Abstract
Allelic heterogeneity in disease-causing genes presents a substantial challenge to the translation of genomic variation to clinical practice. Few of the almost 2,000 variants in the cystic fibrosis transmembrane conductance regulator (CFTR) gene have empirical evidence that they cause cystic fibrosis. To address this gap, we collected both genotype and phenotype data for 39,696 cystic fibrosis patients in registries and clinics in North America and Europe. Among these patients, 159 CFTR variants had an allele frequency of ≥0.01%. These variants were evaluated for both clinical severity and functional consequence with 127 (80%) meeting both clinical and functional criteria consistent with disease. Assessment of disease penetrance in 2,188 fathers of cystic fibrosis patients enabled assignment of 12 of the remaining 32 variants as neutral while the other 20 variants remained indeterminate. This study illustrates that sourcing data directly from well-phenotyped subjects can address the gap in our ability to interpret clinically-relevant genomic variation.
The utility of genetic testing for both Mendelian and polygenic disorders is limited by the substantial number of DNA variants of uncertain significance (VUS)1-4. Next-generation sequencing in clinical laboratories will dramatically increase the number of variants of potential medical relevance5. Thus, an ever-widening gap is likely to occur between our ability to identify DNA variation and our ability to interpret its consequence6. One approach to address this gap is to aggregate variants identified by clinical and research laboratories into central repositories7, 8. Observation of the same variant in individuals with the same phenotype supports that the variant may be deleterious. However, physicians request clinical testing for a number of reasons including confirmation or exclusion of a specific diagnosis. Aggregation of variants from testing facilities without robust phenotype and functional annotation can diminish the potential clinical value of repositories9, 10.
A prime example of the challenge of allelic heterogeneity is the gene responsible for cystic fibrosis, the cystic fibrosis transmembrane conductance regulator (CFTR; NM_000492.3). Almost 2,000 variants have been reported in the CFTR coding and flanking sequences, but the disease liability of only a few dozen has been ascertained11. Consequently, sequence analysis of the CFTR gene for diagnostic purposes frequently uncovers VUS. The clinical implications of incomplete annotation of CFTR sequence variation extend well beyond the ~70,000 cystic fibrosis patients worldwide, particularly since CFTR genetic testing is frequently part of newborn screening12-15. Furthermore, population-based carrier screening for cystic fibrosis has become progressively more common with an estimated 1.2 million individuals tested each year in the U.S.16, 17. In cases where one member of a couple is discovered to carry a known cystic fibrosis-causing variant, extensive CFTR analysis is often performed on the other member that reveals VUS18. Finally, the large number of non-experimentally verified disease-associated variants hampers understanding of how structural changes in CFTR lead to dysfunction and produce the cystic fibrosis phenotype. The gap in our understanding of disease versus neutral alleles presents a major challenge in the genomic sequencing era.
A central repository for CFTR variants termed the Cystic Fibrosis Mutation Database (CFMD; http://www.genet.sickkids.on.ca/cftr/app) began in 1990 shortly after CFTR was identified. CFMD content was generated from discoveries in research laboratories with additional contributions from genetic testing facilities. While providing an extensive collection of variation in CFTR, CFMD has little phenotypic annotation, and functional consequences are primarily derived from predictions based on the nature of the nucleotide changes. Assessing disease liability of CFTR variants with predictive algorithms has proven to be of limited utility19, 20. A key weakness in the development of more accurate algorithms is the paucity of variants with well-defined functional consequences21.
As the CFMD constituted an excellent existing repository of nucleotide variation in CFTR, a new approach was taken to comprehensively address phenotypic and functional implications of CFTR variants. The Clinical and Functional TRanslation of CFTR (CFTR2) project assembled clinical data and accompanying CFTR variants from cystic fibrosis patients enrolled in national registries and large clinical centers from twenty-four countries. By focusing on variants present in individuals with a diagnosis of cystic fibrosis ascertained by expert clinicians, the project used a ‘phenotype-driven’ approach to data collection rather than the laboratory-based ‘genotype-driven’ approach. Secondly, microattribution recognition was used to identify the source and credit the contributors of the clinical and genetic data that constitute the CFTR2 database22, 23. To prioritize evaluation, the CFTR2 project started with the subset of CFTR variants exceeding an allele frequency of 0.01% in the collected cystic fibrosis patients. Clinical features of patients and functional assessment of each variant were used to define disease-causing variants. Variants not meeting clinical or functional thresholds were evaluated for disease penetrance using a population-based approach. The phenotype-driven approach presented here could be used to inform the assignment of disease liability in a wide range of genetic disorders.
RESULTS
159 CFTR variants represent 96% of cystic fibrosis alleles
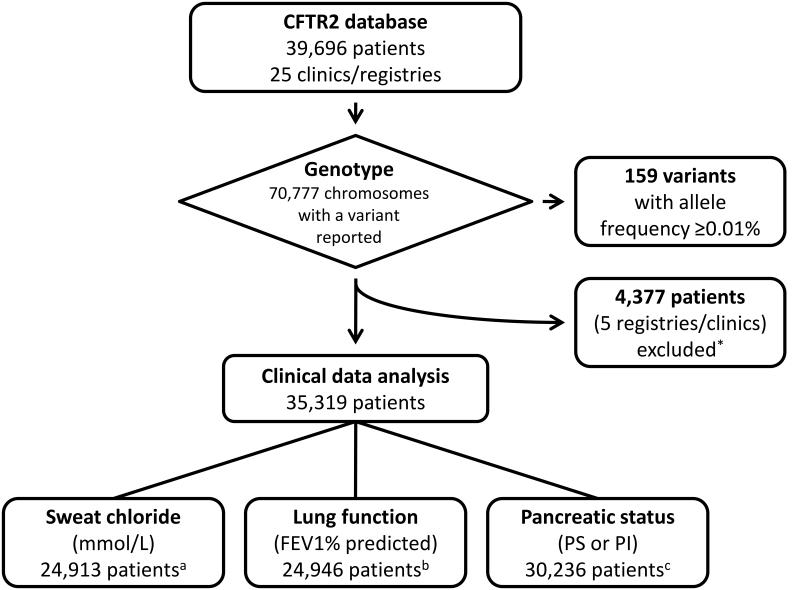
Data from the 39,696 cystic fibrosis patients in CFTR2 (Figure 1) were collected from national cystic fibrosis patient registries or cystic fibrosis specialty clinics (Supplementary Table 1) and represent 57% of the estimated 70,000 patients with cystic fibrosis24. The vast majority (95% of the 31,727 patients with ethnicity data) are listed as Caucasian. One thousand forty-four distinct CFTR variants were seen in these patients. The most common variant, p.Phe508del, accounted for 70% of the identified alleles in these patients. Twenty-two additional variants previously defined as cystic fibrosis-causing and reported to occur at a frequency of 0.1% or higher in cystic fibrosis patients by the American College of Medical Genetics represented 17.5% of the alleles11. Another 136 variants occurred at a frequency exceeding 0.01% and were reported on at least 9 alleles in the CFTR2 database (Supplementary Table 2). Together, these 159 variants accounted for 96.4% of the identified cystic fibrosis alleles in CFTR2. Our efforts focused on evaluation of the disease liability of these 159 variants to maximize clinical sensitivity for cystic fibrosis genetic testing.
Figure 1. Data collected for the CFTR2 project.
The 159 variants seen in 9 or more alleles with an allele frequency of ≥0.01% in CFTR2 were prioritized for further analysis. *Incomplete clinical information available from submitting registry at the time of analysis. aSweat chloride data was not reported for 10,170 patients. 236 patients had sweat chloride values outside physiologic range (>150 mmol/L or <5 mmol/L) and were excluded. bLung function data was not reported for 10,197 patients. 5,633 patients were under the age of 6 years and were excluded, if measurements were present. 46 patients had lung function measurements outside physiologic range (<3% or >150% predicted) and were excluded. cPancreatic status was characterized as sufficient (PS) or insufficient (PI). Data was not reported on 5,083 patients.
