controlled data are lacking for this sub-population.6
Dramatic advances in cardiac genetics have paralleled improvements in the treatment of CHD. Over the past two decades, 50 to 70% of the genetic causes of inherited cardiomyopathies were established through increasingly powerful methods of DNA analysis. Discovery of the genetics of nonsyndromic congenital heart disease lags behind the inherited cardiomyopathies. Sequencing initially identified mutations in transcription factor genes involved in heart development in familial cases, and the large majority of sporadic CHD have remained unexplained. This is likely due to the complexity of the underlying genetic causes; multiple mutations may increase the risk of CHD, in an additive fashion, or manifest in the presence of environmental factors and result in cardiac malformation. Current research emphasizes the important role of gene-environment interactions and epigenetics in CHD.
As the molecular signatures of CHD and HF are being characterized with the use of available technology, it is increasingly appreciated that at least some disease pathways are shared. Determining the molecular basis of HF in ACHD will play a crucial role in developing treatment strategies for this growing population. This review will summarize the limited current knowledge of the genetics of HF in ACHD, and discuss how innovation may discover therapies for this population.
Routes to Heart Failure in CHD Patients
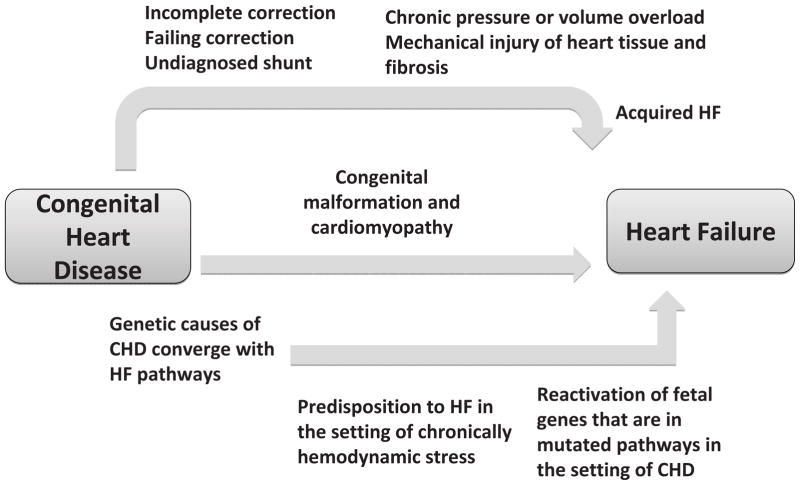
The progression to HF in patients with CHD involves proven and hypothesized mechanisms, which we classify into three routes (Figure 1). The first route is purely acquired and mechanical with no genetic element. This includes incomplete or palliative correction of a lesion leading to a chronic state of hemodynamic stress and subsequent heart failure.6 The probability of heart failure in CHD lesions such as tetralogy of Fallot (TOF), and transposition of the great arteries (TGA) can be as high as 80% at 50 years of age, while it is around 20–30% for isolated valvular disease or defects that result in left-to-right shunt.6 Additional myocardial insults can complicate surgery, including injury to the myocardium, coronary arteries, and conduction system.6 Post surgical conduction disease may require permanent ventricular pacing, which can lead to progressive contractile dysfunction.6 Since these insults often occur in the first years of life, the effects of altered hemodynamics or tissue injury accumulate over years, resulting in early development of HF.
Figure 1.
Schematic of the hypothesized mechanisms linking congenital heart disease with heart failure. We propose three putative routes which lead to HF in ACHD: rare monogenic entities that cause both CHD and HF (middle arrow), severe CHD lesions in which acquired hemodynamic effects of CHD or surgery result in HF (top arrow), and most commonly a combined effect of complex genetics in overlapping pathways and acquired stressors caused by the lesion (bottom arrow).