Phenotypic analysis
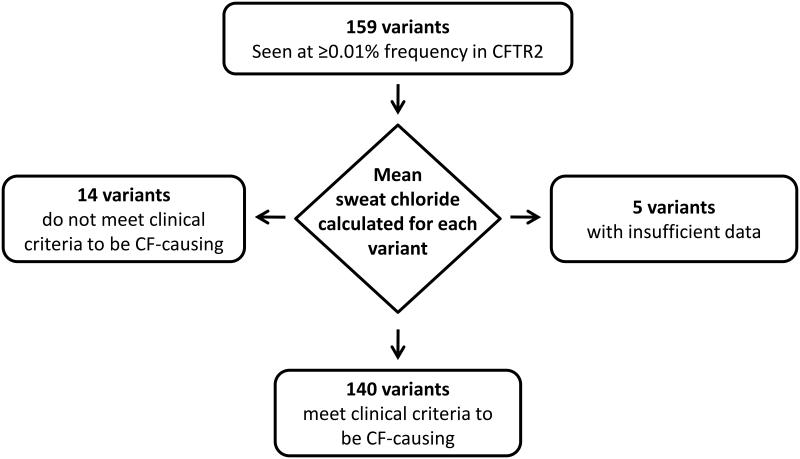
All patients in the CFTR2 database were clinically diagnosed with cystic fibrosis; however, cystic fibrosis is a highly variable disorder25. To evaluate patients across the spectrum of cystic fibrosis severity, a biochemical measure integral to the diagnosis of cystic fibrosis was used to establish whether a CFTR variant caused cystic fibrosis based on phenotypic evidence. Determination of sweat chloride concentration provides a measure of CFTR function in vivo that is widely performed in a standardized fashion and has well-defined differences from values in the non-cystic fibrosis population26-29. A variant was deemed disease-causing by clinical criteria if the mean sweat chloride concentration derived from at least 3 patients carrying the variant was ≥60 mmol/L28, 30. The use of an average measure enabled accommodation of individual variability in sweat chloride concentration due to non-CFTR factors (Supplementary Figure 1). When data was only available from two patients, both sweat chloride concentrations had to exceed 90 mmol/L. To attribute sweat chloride concentration to the variant under study, we analyzed patients who carried a variant in their other CFTR gene that was known to cause complete or near-complete loss of CFTR function (Methods). Of the 159 variants under study, 140 met clinical criteria (Figure 2), of which 138 had sweat chloride concentrations derived from three or more patients while two variants each had measures exceeding 90 mmol/L in two patients (Supplementary Table 2). Thirteen of the fourteen variants not meeting clinical criteria were associated with mean sweat chloride concentrations in the clinical “intermediate” range from 40–58 mmol/L; the remaining variant had an average measure of 39 mmol/L. Individual variant data and other cardinal phenotypes of cystic fibrosis are shown in Supplementary Table 2.
Figure 2. The process used to assign CFTR variants as cystic fibrosis-causing based on a biochemical measure.
The mean sweat chloride concentration was evaluated for patients with a given variant in trans with a known cystic fibrosis-causing variant (16 commonly occurring pancreatic insufficient variants among the 23 originally identified as cystic fibrosis-causing in the ACMG panel)65. Five variants did not have sufficient sweat chloride values from patients with the variant of interest in trans with a cystic fibrosis-causing variant.
Functional analysis
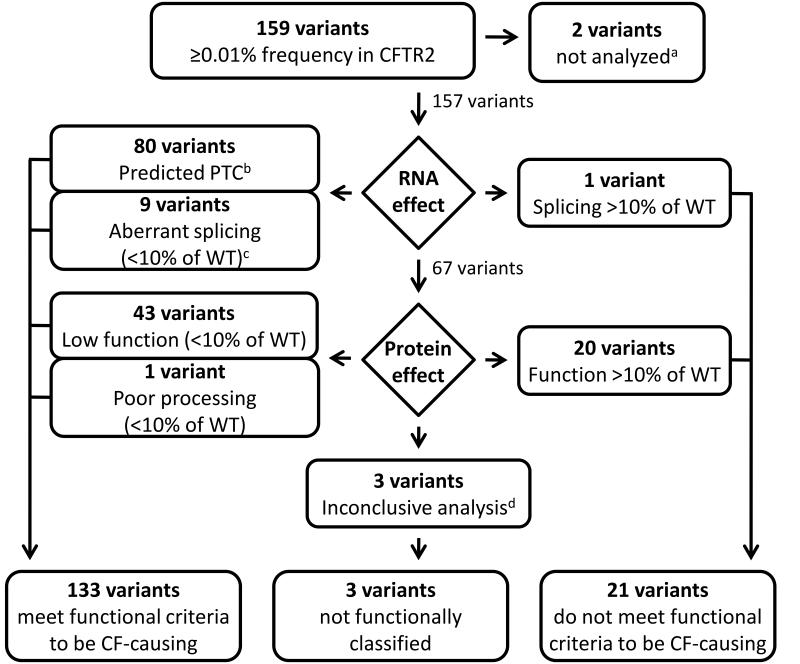
Two common variants (>5% frequency in the general population) in the length of a polythymidine region of intron 9 (c.1210-12T[5] and c.1210-12T[7]; legacy names 5T and 7T, respectively) that have been extensively studied were not reanalyzed here (Supplementary Note). Eighty of the remaining 157 variants are predicted to introduce a premature termination codon (PTC) to the CFTR mRNA (nonsense variants [n=35], variants in the canonical nucleotides of the splice donor/acceptor sites [‘GT-AG’, n=15], or insertion/deletion variants causing frameshifts [n=30]; Figure 3). A common consequence of a PTC variant is nonsense mediated decay (NMD) of mRNA resulting in severe reduction of RNA and no protein produced31, 32. In rare cases, variants affecting splicing can create stable in-frame transcripts due to skipping of in-frame exons, however; the translated protein is almost invariably non-functional (e.g. c.1393-1G>A [legacy name 1525-1G->A])33. Thus, these 80 CFTR variants were predicted to be clinically deleterious4 and cystic fibrosis-causing (Supplementary Figure 2). Ten variants occurred within or near splice sites but did not alter canonical splice donor/acceptor sites (Figure 3). Five of these variants have been previously evaluated and shown to express aberrant alternatively-spliced transcripts in relevant tissues leading to severe reduction in the level of full length CFTR mRNA (0-8% of wild-type level)34-39 (Supplementary Table 3a). The remaining five putative splice variants were studied using minigene analysis (Methods and Supplementary Table 3b). Aberrant splicing (<10% wild type [WT] CFTR transcript) and reduced mature CFTR protein (<10% WT-CFTR) were observed for four of the five variants (Supplementary Table 3c). While there is no firmly established level of essential function, less than 10% of WTCFTR function has been generally accepted as a conservative threshold for the presence of cystic fibrosis features in the exocrine pancreas, sweat gland and lungs38, 40, 41. Together, nine of the ten variants that affect splicing had evidence of deleterious consequence consistent with disease (Figure 3).
Figure 3. The process used to assign CFTR variants as cystic fibrosis-causing based on functional analysis.
Variants were sorted by their predicted effect. Those expected to disrupt the amount or quality of RNA included variants that cause a premature termination codon (PTC) and therefore no protein (introduction of a stop codon, variants that affect splice donor-acceptor sites, insertion or deletion changes that introduce a frameshift) and variants predicted to result in altered mRNA splicing efficiency and therefore reduced full-length CFTR protein produced. Variants predicted to produce full-length CFTR protein, but with an amino acid substitution, insertion, or deletion (missense and insertion or deletion changes that do not introduce a frameshift) were evaluated to determine protein level (defined as percentage of mature protein present) or function (defined as percentage of chloride current). Variants were considered disease-causing if they resulted in less than 10% of the level of WT-CFTR mRNA transcript, WT-CFTR protein or WT-CFTR chloride current. aTwo common variants in intron 9 that have a complex effect upon the cystic fibrosis phenotype and have been extensively studied were excluded from further analysis (see text). b Variants known to cause additional consequences include: c.1393-1G>A (legacy name 1525-1G->A), which skips an in-frame exon; p.Glu831X, which results in an alternatively spliced mRNA in addition to synthesis of a truncated protein; and p.Glu1418ArgfsX14 in which the deletion in the final exon would not be expected to cause nonsense mediated decay (NMD). Each of these variants is associated with a mean sweat chloride concentration above 60mmol/L (Supplementary Figure 2). cFive variants previously reported in the literature to have aberrant splicing; four variants found to have aberrant splicing by minigene analysis. dVariants p.[Gln359Lys;Thr360Lys], p.Leu558Ser, and p.Arg1070Gln.
Sixty-seven variants predicted either an amino acid substitution (missense, n=65) or omission of a single amino acid (in-frame deletions, n=2). As these variants permit synthesis of stable RNA and full length protein, experimental studies were performed on each variant in isolation to determine consequence upon CFTR biogenesis and function. CFTR bearing missense or in-frame changes was expressed in HeLa and Fischer Rat Thyroid (FRT) cells to assess glycosylation status with Western blotting, a well-established method to monitor CFTR maturation (Supplemental Note)42, 43. Sixty-three (61 missense and 2 in-frame deletions) of the 67 variants were tested in both cell lines for their effect upon CFTR processing. Results of the two cell lines largely agreed (r2=0.94, p<0.001; Supplementary Figure 3). The variants fell into three groups: those with minimal disruption in processing (>80% of CFTR protein in mature form in both cell lines; n=32), those with intermediate disruption in processing (10% to 80% mature in at least one cell line; n=21), and those with a dramatic negative effect on processing (≤10% mature in both cell lines; n=10). Among the intermediate group, eleven variants caused a severe defect in processing in one cell line but not the other; the remaining ten variants caused an intermediate defect in both cell lines.