Although the increased prevalence of HF in ACHD is primarily viewed as a result of a volume or pressure overload, whereby the starting point is an abnormal heart, an independent genetic component is also present. This second route depicted in Figure 1 delineates a purely genetic component that causes both cardiac malformation and a cardiomyopathy that result in HF, unrelated to hemodynamic stress. Many of the pathways involved in cardiac development in utero are also involved in myocardial structure and stability. Therefore, it is not surprising that certain molecular perturbations can cause both a cardiac defect at birth, and a cardiomyopathy that can present later in life, often in childhood. As described in greater detail in following sections, Noonan Syndrome (NS) is the second most frequent syndromic form of CHD and can cause both CHD and cardiomyopathy. Similarly, another example of congenital cardiomyopathy is left ventricular non-compaction (LVNC), a heterogeneous disorder that often results in HF. LVNC has been associated with CHD including atrial and ventricular septal defects, Ebstein anomaly and outlflow tract lesions, and is caused by genes such as MYH7 and the transcription factor NKX2-5 among others.7–10
In a Dutch registry of 10,808 CHD patients followed for 21 years (median), the incidence of HF-admissions was 1.2 per 1000 patient years and median age at first HF-admission was 46.7 years. This incidence of HF admissions in ACHD patients is more than ten times what is reported in an age matched cohort without CHD (0.1 per 1000 person years).11 The true prevalence of HF is thought to be higher as not all HF events in CHD patients resulted in hospital admission. Low prevalence CHD lesions such as TGA and TOF that result in HF via markedly abnormal hemodynamics do not solely account for the epidemic of HF in CHD. Moreover, gene mutations that cause both cardiomyopathy and CHD are extremely rare. Therefore, it is unlikely that these two routes alone explain the high incidence of HF in CHD. This suggests additional mechanisms through which CHD patients can develop HF. We present this as a third route in figure 1, which is a combination of congenital genetic risk and acquired hemodynamic stressors. There is significant overlap in the molecular pathways that result in CHD during development and those that are responsible for the integrity of the postnatal myocardium. This overlap suggests that molecular perturbations that result in abnormal cardiac development can increase the risk for heart failure in adulthood, especially in the presence of chronically perturbed hemodynamics. Heart failure also involves the reactivation of many fetal genes. One can expect the reactivation of a mutated pathway to exacerbate the progression to HF in the setting of CHD. With the current era of high throughput DNA, RNA, protein and metabolic analysis this hypothesis can be studied using a systems biology approach to investigate the development of HF in CHD patients, particularly those with mild phenotypes whose progression to HF might not be justified by the degree of volume or pressure overload caused by the lesion.
Monogenic Causes of Heart Failure in Cardiomyopathy and CHD
The genetic basis of HCM was first described in 1990.12,13 Following this discovery, the principal genetic causes of hypertrophic, dilated and arrythmogenic cardiomyopathies have been identified. Depending on phenotype, a disease causing mutation can be found in 30–70% of patients. Culprit genes encode structural proteins such as those that form the sarcomere, cellular cytoskeleton, or ion channels.14 (Table 1) Genetic testing for inherited cardiomyopathies is clinically available and improves the efficiency of family screening, enabling early diagnosis and timely clinical management in at-risk individuals.15
Table 1.
Comparison of differences and similarities between congenital heart disease and cardiomyopathy
| Congenital Heart Disease | Cardiomyopathy | |
|---|---|---|
| Differences | ||
| Phenotype | Wide spectrum of cardiac malformations including the valves, septa, and great vessels | Dilation or hypertrophy of the myocardium +/− disease of the conduction system; results in HF or SCD |
| Expression | Present at birth by definition | Typical disease onset is in adolesence or adulthood, but can express at any age |
| Genetics | LoF mutations in transcription factor genes and other signaling molecules that disturb molecular pathways during cardiac development | Mutations in structural genes involving proteins of the sarcomere, cellular cytoskeleton, or ion channels |
| Population Attributable Risk of Genetic Causes | 5–10% | 50–70% |
| Clinical Genetic Testing | Not recommended except in rare syndromic cases | Recommended in most familial cases |
| Similarities | ||
| Phenotype | Several syndromes with CHD and congenital CMP; Co-occurrence of LVNC with CHD | |
| Expression | Congenital cardiomyopathies occurring in the setting of CHD | |
| Genetics | Multiple shared pathways between heart development and myocardial disease; Few genes that cause both CHD and congenital CMP; Multiple genes that cause CHD have altered expression in the setting of heart failure | |
HF Heart Failure, SCD Sudden Cardiac Death, LoF Loss of Function, CHD Congenital Heart Disease, CMP Cardiomyopathy, LVNC Left Ventricular Non-Compactum
Alternatively, the clinical application of CHD genetics remains limited. Monogenic causes of CHD are estimated to account for only 5–10% of disease, usually related to loss of function mutations in transcription factor genes and other signaling molecules that disturb molecular pathways during cardiac development.16 (Table 1) Monogenic causes have predominantly been identified in syndromic cases where CHD occurs with non-cardiac congenital malformations.16 In only a minority of presentations is genetic testing available and useful. Genetic testing is available for most syndromic cases. To name few examples, Holt-Oram Syndrome, which causes CHD and upper limb malformations, is caused by mutations in TBX5 and SALL4 genes.16 Alagille Syndrome causing CHD along with liver disease, is caused by mutations in the JAG1 or NOTCH2 genes.16 Among the non-syndromic genes, GATA4 and Nkx2-5 are two well-established genes in familial CHD,16 and are also available for clinical genetic testing. The genetic cause of non-syndromic CHD is less well understood, and clinical genetic testing is not routinely advised for such patients.17,18 However, there are several genetically characterized syndromes where CHD and HF are common morbidities; including Noonan, Williams-Beuren and 22.q11.2 deletion syndromes.