To assess the effect of the missense variants upon function, chloride current measurements were performed on FRT cells expressing CFTR bearing each of 63 variants individually (61 missense and 2 in-frame deletions). Chloride conductance was not determined for 4 missense variants. Functional analysis of primary airway cells obtained from cystic fibrosis patients bearing nine different CFTR genotypes composed of established disease-causing variants was consistent with a threshold of 10% CFTR function being associated with cystic fibrosis (Supplementary Figure 4). Forty-three variants (41 missense and two in-frame deletions) conducted chloride at a level less than 10% of WT-CFTR and were deemed disease-causing (Figure 3). CFTR bearing each of the remaining 20 missense changes generated chloride conductance that ranged from 10.5% to 147% of WT-CFTR. As such, the effects of these 20 variants upon CFTR function were classified as inconsistent with cystic fibrosis although they could contribute to other phenotypes. Comparison of CFTR processing and chloride current revealed that a severe processing defect in HeLa or FRT cells (C/(B+C) <0.1) was consistently associated with CFTR chloride channel function less than 10% (Supplementary Note). Of the four variants that did not have chloride conduction measured, one (p.His199Tyr) exhibited a severe processing defect in HeLa cells (<0.01) and was categorized as functionally deficient (Figure 3). The remaining three variants (p.[Gln359Lys;Thr360Lys], p.Leu558Ser and p.Arg1070Gln) exhibited processing greater than 10% of WT and were not functionally classified.
Penetrance analysis
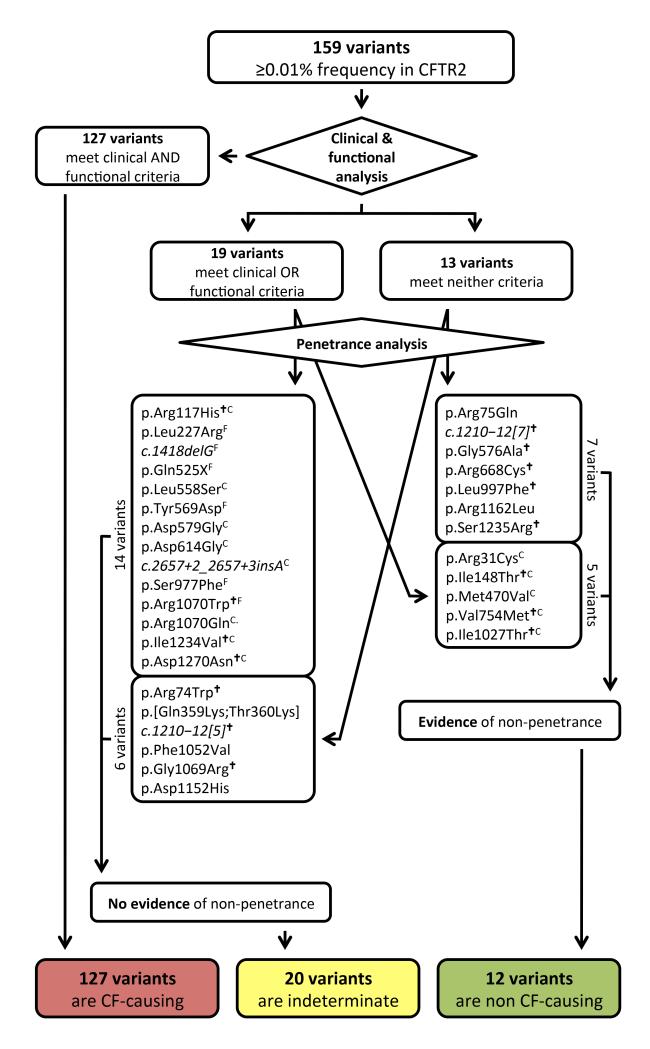
Among the 159 variants studied, 127 met clinical and functional criteria and were classified as cystic fibrosis-causing variants (Figure 4 and Supplementary Table 2). To aid classification of variants not meeting clinical or functional criteria, a penetrance study was performed using 2,188 fathers of cystic fibrosis patients recruited from North America and in Europe (Supplementary Table 4). The presence of a normally-functioning CFTR gene is required in fathers of cystic fibrosis patients, as reduced CFTR function is associated with male infertility due to congenital bilateral absence of the vas deferens (CBAVD)44. Male infertility due to CBAVD affects 97-98% of males with cystic fibrosis45, 46. Fathers of naturally-conceived cystic fibrosis offspring will transmit one pathogenic allele to their affected children. As those fathers are fertile, the non-transmitted allele should not contain a deleterious variant. Thus, any CFTR variants occurring on the non-transmitted allele in a fertile father was deemed non-penetrant for cystic fibrosis and CBAVD. To exclude errors that could have occurred during sample processing or if assisted reproductive technologies were used without our knowledge, a variant had to be observed on the non-transmitted CFTR allele in at least two fathers.
Figure 4. Assignment of disease liability to the 159 most frequent CFTR variants using three criteria.
127 variants deemed cystic fibrosis-causing met clinical and functional criteria and had no evidence of non-penetrance. Of 19 variants meeting clinical or functional criteria but not both, 14 had no evidence of non-penetrance and were classified as indeterminate; 5 variants were seen on the non-transmitted CFTR allele in fathers of cystic fibrosis offspring and were classified as non-cystic fibrosis causing. Thirteen remaining variants met neither clinical nor functional criteria, 7 of which were observed to be non-penetrant and were classified as non-cystic fibrosis causing. The remaining 6 variants had no evidence of non-penetrance but insufficient evidence to be classified as non cystic fibrosis-causing and so were classified as indeterminate. CVariants that met clinical criteria but not functional criteria. FVariants that met functional criteria but not clinical criteria. †Variant known to be part of a complex allele or found in cis with another variant.
Genotyping for the 159 CFTR variants yielded 2,062 samples suitable for penetrance analysis, of which 185 had two or more variants identified (Supplementary Figures 6 and 7). After additional filtering, 100 fathers were found to carry at least one of the 159 variants in trans with a previously-accepted cystic fibrosis-causing variant (Supplementary Figure 6). Among these 100 fathers were ten variants deemed nonpenetrant, as each occurred in the non-transmitted “healthy” CFTR gene of at least two fathers (Table 1). To assess the validity of labeling these variants non-penetrant, we compared the frequency of each variant in the fathers with frequency data available in the 1000 Genomes Project47. Our first premise was that non-penetrant and phenotypically irrelevant variants should occur in healthy cystic fibrosis carrier fathers on the non-transmitted allele at the same frequency as observed in the general population. This was the case for all non-penetrant variants (Table 1). The second premise was that non-penetrant variants should occur at a much lower frequency in patients with cystic fibrosis than observed in the general population. Indeed, the frequency of each non-penetrant variant in cystic fibrosis patients enrolled in CFTR2 was at least 10-fold lower than the frequency in the general population. In addition to these ten variants, c.1210-12[7] (legacy name 7T) had already been reported to be nonpenetrant48 and was identified as a second variant in numerous fathers and a twelfth variant, p.Ile1027Thr, was deemed non-penetrant as it was observed exclusively in cis with the p.Phe508del change. The presence of non-penetrant variants in the CFTR2 database is likely due to incomplete genotyping and/or lack of analysis of allele assortment. Analysis of assortment is essential as multiple examples of complex alleles were disclosed in the penetrance study (Supplementary Note)49.
Table 1.
Variants associated with incomplete penetrance
| Variant | # alleles in CFTR2 |
Frequency in CFTR2 (out of 70,777 known alleles) |
# that occur in fathers with a CF- causing variant |
# that occur in trans with a CF- causing variant in fathers |
Frequency in fathers (out of 4,296 total alleles) |
Allele freq. in 1000 Genomes |
Notes |
|---|---|---|---|---|---|---|---|
| Variants that met clinical criteria, but did not meet functional criteria | |||||||
| p.Arg31Cys | 13 | 0.0002 | 4 | 4 | 0.00093 | 0.001- 0.004 |
|
| p.Ile148Thr | 99 | 0.0014 | 5 | 4 | 0.00209 | 0.013 (Applera data) |
Always seen in CF patients with p.Ile1023_Val1024del* |
| p.Met470Val | 41 | 0.0006 | 1185 | Not analyzed |
0.35196 | 0.087- 0.647 |
|
| p.Val754Met | 9 | 0.0001 | 6 | 4 | 0.00163 | 0-0.003 | |
| Variants that did not meet clinical nor functional criteria | |||||||
| p.Arg75Gln | 28 | 0.0004 | 57 | 48 | 0.01723 | 0.009- 0.033 |
|
| p.Gly576Ala | 42 | 0.0006 | 17 | 12 | 0.00466 | 0.004- 0.009 |
In 1000 genomes and in fathers, always seen in cis with p.Arg668Cys |
| p.Arg668Cys | 49 | 0.0007 | 24 | 16 | 0.00675 | 0.004- 0.009 |
In 1000 genomes, always seen in cis with p.Gly576Ala; seen without p.Gly576Ala fathers |
| p.Leu997Phe | 28 | 0.0004 | 7 | 5 | 0.00209 | 0.001- 0.003 |
|
| p.Arg1162Leu | 9 | 0.0001 | 3 | 2 | 0.00140 | 0.001 | |
| p.Ser1235Arg | 54 | 0.0008 | 18 | 15 | 0.00489 | 0.005- 0.016 |
|
The variant p.Ile148Thr occurs on multiple haplotypes, but only causes cystic fibrosis when appearing in cis with p.Ile1023_Val1024del66.