Noonan Syndrome (NS), in which HCM is the second most common cardiac manifestation, is a representative example. NS results from abnormal RAS-MAPK signaling, which normally regulates cellular proliferation, differentiation, and survival.19 Mutations in different components of the RAS-MAPK pathway have been described, including PTPN11, KRAS, SOS1, NRAS, RAF1, SHOC2 and CBL;19,20 which typically result in constitutive activation.19 The 20% of NS patients who develop HCM present with a wide phenotypic spectrum, with approximately 25% dying of heart failure in the first year of life.19 The CHD manifestations of NS are pulmonary stenosis, present in more than 50% of cases and secundum atrial septal defect in 6–10%. Other less common malformations include ventricular septal defect, atrioventricular canal, aortic stenosis and coarctation.19 Common extra-cardiac manifestations of NS include short stature and facial dysmorphism.20 Adults with NS suffer from cardiac complications of the syndrome that result frequently in HF, whether due to the cardiomyopathy, the associated valvular disease, or both.21 The limited natural history data in NS suggests that cardiovascular complications are a major cause of mortality. 22 Noonan syndrome with Multiple Lentigenes (formerly known as LEOPARD syndrome) is similar to NS, is caused by loss of function mutations inthe PTPN11 gene and has a high frequency (>70%) of associated HCM.23 Additional manifestations of this rare syndrome include multiple lentigines and sensorineural deafness.24
Williams-Beuren Syndrome (WBS) is caused by a microdeletion on chromosome 7q11.23, a region that includes 26 to 28 genes.25 The syndrome is characterized in most cases by supravalvular aortic stenosis (SVAS), mental retardation, and distinctive facial features.25 Loss of one allele of the ELN gene, which encodes elastin and resides on the 7q11.23 locus, causes familial SVAS without the other manifestations of WBS.25 Severe SVAS in children leads to cardiac hypertrophy and heart failure unless corrected surgically. Adults with SVAS suffer from high risk of cardiovascular complications; of 113 patients with median 6 year follow up, 7.3% developed new-onset heart failure26
The 22q11.2 microdeletion syndrome (also known as DiGeorge or velocardiofacial syndrome) has a diverse phenotypic spectrum. Common manifestations include ventricular outflow tract defects, facial dysmorhpism, hypocalcemia and immunodeficiency.21 The syndrome is caused by a 1.5 to 3 Mb hemizygous deletion on chromosome 22q11.2 and most clinical manifestations, especially the cardiac malformations, are explained by the loss of one allele of the TBX1 gene.21 The most common outflow tract defects seen in the syndrome are TOF, type B interrupted aortic arch (IAA), and truncus arteriosus (TA). Microdeletion in the same region may cause isolated CHD without obvious extra-cardiac syndromic manifestations.21 Complications of these malformations in adulthood are common and include re-coarctation of an IAA that was previously surgically reconstructed, residual or acquired obstruction of the left ventricular outflow tract, or arrhythmias that arise after repair of TOF.21 Genetic testing for 22q11.2 deletion is readily available and recommended for CHD lesions typical of the 22q11.2 syndrome, including aortic arch abnormalities.27 Genetic diagnosis may have clinical implications, as in patients with isolated TOF and 22q11.2 deletion, there is evidence for an increased prevalence of aortic root dilation.28
Several other genes have been associated with familial forms of non-syndromic CHD. Evidence is most robust for loss of function (LOF) mutations in GATA4, TBX5, and NKX2-5, which are transcription factors expressed in the developing heart,16 although other transcription factors and signaling molecules have been implicated. The transcripts of these three genes form a complex that regulates downstream targets involved in the regulation of cardiac development. LOF mutations in these genes can be a result of nonsense or frameshift mutations that cause a truncated protein or due to CNVs, which result in loss or gain of a copy of a gene. For example, at least 10% of TOF are caused by de novo CNVs.29 In addition to identifying the role of CNVs in CHD, the advent of exome sequencing is starting to yield a better understanding of novel molecular causes of CHD. A recent study showed that de novo LOF mutations in histone-modifying genes contribute to 10% of severe CHD, highlighting a unique role for epigenetic mechanisms involved in the molecular pathophysiology of CHD.18 Identification of CHD as part of a genetic syndrome would allow prediction of not only cardiac but also extra-cardiac outcomes as well as interventions to modify them e.g. aggressive monitoring and treatment of hypertension and of coronary artery lesions in WBS that may contribute to cardiac hypertrophy and dysfunction. This may help personalize the care of the adult CHD patient at risk for HF.