One-hundred and forty seven variants had no evidence of non-penetrance in the fathers screen (all 127 that met clinical and functional criteria; 8 variants meeting only clinical criteria, 6 only functional criteria, and 6 neither criteria; Figure 4). Included among the variants meeting neither clinical nor functional criteria are those that have previously been associated with variable penetrance (such as p.Asp1152His), variants that have been reported as part of complex alleles in which the disease liability of each variant individually could not be determined (such as the pair p.Arg74Trp and p.Asp1270Asn) and variants with incomplete clinical or functional analysis.
DISCUSSION
Genetic testing of CFTR is widely employed for diagnosis in symptomatic individuals29, for carrier status in the general population17, increasingly as part of newborn screening50, 51 and most recently for selection for treatment with variant-specific molecular therapy52. The primary goal of the CFTR2 project was to increase the fraction of variants in the CFTR gene that have been assessed for propensity to cause disease. At the initiation of the project, 23 variants were defined as disease-causing11. Combining phenotypic evidence with functional analysis enabled unambiguous assignment of pathogenicity to an additional 104 variants. Testing for all 127 variants is estimated to account for 95.4% of cystic fibrosis alleles in our sample, leaving only 0.21% of patients in our sample without at least one pathologic CFTR variant identified. Couples undergoing carrier screening will also benefit as the sensitivity for detection of couples at 1 in 4 risk of having a child with cystic fibrosis should increase from 72% to ~91% when screening for the 127 variants. It should be noted that these estimated detection rates are subject to regional and ethnic variability of variant distribution and frequency. This project illustrates the feasibility of translating allelic diversity to clinical application but also the challenges in interpreting the disease implications of rare DNA changes.
The CFTR2 project gathered both genotype and phenotype data on patients that were enrolled in registries and clinics. While this approach enriched the subject pool for affected individuals, additional objective measures were used to differentiate variants causing life-shortening cystic fibrosis from those causing less severe disease44. Notably, 19 of 159 variants (12%) studied did not meet our clinical threshold despite being reported in patients diagnosed with cystic fibrosis by a medical professional familiar with the disease. This finding revealed the degree of phenotypic heterogeneity existing even among well-annotated clinical data collections. Of the 140 variants assigned as disease-causing using clinical criteria, 13 (9.3%) did not meet functional criteria. Our study emphasizes the importance of phenotypic and functional analysis to clinically annotate variants found in patients and demonstrates that presence of a rare variant, even if reported in multiple unrelated affected individuals, does not assure that it is deleterious or pathogenic.
Phenotypic and functional criteria for disease can be based on metrics that already exist for many genetic diseases. For this study, sweat chloride concentration was chosen to define the phenotype because it is dependent on CFTR function, correlates with disease severity, is performed frequently in a standardized fashion, and has well validated cut-offs between normal and disease29, 30. Similarly, assessment of functional effects of variants can follow established guidelines4. For example, the assumption that variants predicted to introduce a PTC are deleterious is commonly accepted practice4. As noted here, the clinical features of patients carrying predicted PTC variants are consistent with disease (Supplementary Figure 2). Evaluation of the effect of missense variants poses the greatest hurdle; however, relatively straightforward assays such as Western blotting can disclose processing defects, a common consequence of amino acid substitutions. Expression of mutated protein in multiple cell lines, as employed here, minimizes cell-type specific effects. Perhaps the most challenging issue is the establishment of thresholds for both phenotypic and functional measures. In this study, the adopted thresholds were vetted by experts in the clinical and functional domains of cystic fibrosis research. The 10% threshold for protein expression and chloride conductance (both in comparison to WT-CFTR) is not an absolute demarcation between disease and health, but is a conservative threshold consistent with prior research correlating CFTR function with disease38, 40, 41. Provided that it is acknowledged that consensus opinions represent the current understanding of pathogenesis, thresholds can be modified if warranted by future studies, as some variants may influence CFTR in a manner not captured by our methods.
The 127 variants that met both clinical and functional criteria were designated as cystic fibrosis-causing; however, 32 remaining variants (20%) required further analysis to determine if they were neutral with respect to disease, or associated with milder phenotype or partial penetrance. Demonstration that a variant occurs in a sample of normal controls at the same frequency as observed in patients has been a long-accepted method to determine neutrality53, 54. In recessive conditions, this test can only be performed in ‘control’ individuals known to carry a deleterious allele in trans. Fathers of cystic fibrosis patients provide an ideal group to assess neutrality as they carry a functional CFTR gene by virtue of their fertility and a disease-causing variant transmitted to their affected offspring. Demonstration that a variant under study occurs in the ‘healthy’ (non-transmitted) CFTR genes of fertile fathers provides compelling evidence of neutrality or non-penetrance for cystic fibrosis. The power of this approach depends upon the frequency of the alleles in the population and the number of ‘controls’ tested. In the test of non-transmitted (non-cystic fibrosis) chromosome of fertile fathers, confidence that a given variant was not found because it is fully penetrant declines as the allele frequency declines. Therefore we are more confident that more frequent variants such as p.Gly551Asp are fully penetrant compared to variants such as p.[Gln359Lys;Thr360Lys], p.Phe1052Val, and p.Gly1069Arg, which were seen with an allele frequency of less than 0.0002. Additional confidence in the assignment of variants is derived from the observation that variants that were non-penetrant for cystic fibrosis occurred at similar frequencies to those reported in Caucasian subjects in the 1000 Genomes Project. Penetrance analysis should become more useful for clinical applications as the frequencies of rare variants in the healthy population become more robust and complete (e.g. 100,000 genomes) and with more complete delineation of ethnic/geographic cohorts, to which we did not have access.
The instances in which the apparent disease liability determined by clinical, functional and penetrance criteria were discordant deserve special attention. For the 13 variants meeting clinical but not functional criteria, a common finding was the presence of additional variants in cis that ablated or modified CFTR function, thereby explaining the presence of these variants in cystic fibrosis patients. Recognizing that complex alleles may account for discordance between phenotype and genotype is critical in the clinical arena as misidentification can lead to inappropriate medical actions. These findings emphasize that complete sequencing of the coding regions of genes bearing rare or novel alleles should be undertaken to identify all potentially deleterious alleles. Finally, penetrance analysis was helpful in distinguishing variants that might contribute to disease from those that were neutral. Included in the non cystic fibrosis-causing group are known polymorphic variants such as p.Met470Val55, 56 that appear to be entered into patient registries due to incomplete genotyping of CFTR. Patients carrying non cystic fibrosis-causing variants such as p.Met470Val with symptoms indicative of cystic fibrosis may benefit from being re-genotyped. Conversely, patients diagnosed with cystic fibrosis based on genetic findings with one or more variants now considered not to cause cystic fibrosis should be re-evaluated.
While this work establishes the disease liability for most of the alleles found in cystic fibrosis patients, 20 variants remain indeterminate. The ACMG has issued recommendations for classification of unknown variants beyond those used in this study4, 57; however, probabilistic estimation may not be appropriate for all variants deemed indeterminate after extensive clinical, functional and penetrance analysis. As a purpose of this project is to definitively place CFTR variants into well-defined categories, further classification of indeterminate variants will require additional analysis to quantify the probability of causing or not causing disease. For example, it is possible that one or more of the indeterminate variants cause dysfunction of CFTR in a manner unique from functional assessments used in this study, as has been shown for two missense changes that also affect RNA splicing (p.Gly576Ala58 and p.Ile1234Val; work in publication review).
The indeterminate variants as well as over 1,600 CFTR variants that are unclassified remain a diagnostic dilemma. Computational approaches predicting disease liability have been applied to splice site59 and missense changes19-21 to classify CFTR variants. These approaches lack specificity needed for definitive clinical classification. Algorithms that predict splicing are useful for highly conserved sequences but experimental studies are needed for changes in less-well conserved nucleotides or nucleotides outside of consensus splice sites, as shown in the Supplementary Note59, 60. Given the large and diverse structure of the full-length CFTR protein (1480 residues), annotation of more variants for algorithmic training should substantially improve the predictive performance of the classifier. To that end, machine learning approaches could prioritize future experimental testing.
Increased use of sequencing in the clinical setting has emphasized the medical challenges posed by rare variants. While it appears a daunting task to determine the disease liability of all variants accounting for a Mendelian disease, the CFTR2 project demonstrates the feasibility of the task using a phenotype-driven approach. Patient registries have been assembled for many genetic disorders61-63 that should enable collation of patient genotypes and associated phenotype data for detailed analysis. Microattribution can identify the data source and composition while acknowledging the contributor and data integrity64. For recessive disorders, the number of alleles in the human population in each gene is finite and stable (excepting extremely rare de novo variants). Thus, careful assignment of disease liability to the variants responsible for these disorders will be valuable for current and future generations of patients and their family members.
METHODS (online)
Patients
Anonymized genotype and cross-sectional clinical information were collected from 25 national cystic fibrosis registries and major clinical centers in countries without a registry (listed in Acknowledgements and Supplementary Table 1). Genotype was recorded from the clinical record. For 13 datasets, (7.5% of patients), clinical information was provided only for patients carrying at least one variant not included in the American College of Medical Genetics panel for cystic fibrosis screening11. Sweat chloride concentration, obtained at the time of diagnosis and averaged if performed more than once, was recorded in mmol/L (mEq/L). Results from 236 patients (1% of measurements) were dropped because they were not within the physiologic range of 5-150 mmol/L29. Pancreatic status, defined differently by registry, was recorded from the submitting registry. Raw FEV1 in liters was converted to % predicted using patient age, gender, race (if known), and height using the Wang equation (for individuals under 18 years old) or the Hankinson equation (18 years and older)67, 68. Otherwise, FEV1% predicted was used as provided. The most recent measurement within the last recorded year was used. Clinical features ascribed to a CFTR variant were derived from patients bearing the variant in trans with a cystic fibrosis-causing variant previously shown to have minimal residual function65 and averaged across patients with that particular genotype (variant of interest/known cystic fibrosis-causing variant). All data collection was approved by the Institutional Review Board at Johns Hopkins University and by the Registry Advisory Committee for the US Cystic Fibrosis Foundation.
Analysis of variants expected to affect RNA splicing
CFTR variants predicted to alter splicing efficiency that were not previously studied (Supplementary Table 2) were examined to confirm their deleterious nature using minigene constructs as previously described with some modifications69. Briefly, a five-step strategy was employed; (i) Amplification of the 5′ acceptor and 3′ donor splice site sequences of the intronic region of interest along with flanking exons from genomic DNA using KOD Hot Start DNA polymerase (Novagen). Primer sequences are available on request. (ii) Fusion PCR was performed on the amplicons generated in the first step using the exonic primers only creating a fusion amplicon with 5′ acceptor and 3′ donor splice site sequences with respective exons on either side. (iii) Sticky feet mutagenesis of the pcDNA5/FRT/CFTR using the fusion PCR amplicon as the primer to create pcDNA5/FRT/CFTR minigene70. (iv) Site directed mutagenesis (QuikChange II XL, Agilent Technologies) to create c.579+3A>G (legacy name 711+3A>G), c.579+5G>A (legacy name 711+5G>A), c.1585-8G>A (legacy name 1717-8G>A), c.2657+2_2657+3insA (legacy name 2789+2insA), and c.2988G>A (legacy name 3120G>A) variants (Supplementary Table 3c) in the respective minigenes. The additional splice sites variants c.579+1G>T (legacy name 711+1G>T) and c.2988+1G>A (legacy name 3120+1G>A) were created as positive controls in the assay. Primer sequences are available on request. (v) Finally, re-cloning of the full length CFTR WT and mutant minigene constructs into pcDNA5FRT vector to omit chances of nucleotide errors introduced during mutagenesis steps71. Sequence confirmation of the WT and mutant minigene was performed on ABI 3100 Genetic Analyzer (Applied Biosystems). WT and mutant minigene plasmids were transfected into Human Embryonic Kidney (HEK) 293 (ATCC) and Cystic Fibrosis Bronchial Epithelial (CFBE 41o-) cells (a generous gift from Prof. D. Gruenert, University of California-San Francisco, San Francisco, CA) cells. All cell lines used were tested for mycoplasma contamination at the Cell Core Center and Biorepository, Johns Hopkins University, Baltimore, MD. Forty-eight hours post transfection total RNA and whole cell lysates were prepared. First strand cDNA was synthesized using i-Script cDNA synthesis kit (BioRad, USA) or SuperScript RT III reverse transcriptase and random hexamers (Invitrogen, UK). The resulting cDNA product was used directly for PCR amplification using exonic primers from the regions of interest. Primer sequences are available on request. Agarose gel (1.5%) electrophoresis was performed to analyze the RT-PCR products and transcripts were sequenced after the gel extraction. The quality of RNA in all the samples was verified by amplification of transcript encoding the TATA box binding protein (TBP). Controls without reverse transcriptase and without RNA were included. The amount of correctly spliced product from each CFTR mutant minigene relative to respective WT minigene was calculated from the sequencing data as described before72. Western blot was performed to evaluate the amount of complex glycosylated (C-band) CFTR. Mouse monoclonal antibody 570 (R domain or 590 (NBD2; UNC antibody distribution program sponsored by Cystic Fibrosis Foundation Therapeutics, USA) and/or MM13-4 (N-terminal; Chemicon, USA) were used to detect CFTR. GAPDH or tubulin was used as loading control. The blots were quantified using Image J software (NIH) to determine the amount of processed CFTR (C-band) for each experimental sample relative to WT minigene.
Analysis of variants expected to alter protein processing and/or function
Variants causing an amino acid substitution or an in-frame deletion were introduced individually into CFTR cDNA using site directed mutagenesis as previously described73. The WT-CFTR clone contained was obtained from a non-cystic fibrosis individual and contained the known neutral variant p.Val1475Met. Transient expression of CFTR in HeLa cells (Clontech) was achieved as described previously43. Stable expression of CFTR in Fischer Rat Thyroid cells (FRT; a kind gift from Michael Welsh, University of Iowa, Iowa City, IA) was achieved by integrating each mutated CFTR cDNA as a single copy into the same genomic location using the Invitrogen Flp-In system™ as described previously73, 74. Following selection and confirmation of CFTR cDNA with the desired variant, the level of the heterologous human CFTR RNA was determined for each cell line. Cell lines with mRNA levels >0.5 or <3 fold the average level of four independent FRT cell lines expressing WT-CFTR were tested. CFTR maturation and protein expression level was quantified using the ratio of C-band CFTR to B-band + C-band CFTR (normalized to WT-CFTR as described previously)43. Forskolin-activated, CFTR-dependent chloride secretion was measured on confluent FRT cells by short circuit current (Isc) in Ussing chambers. Isc measurements were repeated 3 to 14 times per cell line and averaged. All readings were reported as a percentage of the average Isc of wild-type CFTR-expressing FRT cell lines75. Measurements of four separate FRT cell lines were used to establish the mean current and variance of wild-type CFTR. Human bronchial epithelial (HBE) cells were isolated from non-cystic fibrosis and cystic fibrosis subjects and short-circuit current was measured as previously described76.
MassArray Assay
An assay to screen for the 159 CFTR variants seen in ≥0.01% of patients was developed using an open platform matrix-assisted laser desorption/ionization time of flight (MALDI-ToF) mass spectroscopy77. The assay was validated using genomic DNA (gDNA) controls obtained from available study stocks from the US (n=99), France (n=18), Canada (n=22), Czech Republic (n=2) and Serbia (n=1). When gDNA was not available (n=11), plasmid DNAs were created using QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies). Two variants were unable to be confirmed because of a problem with the extension primer (p.Gly330X) and inability to derive a positive control (p.Glu1418ArgfsX14). The assay was validated with Sanger sequencing. In validation studies, 35/35 variants were identified and 29/29 WT chromosomes were confirmed.
The multiplex assay design was initially accomplished using Assay Designer Software, Version 4.0 (Sequenom Inc., San Diego, CA) to construct both amplification and extension primers and subsequently optimized based on the results of the positive controls. Multiplex PCR amplification of regions up to 300nt from genomic DNA, whole genome amplified (WGA) DNA, or plasmid DNA at concentration of 25 ng/ul, 50 ng/ul or 5 ng/ul respectively was performed in 384-well plates. The reaction contained reagents from the iPLEX Gold SNP Genotyping Kit (Sequenom). Because of variation in amplicon length and the likelihood of primer-dimer formation, differential primer concentrations were used. Primer multiplexes were evaluated for possible dimers and hairpins using NIST primer tools
(http://yellow.nist.gov:8444/dnaAnalysis/primerToolsPage.do) and subsequently adjusted. Primer sequences and concentrations are available on request. Unincorporated dNTPs were neutralized using reagents from the iPLEX gold reagent kit (Sequenom).
Single base extension (SBE) products were generated from each purified PCR reaction using reagents from the iPLEX Gold Reagent Kit (Sequenom). The resulting SBE PCR products were prepared for mass spectrometry and dispensed to a SpectroCHIP (Sequenom). Data was analyzed using a MALDI-ToF spectrometer (Sequenom). Data was generated with SpectroACQUIRE software version 3.0 on the MassARRAY spectrometer (Sequenom) and then analyzed using Typer software, version 4.0.22 (Sequenom). This assay was employed to test DNA from fathers of cystic fibrosis offspring for variants suspected of being cystic fibrosis-causing. Fathers provided informed consent for genetic study and were anonymized for analysis. Each father should carry only the deleterious CFTR variants passed to his offspring. Additional variants detected in fathers represent either a complex allele, in which two CFTR variants are present on the same chromosome, or a CFTR variant in trans with the transmitted variant that is insufficiently deleterious to make the father infertile. Fathers with a cystic fibrosis-causing variants and a second variant had their offspring genotyped when available (n=145) to delineate phase.