Overlapping Pathways in CHD and HF Genetics
While the genetics of CHD and HF in humans has been studied separately, with the few directly shared disease genes described to date, there is significant evidence from model organisms and in-vitro studies that similar molecular pathways are involved (Table 2). Several genes involved in embryonic cardiac development continue to be expressed in the adult heart where they play a role in maintaining cardiomyocyte survival and integrity. GATA4 and GATA6 are two transcription factor genes that are mutated in familial cases of CHD.16 In addition to their role in cardiac development, they are potent activators of cardiac promoters such as atrial natriuretic factor (ANF) and B-type natriuretic peptide (BNP) in the adult heart.30 They also regulate the transcription of cardiac sarcomere genes, namely alpha- and beta-myosin heavy chain.30 Moreover, GATA4 is a survival factor for adult cardiomycytes as it also regulates the antiapoptotic gene BCL-X.31 Myocyte apoptosis is a major element in the pathophysiology of heart failure, and it has been established that genetic or pharmacologic enhancement of GATA4 can prevent cardiomyocyte toxicity and drug-induced cardiotoxicity such as with doxorubicin treatment, where GATA4 depletion is an early event.31 This data collectively suggest that GATA4 is a potential target in heart failure treatment, and that CHD patients with LOF mutations in their GATA4 or GATA6 genes might have increased apoptosis of their cardiomyocytes predisposing them to heart failure in the setting of other insults such as volume overload caused by CHD. GATA4 also plays a role in calcium-calcineurin signaling responsible for cardiac hypertrophy and heart failure.32 Calcineurin dephosphorylates Nuclear Factor of Activated T Cells (NFATC), which results in its translocation to the nucleus where it binds GATA4 and activates transcription of a hypertrophic gene program.32 NFATC transcription factor is essential for valve formation in mice,33 and mutations in NFATC1 have been associated with CHD in humans.34,35 Recent murine data also suggests an important role for NFATC transcription factors in the cardiomyocyte response to stress.36 Calcineurin also activates CaMK and results in phosphorylation of histone deacetylase (HDAC), a nuclear protein causing its dissociation from the transcription factor Myocyte Enhancer Factor (MEF2) and its externalization from the nucleus.32 MEF2 transcription factors are the second effectors of calcineurin signaling, and also play critical roles in both cardiac development and normal postnatal myocardial survival.37 This calcium-dependent signaling pathway constitutes an excellent potential target in heart failure and/or cardiac hypertrophy.32
Table 2.
Canonical Pathways from IPA Analysis of CHD Candidate Genes that have major roles in HF
| Pathway | Candidate Genes in CHD |
|---|---|
| Wnt/β-catenin Signaling | SOX2,GJA1,CREBBP |
| HIF1α Signaling | CREBBP,HRAS,NAA10 |
| Cardiac hypertrophy signaling | NKX2-5,ADCY2,SOS1,CREBBP,HRAS,GATA4 |
| Noonan pathway | SOS1, PTPN11 |
| BMP Signaling Pathway | NKX2-5,SOS1,CREBBP,HRAS,PITX2 |
| TGF-β Signaling | NKX2-5,SOS1,CREBBP,HRAS,PITX2 |
| JAK/Stat Signaling | PTPN11,SOS1,HRAS |
| RAR Activation | NSD1,ADCY2,CREBBP,PML,CITED2 |
| PPARα/RXRα Activation | ADCY2,SOS1,CREBBP,HRAS,MED12 |
Epigenetics, particularly the role of molecules such as HDAC responsible for DNA methylation and histone modifications represents another shared pathway in CHD and HF. The importance of increased DNA methylation of genes in cardiomyopathy hearts as compared to controls has been described, 38,39 and recently, a large exome sequencing project identified that de novo truncating mutations in similar histone-modifying genes is seen in ~10% of sporadic CHD.18 Heart failure progression involves a pattern of gene expression including re-expression of fetal genes involved in cardiac development, increased expression of genes encoding extracellular matrix proteins, altered calcium signaling, and histone modification.40 These processes involve genes that can be mutated in the setting of CHD. Therefore, one can hypothesize that pre-existing defects in these pathways can potentially worsen the progression to heart failure in CHD patients. Addressing this hypothesis is a methodologic challenge that has only recently become possible with the advent of technologies such as next-generation sequencing. While this review does not intend to explore all potential overlapping molecular pathways in heart failure and CHD, the above examples of epigenetics, transcription factors, and calcium-dependent signaling serve as putative examples. Using IPA (Ingenuity® Systems, www.ingenuity.com), a software toolkit to model and analyze complex biological systems, we performed a rapid analysis of canonical pathways on a set of 146 literature-curated genes known to be involved in CHD in humans or in animal models. This yielded multiple pathways that are known to be involved in heart failure (Table 2). Genes involved in Hypoxia-Inducible Factor (HIF)1-alpha signaling are activated in cardiac hypertrophy,41 while their down-regulation is associated with the development of TGA in mice.42 Similarly, other signaling pathways such as Wnt/β-catenin, TGF-β, and JAK/Stat are involved in heart failure and at the same time harbor CHD candidate genes. Understanding the genomics of HF has crossed many milestones, but most of the work has been focused on identifying the molecular signature of ischemic and non-ischemic cardiomyopathies and trying to target the ensuing HF.43,44 CHD patients with HF constitute a smaller niche that has not been interrogated. Studies looking at gene expression profiles through RNA sequencing of human hearts with CHD and HF and corresponding controls will be extremely valuable to understand the unique molecular causes of HF in CHD patients.