Statistical Analysis
Statistical analyses were performed using Intercooled Stata version 11 (StataCorp, USA) using the METAREG plug in78. A meta-analytic approach was used to compare clinical features in patients with PTC versus non-PTC variants and to perform regression analysis of aggregate data for each cell line. We incorporated the observed variance across subjects for each variant and allowed for heterogeneity of variance across variants to compare groups. Group means (for chloride current) were compared under a random effects model accounting for statistical variance in the measurements both within and across the variant groups. All reported regression coefficients are unstandardized.
Supplementary Material
Acknowledgements
The authors would like to thank the patients who participated in their national/clinic registries. This work was supported by grants from the NIDDK (5R37DK044003 to G.R.C.), from the NIH (DK49835 to P.J.T.), funding from the Cystic Fibrosis Foundation Therapeutics, Inc. (to P.J.T.), the US Cystic Fibrosis Foundation (CUTTING08A, 09A, 10A to G.R.C.; SOSNAY10Q to P.R.S.), from FCTPortugal (PIC/IC/83103/2007 and PEstOE/BIA/UI4046/2011 to M.A and BioFIG). Assistance with statistical analysis was provided by Elizabeth Johnson, M. Brad Drummond, Dave Cutler, and Dan Arking. The authors received considerable guidance from the CFTR2 clinical expert panel: Christiane De Boeck, Peter Durie, Stuart Elborn, Philip Farrell, Michael Knowles, and Isabelle Sermet; and from the CFTR2 functional studies expert panel: Robert Bridges, Gergely Lukacs, and David Sheppard; and from Molly Sheridan for her critical review.
Footnotes
URLs 1000 genomes browser, http://www.1000genomes.org/; ClinVar, http://www.ncbi.nlm.nih.gov/clinvar/; Cystic Fibrosis Mutation Database, http://www.genet.sickkids.on.ca/app; EURODIS, http://www.eurordis.org/; NIH Global Rare Disease Patient Registry and Data Repository, http://www.grdr.info/; Leiden Open Variation Database, http://www.lovd.nl/3.0/home; National Organization for Rare Disorders, http://www.rarediseases.org/.
Contributors of fathers’ DNA samples: Teresa Casals (Bellvitge Biomedical Research Institute, Spain), Garry Cutting (Johns Hopkins University, USA),, Cristina Dechecchi (University Hospital of Verona, Italy), Ruslan Dorfman (The Hospital for Sick Children, Canada), Claude Ferec (Centre Hospitalier Universitaire, France), Enmanuelle Girodon (GH Henri Mondor, France), Milan Macek, Jr. (Charles University, Prague), Dragica Radojkovic (Institute of Molecular Genetics and Genetic Engineering, Serbia), Martin Schwarz (St. Mary’s Hospital, United Kingdom), Manuela Seia (Fondazione IRCCS Cà Granda Ospedale Maggiore Policlinico, Italy), Manfred Stuhrmann (Medical School Hannover, Germany), Maria Tzetis (National Kapodistrian University of Athens, Greece), and Julian Zielenski (The Hospital for Sick Children, Canada, with partial support from Genome Canada, through the Ontario Genomics Institute per research agreement 2004-OGI-3-05)
Contributors of CFTR2 patient data: Celeste Barreto (Hospital Santa Maria, Portugal), Diana Bilton (Royal Brompton and Harefield Hospital, UK), Joseph Borg (University of Malta), Carla Colombo (University of Milan, Italy), Stavros Doudounakis (Aghia Sophia Children’s Hospital, Greece), Helmut Ellemunter (Innsbruck Medical University, Austria), Godfrey Fletcher (Cystic Fibrosis Registry of Ireland), Ivanka Galeva (University Hospital Aleksandrovska, Bulgaria), Silvia Gartner (Hospital Vall de Hebron Unidad de Fibrosis Quistica, Spain), Vincent A.M. Gulmans (Dutch Cystic Fibrosis Foundation, Netherlands), Elpis Hatziagorou (Aristotle University, Greece), Lena Hjelte (Karolinska Institutet, Sweden), Tiina Kahre (University of Tartu, Estonia), Nataliya Kashirskaya (Russian Academy of Medical Sciences), Anna Katelari (Aghia Sophia Children’s Hospital, Greece), Paul Laissue (Universidad del Rosario, Colombia), Lydie Lemonnier (Association Vaincre La Mucoviscidose, France), Anders Lindblad (Sahlgrenska University Hospital, Sweden), Vincenzina Lucidi (Ospedale Bambino Gesù, Italy), Milan Macek, Jr. (Charles University Prague, Czech Republic), Halyna Makukh (Ukrainian Academy of Medical Sciences), Bruce Marshall (United States Cystic Fibrosis Foundation), Ian McIntosh (Cystic Fibrosis Canada), Meir Mei-Zahav (Tel Aviv University, Israel), Predrag Minic (Mother and Child Health Institute of Serbia), Hanne Vebert Olesen (Aarhus University Hospital, Denmark), Nika Petrova (Russian Academy of Medical Sciences), Tania Pressler (University of Copenhagen, Denmark), Danijela Radivojevic (Mother and Child Health Institute of Serbia), Sophie Ravilly (Association Vaincre La Mucoviscidose, France), Nicolas Regamey (University Hospital Bern, Switzerland), Gabriela Repetto (Universidad del Desarrollo, Chile), Maria Teresa Sanseverino (Hospital de Clinicas de Porto Alegre, Brazil), Christian Scerri (University of Malta), Anne Stephenson (Cystic Fibrosis Canada), Martin Stern (University of Tübingen, Germany), Vija Svabe (Riga Stradins University, Latvia), Muriel Thomas (Belgian Cystic Fibrosis Registry), John Tsanakas (Aristotle University, Greece), Vera Vavrova (Charles University and University Hospital Motol, Czech Republic), and Paul Wenzlaff (Centre for Quality and Management in Health Care, Germany)
Data Access All data can be accessed via the CFTR2 website. Individual variant rsIDs (created by the Single Nucleotide Polymorphism Database [dbSNP]) are listed in Supplementary Table 2. Variants have been submitted to ClinVar and the Leiden Open Variant Database (LOVD) and are searchable under the NCBI CFTR gene ID 1080. The RefSeq accession number for CFTR is NM_000492; the UniProt accession number for CFTR is P13569.
AUTHOR CONTRIBUTIONS P.R.S. jointly supervised research, collected and curated clinical data, performed statistical analysis, analyzed the data, and wrote the manuscript. K.R.S. curated clinical data, analyzed the data, and wrote the manuscript. F.V.G. and H.Y. conceived, designed, and performed chloride conduction experiments and analyzed the data. K.K. conceived, designed, and performed the penetrance analysis and analyzed the data. N.S., A.S.R. and M.D.A conceived, designed, and performed splice analysis and analyzed the data. R.D. and J.Z. curated variant data for CFMD. D.L.M. and R.K. performed algorithm analysis. L.M. and P.T. conceived, designed, and performed the CFTR processing experiments and analyzed the data. G.P.P. advised and aided with the design and implementation of the microattribution process. M.C. jointly supervised research and analyzed the data. M.H.L. jointly supervised research and analyzed the data. J.M.R. curated data for CFMD, jointly supervised research and analyzed the data. C.C. coordinated the collection of clinical data, jointly supervised research, and analyzed the data. C.M.P. jointly supervised research and analyzed the data. G.R.C. supervised the research, conceived and designed experiments, analyzed the data, and wrote the manuscript.
Competing financial interests F.VG. is employed by Vertex Pharmaceuticals. H.Y. is employed by Vertex Pharmaceuticals and owns stock in the company. P.J.T. has financial interest in and sponsored research from Reata Pharmaceuticals. G.R.C. is a consultant for the Cystic Fibrosis Foundation, Vertex Pharmaceuticals, Illumina, aTyr Pharma, and Canon Biosciences.