The Future of Personalized Treatment of HF in ACHD
Current treatment of HF in ACHD involves therapies which limit neurohumoral activation, and are based on extrapolation from adults with HF unrelated to CHD. While using beta-adrenergic blockers, aldosterone antagonism, and angiotensin inhibition has intuitive appeal, there are limited data to support the use of these drugs in the ACHD population with HF. Indeed, emerging data challenges the notion that we can extrapolate conventional HF therapies to ACHD patients with HF. For example, a recent trial of valsartan in patients with a systemic right ventricle failed to show any beneficial effect on the primary end point of right ventricular ejection fraction, and most secondary end points.45 This trial suggests that the ACHD population might have unique genetic and acquired factors that would make medications used in common causes of HF ineffective. Pharmacogenomic studies have established that SNPs in adrenergic receptors could modify the response to beta-blockers.46 Similarly in CHD patients, SNPs in HF pathways targeted by pharmacotherapy can potentially result in resistance to drug therapy. Such studies will be needed for CHD patients to have better tailored therapies. In one successful example, polymorphisms in the renin-angiotensisn-aldosterone system (RAAS) genes were used to identify a subgroup of high-risk patients with single ventricle that do not show reverse remodeling after staged surgery.47 This high-risk subset may benefit from earlier volume unloading surgery before injury becomes irreversible. A study in surgically repaired TOF patients identified variations in HIF1-alpha genes were associated with RV dilation and dysfunction during follow-up likely related to maladaptive response to hypoxia and hemodynamic load.48 It raised the possibility that earlier repair of TOF may limit hypoxic injury of the myocardium and promote better long-term adaptation. These studies show the power of genomics in guiding clinical decisions including surgical decisions. These studies need to be done in adult CHD patients to determine if the effect of genetic variations on adverse ventricular remodeling persists into adulthood.
Another promising aspect of how genomics can help tailor therapy in HF in ACHD is targeted therapy of particular pathways known to be involved in the progression to heart failure. While it seems early to discuss therapeutics because most of the complex genetic aspects of HF in CHD remains incompletely understood, there has been at least one example with inhibition of the pathways that leads to HCM in NS, where a clinical trial using a MEK1 inhibitor is underway (www.clinicaltrials.gov NCT01556568). Once other pathways are clearly defined, similar targeted therapy for the prevention of HF in other forms of ACHD may follow. Progress towards this goal has been enabled by high throughput sequencing technologies and with the availability of patient-specific induced pluripotent stem cells (iPS) from patients with cardiomyopathies where drug specific cardiotoxicity was demonstrated at the single-cell level.49 These cells could be used for rapid screening of cardioactive drugs with the patient’s genetic background, an approach being assesed in primary cardiomyopathy.50,51
Conclusions
Heart failure is a major cause of morbidity and mortality in ACHD and the current approach for its treatment is rarely evidence-based. Rare monogenic causes of CHD and HF include Noonan Syndrome, Williams-Beuren Syndrome, and 22q11 Syndrome. Overlapping molecular pathways between CHD and HF exist, and their interrogation will pave the way for a better understanding of the development of HD in CHD. HF in ACHD is most commonly a result of a confluence between inherited complex genetic factors and acquired stressors caused by the defect. Current technologies such as next-generation sequencing and iPS will contribute to the understanding of this confluence and pave the way for novel therapeutics.
Figure 2.
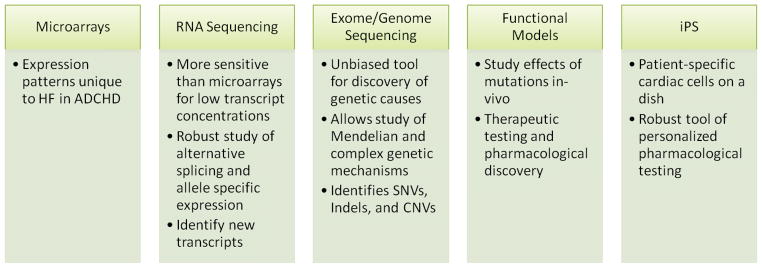
Current genomic technologies and proposed mechanisms of contribution to the understanding of HF in ADCHD.
Key Points.
Heart failure is a major cause of morbidity and mortality in ACHD and the current approach for its treatment is rarely evidence-based.
Rare monogenic causes of CHD and HF include Noonan Syndrome, Williams-Beuren Syndrome, and 22q11 Syndrome.
Overlapping molecular pathways between CHD and HF exist, and their interrogation will pave the way for a better understanding of the development of HD in CHD.
HF in ACHD is most commonly a result of a confluence between inherited complex genetic factors and acquired stressors caused by the defect. C
urrent technologies such as next-generation sequencing and iPS will contribute to the understanding of this confluence and pave the way for novel therapeutics.
Footnotes
The authors have no conflicts of interest to disclose
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Montana E, Khoury MJ, Cragan JD, Sharma S, Dhar P, Fyfe D. Trends and outcomes after prenatal diagnosis of congenital cardiac malformations by fetal echocardiography in a well defined birth population, Atlanta, Georgia, 1990–1994. Journal of the American College of Cardiology. 1996 Dec;28(7):1805–1809. doi: 10.1016/S0735-1097(96)00381-6. [DOI] [PubMed] [Google Scholar]
- 2.Webb CL, Jenkins KJ, Karpawich PP, et al. Collaborative care for adults with congenital heart disease. Circulation. 2002 May 14;105(19):2318–2323. doi: 10.1161/01.cir.0000017557.24261.a7. [DOI] [PubMed] [Google Scholar]
- 3.Greutmann M, Tobler D. Changing epidemiology and mortality in adult congenital heart disease: looking into the future. Future cardiology. 2012 Mar;8(2):171–177. doi: 10.2217/fca.12.6. [DOI] [PubMed] [Google Scholar]
- 4.Khairy P, Ionescu-Ittu R, Mackie AS, Abrahamowicz M, Pilote L, Marelli AJ. Changing mortality in congenital heart disease. Journal of the American College of Cardiology. 2010 Sep 28;56(14):1149–1157. doi: 10.1016/j.jacc.2010.03.085. [DOI] [PubMed] [Google Scholar]
- 5.Norozi K, Wessel A, Alpers V, et al. Incidence and risk distribution of heart failure in adolescents and adults with congenital heart disease after cardiac surgery. The American journal of cardiology. 2006 Apr 15;97(8):1238–1243. doi: 10.1016/j.amjcard.2005.10.065. [DOI] [PubMed] [Google Scholar]
- 6.Parekh DR. A review of heart failure in adults with congenital heart disease. Methodist DeBakey cardiovascular journal. 2011 Apr-Jun;7(2):26–32. doi: 10.14797/mdcj-7-2-26. [DOI] [PubMed] [Google Scholar]
- 7.Cavusoglu Y, Ata N, Timuralp B, et al. Noncompaction of the ventricular myocardium: report of two cases with bicuspid aortic valve demonstrating poor prognosis and with prominent right ventricular involvement. Echocardiography. 2003 May;20(4):379–383. doi: 10.1046/j.1540-8175.2003.03045.x. [DOI] [PubMed] [Google Scholar]
- 8.Ichida F, Tsubata S, Bowles KR, et al. Novel gene mutations in patients with left ventricular noncompaction or Barth syndrome. Circulation. 2001 Mar 6;103(9):1256–1263. doi: 10.1161/01.cir.103.9.1256. [DOI] [PubMed] [Google Scholar]
- 9.Puley G, Siu S, Connelly M, et al. Arrhythmia and survival in patients >18 years of age after the mustard procedure for complete transposition of the great arteries. The American journal of cardiology. 1999 Apr 1;83(7):1080–1084. doi: 10.1016/s0002-9149(99)00019-3. [DOI] [PubMed] [Google Scholar]
- 10.Wessels MW, De Graaf BM, Cohen-Overbeek TE, et al. A new syndrome with noncompaction cardiomyopathy, bradycardia, pulmonary stenosis, atrial septal defect and heterotaxy with suggestive linkage to chromosome 6p. Human genetics. 2008 Jan;122(6):595–603. doi: 10.1007/s00439-007-0436-x. [DOI] [PubMed] [Google Scholar]
- 11.Zomer AC, Vaartjes I, van der Velde ET, et al. Heart failure admissions in adults with congenital heart disease; risk factors and prognosis. International journal of cardiology. 2013 Apr 18; doi: 10.1016/j.ijcard.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 12.Jarcho JA, McKenna W, Pare JA, et al. Mapping a gene for familial hypertrophic cardiomyopathy to chromosome 14q1. The New England journal of medicine. 1989 Nov 16;321(20):1372–1378. doi: 10.1056/NEJM198911163212005. [DOI] [PubMed] [Google Scholar]
- 13.Geisterfer-Lowrance AA, Kass S, Tanigawa G, et al. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 1990 Sep 7;62(5):999–1006. doi: 10.1016/0092-8674(90)90274-i. [DOI] [PubMed] [Google Scholar]
- 14.Teekakirikul P, Kelly MA, Rehm HL, Lakdawala NK, Funke BH. Inherited cardiomyopathies: molecular genetics and clinical genetic testing in the postgenomic era. The Journal of molecular diagnostics: JMD. 2013 Mar;15(2):158–170. doi: 10.1016/j.jmoldx.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Ingles J, McGaughran J, Scuffham PA, Atherton J, Semsarian C. A cost-effectiveness model of genetic testing for the evaluation of families with hypertrophic cardiomyopathy. Heart. 2012 Apr;98(8):625–630. doi: 10.1136/heartjnl-2011-300368. [DOI] [PubMed] [Google Scholar]