Reference List
- 1.Chenevix-Trench G, et al. Genetic and histopathologic evaluation of BRCA1 and BRCA2 DNA sequence variants of unknown clinical significance. Cancer Res. 2006;66:2019–2027. doi: 10.1158/0008-5472.CAN-05-3546. [DOI] [PubMed] [Google Scholar]
- 2.Easton DF, et al. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum. Genet. 2007;81:873–883. doi: 10.1086/521032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Plon SE, et al. Sequence variant classification and reporting: recommendations for improving the interpretation of cancer susceptibility genetic test results. Hum. Mutat. 2008;29:1282–1291. doi: 10.1002/humu.20880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richards CS, et al. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- 5.Bamshad MJ, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet. 2011;12:745–755. doi: 10.1038/nrg3031. [DOI] [PubMed] [Google Scholar]
- 6.Kricka LJ, Di RC. Translating genes into health. Nat. Genet. 2013;45:4–5. doi: 10.1038/ng.2510. [DOI] [PubMed] [Google Scholar]
- 7.Samuels ME, Rouleau GA. The case for locus-specific databases. Nat. Rev. Genet. 2011;12:378–379. doi: 10.1038/nrg3011. [DOI] [PubMed] [Google Scholar]
- 8.Celli J, Dalgleish R, Vihinen M, Taschner PE, den Dunnen JT. Curating gene variant databases (LSDBs): toward a universal standard. Hum. Mutat. 2012;33:291–297. doi: 10.1002/humu.21626. [DOI] [PubMed] [Google Scholar]
- 9.Vihinen M, den Dunnen JT, Dalgleish R, Cotton RG. Guidelines for establishing locus specific databases. Hum. Mutat. 2012;33:298–305. doi: 10.1002/humu.21646. [DOI] [PubMed] [Google Scholar]
- 10.Xue Y, et al. Deleterious- and Disease-Allele Prevalence in Healthy Individuals: Insights from Current Predictions, Mutation Databases, and Population-Scale Resequencing. Am. J. Hum. Genet. 2012;91:1022–1032. doi: 10.1016/j.ajhg.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watson MS, et al. Cystic fibrosis population carrier screening: 2004 revision of American College of Medical Genetics mutation panel. Genet. Med. 2004;6:387–391. doi: 10.1097/01.GIM.0000139506.11694.7C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Southern KW, et al. A survey of newborn screening for cystic fibrosis in Europe. J Cyst. Fibros. 2007;6:57–65. doi: 10.1016/j.jcf.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Krulisova V, et al. Prospective and parallel assessments of cystic fibrosis newborn screening protocols in the Czech Republic: IRT/DNA/IRT versus IRT/PAP and IRT/PAP/DNA. Eur. J. Pediatr. 2012;171:1223–1229. doi: 10.1007/s00431-012-1747-z. [DOI] [PubMed] [Google Scholar]
- 14.Vernooij-van Langen AM, et al. Novel strategies in newborn screening for cystic fibrosis: a prospective controlled study. Thorax. 2012;67:289–295. doi: 10.1136/thoraxjnl-2011-200730. [DOI] [PubMed] [Google Scholar]
- 15.Massie RJ, Curnow L, Glazner J, Armstrong DS, Francis I. Lessons learned from 20 years of newborn screening for cystic fibrosis. Med. J. Aust. 2012;196:67–70. doi: 10.5694/mja11.10686. [DOI] [PubMed] [Google Scholar]
- 16.Amos JA, Bridge-Cook P, Ponek V, Jarvis MR. A universal array-based multiplexed test for cystic fibrosis carrier screening. Expert. Rev. Mol. Diagn. 2006;6:15–22. doi: 10.1586/14737159.6.1.15. [DOI] [PubMed] [Google Scholar]
- 17.Strom CM, et al. Cystic fibrosis testing 8 years on: lessons learned from carrier screening and sequencing analysis. Genet. Med. 2011;13:166–172. doi: 10.1097/GIM.0b013e3181fa24c4. [DOI] [PubMed] [Google Scholar]
- 18.Grody WW, Cutting GR, Watson MS. The Cystic Fibrosis mutation “arms race”: when less is more. Genet Med. 2007;9:739–744. doi: 10.1097/gim.0b013e318159a331. [DOI] [PubMed] [Google Scholar]
- 19.Dorfman R, et al. Do common in silico tools predict the clinical consequences of amino-acid substitutions in the CFTR gene? Clin. Genet. 2010;77:464–473. doi: 10.1111/j.1399-0004.2009.01351.x. [DOI] [PubMed] [Google Scholar]
- 20.Rishishwar L, et al. Relating the disease mutation spectrum to the evolution of the cystic fibrosis transmembrane conductance regulator (CFTR) PLoS One. 2012;7:e42336. doi: 10.1371/journal.pone.0042336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masica DL, Sosnay PR, Cutting GR, Karchin R. Phenotype-optimized sequence ensembles substantially improve prediction of disease-causing mutation in cystic fibrosis. Hum. Mutat. 2012;33:1267–1274. doi: 10.1002/humu.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giardine B, et al. Systematic documentation and analysis of human genetic variation in hemoglobinopathies using the microattribution approach. Nat. Genet. 2011;43:295–301. doi: 10.1038/ng.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patrinos GP, et al. Microattribution and nanopublication as means to incentivize the placement of human genome variation data into the public domain. Hum. Mutat. 2012;33:1503–1512. doi: 10.1002/humu.22144. [DOI] [PubMed] [Google Scholar]
- 24.Bobadilla JL, Macek M, Fine JP, Farrell PM. Cystic fibrosis: A worldwide analysis of CFTR mutations - Correlation with incidence data and application to screening. Hum. Mutat. 2002;19:575–606. doi: 10.1002/humu.10041. [DOI] [PubMed] [Google Scholar]
- 25.Welsh MJ, Ramsey BW, Accurso FJ, Cutting GR. In: Cystic Fibrosis in The Metabolic and Molecular Bases of Inherited Disease. Scriver CR, Beaudet AL, Valle D, Sly WS, editors. McGraw-Hill, Inc.; New York: 2001. pp. 5121–5188. [Google Scholar]
- 26.DI SANT’AGNESE PA, Darling RC, Perera GA, Shea E. Sweat electrolyte disturbances associated with childhood pancreatic disease. Am. J. Med. 1953;15:777–784. doi: 10.1016/0002-9343(53)90169-7. [DOI] [PubMed] [Google Scholar]
- 27.Gibson LE, Cooke RE. A test for concentration of electrolytes in sweat in cystic fibrosis of the pancreas utilizing pilocarpine by iontophoresis. Pediatrics. 1959;23:545–549. [PubMed] [Google Scholar]
- 28.LeGrys VA, Yankaskas JR, Quittell LM, Marshall BC, Mogayzel PJ., Jr Diagnostic sweat testing: the Cystic Fibrosis Foundation guidelines. J. Pediatr. 2007;151:85–89. doi: 10.1016/j.jpeds.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 29.Farrell PM, et al. Guidelines for diagnosis of cystic fibrosis in newborns through older adults: Cystic Fibrosis Foundation consensus report. J Pediatr. 2008;153:S4–S14. doi: 10.1016/j.jpeds.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilschanski M, et al. Mutations in the cystic fibrosis transmembrane regulator gene and in vivo transepithelial potentials. Am J Respir. Crit Care Med. 2006;174:787–794. doi: 10.1164/rccm.200509-1377OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 32.Bhuvanagiri M, Schlitter AM, Hentze MW, Kulozik AE. NMD: RNA biology meets human genetic medicine. Biochem. J. 2010;430:365–377. doi: 10.1042/BJ20100699. [DOI] [PubMed] [Google Scholar]
- 33.Ramalho AS, et al. Transcript analysis of the cystic fibrosis splicing mutation 1525-1G>A shows use of multiple alternative splicing sites and suggests a putative role of exonic splicing enhancers. J Med. Genet. 2003;40:e88. doi: 10.1136/jmg.40.7.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Highsmith WE, Jr, et al. A novel mutation in the cystic fibrosis gene in patients with pulmonary disease but normal sweat chloride concentrations. N. Engl. J. Med. 1994;331:974–980. doi: 10.1056/NEJM199410133311503. [DOI] [PubMed] [Google Scholar]
- 35.Chillon M, et al. A novel donor splice site in intron 11 of the CFTR gene, created by mutation 1811+1.6kbA-->G, produces a new exon: high frequency in Spanish cystic fibrosis chromosomes and association with severe phenotype. Am J Hum. Genet. 1995;56:623–629. [PMC free article] [PubMed] [Google Scholar]
- 36.Highsmith WE, Jr, et al. Identification of a splice site mutation (2789+5G>A) associated with small amounts of normal CFTR mRNA and mild cystic fibrosis. Hum. Mutat. 1997;9:332–338. doi: 10.1002/(SICI)1098-1004(1997)9:4<332::AID-HUMU5>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 37.Beck S, et al. Cystic fibrosis patients with the 3272-26A-->G mutation have mild disease, leaky alternative mRNA splicing, and CFTR protein at the cell membrane. Hum. Mutat. 1999;14:133–144. doi: 10.1002/(SICI)1098-1004(1999)14:2<133::AID-HUMU5>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 38.Ramalho AS, et al. Five percent of normal cystic fibrosis transmembrane conductance regulator mRNA ameliorates the severity of pulmonary disease in cystic fibrosis. Am. J. Respir. Cell Mol. Biol. 2002;27:619–627. doi: 10.1165/rcmb.2001-0004OC. [DOI] [PubMed] [Google Scholar]