- 16.Fahed AC, Gelb BD, Seidman JG, Seidman CE. Genetics of congenital heart disease: the glass half empty. Circulation research. 2013 Feb 15;112(4):707–720. doi: 10.1161/CIRCRESAHA.112.300853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gelb B, Brueckner M, Chung W, et al. The Congenital Heart Disease Genetic Network Study: rationale, design, and early results. Circulation research. 2013 Feb 15;112(4):698–706. doi: 10.1161/CIRCRESAHA.111.300297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zaidi S, Choi M, Wakimoto H, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013 Jun 13;498(7453):220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts AE, Allanson JE, Tartaglia M, Gelb BD. Noonan syndrome. Lancet. 2013 Jan 26;381(9863):333–342. doi: 10.1016/S0140-6736(12)61023-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tartaglia M, Kalidas K, Shaw A, et al. PTPN11 mutations in Noonan syndrome: molecular spectrum, genotype-phenotype correlation, and phenotypic heterogeneity. American journal of human genetics. 2002 Jun;70(6):1555–1563. doi: 10.1086/340847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin AE, Basson CT, Goldmuntz E, et al. Adults with genetic syndromes and cardiovascular abnormalities: clinical history and management. Genetics in medicine: official journal of the American College of Medical Genetics. 2008 Jul;10(7):469–494. doi: 10.1097/GIM.0b013e3181772111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shaw AC, Kalidas K, Crosby AH, Jeffery S, Patton MA. The natural history of Noonan syndrome: a long-term follow-up study. Archives of disease in childhood. 2007 Feb;92(2):128–132. doi: 10.1136/adc.2006.104547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carcavilla A, Santome JL, Pinto I, et al. LEOPARD Syndrome: A Variant of Noonan Syndrome Strongly Associated With Hypertrophic Cardiomyopathy. Revista espanola de cardiologia. 2013 May;66(5):350–356. doi: 10.1016/j.rec.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 24.Gorlin RJ, Anderson RC, Blaw M. Multiple lentigenes syndrome. Am J Dis Child. 1969 Jun;117(6):652–662. doi: 10.1001/archpedi.1969.02100030654006. [DOI] [PubMed] [Google Scholar]
- 25.Pober BR. Williams-Beuren syndrome. The New England journal of medicine. 2010 Jan 21;362(3):239–252. doi: 10.1056/NEJMra0903074. [DOI] [PubMed] [Google Scholar]
- 26.Greutmann M, Tobler D, Sharma NC, et al. Cardiac outcomes in adults with supravalvar aortic stenosis. European heart journal. 2012 Oct;33(19):2442–2450. doi: 10.1093/eurheartj/ehs206. [DOI] [PubMed] [Google Scholar]
- 27.Peyvandi S, Lupo PJ, Garbarini J, et al. 22q11.2 Deletions in Patients with Conotruncal Defects: Data from 1,610 Consecutive Cases. Pediatric cardiology. 2013 Apr 21; doi: 10.1007/s00246-013-0694-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.John AS, Rychik J, Khan M, Yang W, Goldmuntz E. 22q11.2 deletion syndrome as a risk factor for aortic root dilation in tetralogy of Fallot. Cardiology in the young. 2013 Apr 10;:1–8. doi: 10.1017/S1047951113000309. [DOI] [PubMed] [Google Scholar]
- 29.Greenway SC, Pereira AC, Lin JC, et al. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nature genetics. 2009 Aug;41(8):931–935. doi: 10.1038/ng.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charron F, Paradis P, Bronchain O, Nemer G, Nemer M. Cooperative interaction between GATA-4 and GATA-6 regulates myocardial gene expression. Molecular and cellular biology. 1999 Jun;19(6):4355–4365. doi: 10.1128/mcb.19.6.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aries A, Paradis P, Lefebvre C, Schwartz RJ, Nemer M. Essential role of GATA-4 in cell survival and drug-induced cardiotoxicity. Proceedings of the National Academy of Sciences of the United States of America. 2004 May 4;101(18):6975–6980. doi: 10.1073/pnas.0401833101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Epstein JA, Rader DJ, Parmacek MS. Perspective: cardiovascular disease in the postgenomic era--lessons learned and challenges ahead. Endocrinology. 2002 Jun;143(6):2045–2050. doi: 10.1210/endo.143.6.8910. [DOI] [PubMed] [Google Scholar]
- 33.de la Pompa JL, Timmerman LA, Takimoto H, et al. Role of the NF-ATc transcription factor in morphogenesis of cardiac valves and septum. Nature. 1998 Mar 12;392(6672):182–186. doi: 10.1038/32419. [DOI] [PubMed] [Google Scholar]
- 34.Abdul-Sater Z, Yehya A, Beresian J, et al. Two heterozygous mutations in NFATC1 in a patient with Tricuspid Atresia. PloS one. 2012;7(11):e49532. doi: 10.1371/journal.pone.0049532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yehya A, Souki R, Bitar F, Nemer G. Differential duplication of an intronic region in the NFATC1 gene in patients with congenital heart disease. Genome/National Research Council Canada = Genome/Conseil national de recherches Canada. 2006 Sep;49(9):1092–1098. doi: 10.1139/g06-072. [DOI] [PubMed] [Google Scholar]
- 36.Azzam R, Hariri F, El-Hachem N, et al. Regulation of de novo ceramide synthesis: the role of dihydroceramide desaturase and transcriptional factors NFATC and Hand2 in the hypoxic mouse heart. DNA and cell biology. 2013 Jun;32(6):310–319. doi: 10.1089/dna.2013.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Planavila A, Dominguez E, Navarro M, et al. Dilated cardiomyopathy and mitochondrial dysfunction in Sirt1-deficient mice: a role for Sirt1-Mef2 in adult heart. Journal of molecular and cellular cardiology. 2012 Oct;53(4):521–531. doi: 10.1016/j.yjmcc.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 38.Movassagh M, Choy MK, Goddard M, Bennett MR, Down TA, Foo RS. Differential DNA methylation correlates with differential expression of angiogenic factors in human heart failure. PloS one. 2010;5(1):e8564. doi: 10.1371/journal.pone.0008564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nature reviews. Genetics. 2009 Jan;10(1):32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Creemers EE, Wilde AA, Pinto YM. Heart failure: advances through genomics. Nature reviews. Genetics. 2011 May;12(5):357–362. doi: 10.1038/nrg2983. [DOI] [PubMed] [Google Scholar]
- 41.Krishnan J, Suter M, Windak R, et al. Activation of a HIF1alpha-PPARgamma axis underlies the integration of glycolytic and lipid anabolic pathways in pathologic cardiac hypertrophy. Cell metabolism. 2009 Jun;9(6):512–524. doi: 10.1016/j.cmet.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 42.Amati F, Diano L, Campagnolo L, et al. Hif1alpha down-regulation is associated with transposition of great arteries in mice treated with a retinoic acid antagonist. BMC genomics. 2010;11:497. doi: 10.1186/1471-2164-11-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Piran S, Liu P, Morales A, Hershberger RE. Where genome meets phenome: rationale for integrating genetic and protein biomarkers in the diagnosis and management of dilated cardiomyopathy and heart failure. Journal of the American College of Cardiology. 2012 Jul 24;60(4):283–289. doi: 10.1016/j.jacc.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 44.Lin D, Hollander Z, Meredith A, et al. Molecular signatures of end-stage heart failure. Journal of cardiac failure. 2011 Oct;17(10):867–874. doi: 10.1016/j.cardfail.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 45.van der Bom T, Winter MM, Bouma BJ, et al. Effect of valsartan on systemic right ventricular function: a double-blind, randomized, placebo-controlled pilot trial. Circulation. 2013 Jan 22;127(3):322–330. doi: 10.1161/CIRCULATIONAHA.112.135392. [DOI] [PubMed] [Google Scholar]
- 46.Dorn GW., 2nd Adrenergic signaling polymorphisms and their impact on cardiovascular disease. Physiological reviews. 2010 Jul;90(3):1013–1062. doi: 10.1152/physrev.00001.2010. [DOI] [PubMed] [Google Scholar]
- 47.Mital S, Chung WK, Colan SD, et al. Renin-angiotensin-aldosterone genotype influences ventricular remodeling in infants with single ventricle. Circulation. 2011 May 31;123(21):2353–2362. doi: 10.1161/CIRCULATIONAHA.110.004341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeewa A, Manickaraj AK, Mertens L, et al. Genetic determinants of right-ventricular remodeling after tetralogy of Fallot repair. Pediatric research. 2012 Oct;72(4):407–413. doi: 10.1038/pr.2012.95. [DOI] [PubMed] [Google Scholar]
- 49.Mordwinkin NM, Burridge PW, Wu JC. A review of human pluripotent stem cell-derived cardiomyocytes for high-throughput drug discovery, cardiotoxicity screening, and publication standards. Journal of cardiovascular translational research. 2013 Feb;6(1):22–30. doi: 10.1007/s12265-012-9423-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lan F, Lee AS, Liang P, et al. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell stem cell. 2013 Jan 3;12(1):101–113. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liang P, Lan F, Lee AS, et al. Drug screening using a library of human induced pluripotent stem cell-derived cardiomyocytes reveals disease-specific patterns of cardiotoxicity. Circulation. 2013 Apr 23;127(16):1677–1691. doi: 10.1161/CIRCULATIONAHA.113.001883. [DOI] [PMC free article] [PubMed] [Google Scholar]