- 39.Dujardin G, Commandeur D, Le Jossic-Corcos C, Ferec C, Corcos L. Splicing defects in the CFTR gene: minigene analysis of two mutations, 1811+1G>C and 1898+3A>G. J. Cyst. Fibros. 2011;10:212–216. doi: 10.1016/j.jcf.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Chu C-S, Trapnell BC, Curristin SM, Cutting GR, Crystal RG. Extensive post-translational deletion of the coding sequences for part of nucleotide-binding fold 1 in respiratory epithelial mRNA transcripts of the cystic fibrosis transmembrane conductance regulator gene is not associated with the clinical manifestations of cystic fibrosis. J. Clin. Invest. 1992;90:785–790. doi: 10.1172/JCI115952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson LG, et al. Efficiency of gene transfer for restoration of normal airway epithelial function in cystic fibrosis. Nature Genet. 1993;2:21–25. doi: 10.1038/ng0992-21. [DOI] [PubMed] [Google Scholar]
- 42.Cheng SH, et al. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- 43.Mendoza JL, et al. Requirements for efficient correction of DeltaF508 CFTR revealed by analyses of evolved sequences. Cell. 2012;148:164–174. doi: 10.1016/j.cell.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bombieri C, et al. Recommendations for the classification of diseases as CFTR-related disorders. J Cyst. Fibros. 2011;10 Suppl 2:S86–102. doi: 10.1016/S1569-1993(11)60014-3. [DOI] [PubMed] [Google Scholar]
- 45.Taussig LM, Lobeck C, di Sant ’Agnese PA, Ackerman D, Kattwinkel J. Fertility in males with cystic fibrosis. N. Engl. J. Med. 1972;287:586–589. doi: 10.1056/NEJM197209212871204. [DOI] [PubMed] [Google Scholar]
- 46.Anguiano A, et al. Congenital bilateral absence of the vas deferens - a primarily genital form of cystic fibrosis. JAMA. 1992;267:1794–1797. [PubMed] [Google Scholar]
- 47.Abecasis GR, et al. An integrated map of genetic variation from 1, 092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chu C-S, Trapnell BC, Curristin S, Cutting GR, Crystal RG. Genetic basis of variable exon 9 skipping in cystic fibrosis transmembrane conductance regulator mRNA. Nature Genet. 1993;3:151–156. doi: 10.1038/ng0293-151. [DOI] [PubMed] [Google Scholar]
- 49.Estivill X. Complexity in a monogenic disease. Nature Genet. 1996;12:348–350. doi: 10.1038/ng0496-348. [DOI] [PubMed] [Google Scholar]
- 50.Castellani C, et al. European best practice guidelines for cystic fibrosis neonatal screening. J Cyst. Fibros. 2009;8:153–173. doi: 10.1016/j.jcf.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 51.Wagener JS, Zemanick ET, Sontag MK. Newborn screening for cystic fibrosis. Curr. Opin. Pediatr. 2012;24:329–335. doi: 10.1097/MOP.0b013e328353489a. [DOI] [PubMed] [Google Scholar]
- 52.Ramsey BW, et al. A CFTR potentiator in patients with cystic fibrosis and the G551D mutation. N. Engl. J. Med. 2011;365:1663–1672. doi: 10.1056/NEJMoa1105185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mitchell AA, Chakravarti A, Cutler DJ. On the probability that a novel variant is a disease-causing mutation. Genome Res. 2005;15:960–966. doi: 10.1101/gr.3761405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clarke GM, et al. Basic statistical analysis in genetic case-control studies. Nat. Protoc. 2011;6:121–133. doi: 10.1038/nprot.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cuppens H, Marynen P, De BC, Cassiman JJ. Detection of 98.5% of the mutations in 200 Belgian cystic fibrosis alleles by reverse dot-blot and sequencing of the complete coding region and exon/intron junctions of the CFTR gene. Genomics. 1993;18:693–697. doi: 10.1016/s0888-7543(05)80376-3. [DOI] [PubMed] [Google Scholar]
- 56.Bombieri C, et al. A new approach for identifying non-pathogenic mutations. An analysis of the cystic fibrosis transmembrane regulator gene in normal individuals. Hum. Genet. 2000;106:172–178. doi: 10.1007/s004390051025. [DOI] [PubMed] [Google Scholar]
- 57.Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST. American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet. Med. 2011;13:680–685. doi: 10.1097/GIM.0b013e3182217a3a. [DOI] [PubMed] [Google Scholar]
- 58.Pagani F, et al. New type of disease causing mutations: the example of the composite exonic regulatory elements of splicing in CFTR exon 12. Hum. Mol. Genet. 2003;12:1111–1120. doi: 10.1093/hmg/ddg131. [DOI] [PubMed] [Google Scholar]
- 59.Raynal C, et al. A Classification Model Relative to Splicing for Variants of Unknown Clinical Significance: Application to the CFTR Gene. Hum. Mutat. 2013 doi: 10.1002/humu.22291. [DOI] [PubMed] [Google Scholar]
- 60.Scott A, Petrykowska HM, Hefferon T, Gotea V, Elnitski L. Functional analysis of synonymous substitutions predicted to affect splicing of the CFTR gene. J. Cyst. Fibros. 2012;11:511–517. doi: 10.1016/j.jcf.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mailman MD, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Newcomb PA, et al. Colon Cancer Family Registry: an international resource for studies of the genetic epidemiology of colon cancer. Cancer Epidemiol. Biomarkers Prev. 2007;16:2331–2343. doi: 10.1158/1055-9965.EPI-07-0648. [DOI] [PubMed] [Google Scholar]
- 63.Tuffery-Giraud S, et al. Genotype-phenotype analysis in 2, 405 patients with a dystrophinopathy using the UMD-DMD database: a model of nationwide knowledgebase. Hum. Mutat. 2009;30:934–945. doi: 10.1002/humu.20976. [DOI] [PubMed] [Google Scholar]
- 64.Mons B, et al. The value of data. Nat. Genet. 2011;43:281–283. doi: 10.1038/ng0411-281. [DOI] [PubMed] [Google Scholar]
- 65.Castellani C, et al. Consensus on the use and interpretation of cystic fibrosis mutation analysis in clinical practice. J Cyst. Fibros. 2008;7:179–196. doi: 10.1016/j.jcf.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rohlfs EM, et al. The I148T CFTR allele occurs on multiple haplotypes: A complex allele is associated with cystic fibrosis. Genetics in Medicine. 2002;4:319–323. doi: 10.1097/00125817-200209000-00001. [DOI] [PubMed] [Google Scholar]
- 67.Wang X, Dockery DW, Wypij D, Fay ME, Ferris BG., Jr Pulmonary function between 6 and 18 years of age. Pediatr. Pulmonol. 1993;15:75–88. doi: 10.1002/ppul.1950150204. [DOI] [PubMed] [Google Scholar]
- 68.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir. Crit Care Med. 1999;159:179–187. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- 69.Cooper TA. Use of minigene systems to dissect alternative splicing elements. Methods. 2005;37:331–340. doi: 10.1016/j.ymeth.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 70.Clackson T, Winter G. ‘Sticky feet’-directed mutagenesis and its application to swapping antibody domains. Nucleic Acids Res. 1989;17:10163–10170. doi: 10.1093/nar/17.24.10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ramalho AS, Clarke LA, Amaral MD. Quantification of CFTR transcripts. Methods Mol. Biol. 2011;741:115–135. doi: 10.1007/978-1-61779-117-8_9. [DOI] [PubMed] [Google Scholar]
- 72.Sheridan MB, et al. CFTR transcription defects in pancreatic sufficient cystic fibrosis patients with only one mutation in the coding region of CFTR. J. Med. Genet. 2011;48:235–241. doi: 10.1136/jmg.2010.083287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yu H, et al. Ivacaftor potentiation of multiple CFTR channels with gating mutations. J. Cyst. Fibros. 2012;11:237–245. doi: 10.1016/j.jcf.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 74.Krasnov KV, Tzetis M, Cheng J, Guggino WB, Cutting GR. Localization studies of rare missense mutations in cystic fibrosis transmembrane conductance regulator (CFTR) facilitate interpretation of genotype-phenotype relationships. Hum. Mutat. 2008;29:1364–1372. doi: 10.1002/humu.20866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van Goor F, et al. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18825–18830. doi: 10.1073/pnas.0904709106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Neuberger T, Burton B, Clark H, Van Goor F. Use of primary cultures of human bronchial epithelial cells isolated from cystic fibrosis patients for the pre-clinical testing of CFTR modulators. Methods Mol. Biol. 2011;741:39–54. doi: 10.1007/978-1-61779-117-8_4. [DOI] [PubMed] [Google Scholar]
- 77.Thongnoppakhun W, et al. Simple, efficient, and cost-effective multiplex genotyping with matrix assisted laser desorption/ionization time-of-flight mass spectrometry of hemoglobin beta gene mutations. J. Mol. Diagn. 2009;11:334–346. doi: 10.2353/jmoldx.2009.080151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roger Harbord & Julian Higgins . METAREG: Stata module to perform meta-analysis regression. Boston College Department of Economics. Statistical Software Components; 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.