Abstract
A recent genome-wide association study (GWAS) of subjects from Japan and South Korea reported a novel association between the TP63 locus on chromosome 3q28 and risk of lung adenocarcinoma (p = 7.3 × 10−12); however, this association did not achieve genome-wide significance (p < 10−7) among never-smoking males or females. To determine if this association with lung cancer risk is independent of tobacco use, we genotyped the TP63 SNPs reported by the previous GWAS (rs10937405 and rs4488809) in 3,467 never-smoking female lung cancer cases and 3,787 never-smoking female controls from 10 studies conducted in Taiwan, Mainland China, South Korea, and Singapore. Genetic variation in rs10937405 was associated with risk of lung adenocarcinoma [n = 2,529 cases; p = 7.1 × 10−8; allelic risk = 0.80, 95% confidence interval (CI) = 0.74–0.87]. There was also evidence of association with squamous cell carcinoma of the lung (n = 302 cases; p = 0.037; allelic risk = 0.82, 95% CI = 0.67–0.99). Our findings provide strong evidence that genetic variation in TP63 is associated with the risk of lung adenocarcinoma among Asian females in the absence of tobacco smoking.
Introduction
In our initial genome-wide association study (GWAS) of lung cancer among never-smoking females from East Asia, we found that genetic variation in the CLPTM1L-TERT locus on 5pl5.33 was associated with risk of lung adenocarcinoma (Hsiung et al. 2010). Recently, a GWAS conducted among subjects from Japan and South Korea replicated the association between lung adenocarcinoma and the CLPTM1L-TERT locus in East Asians (Miki et al. 2010). This scan also identified a novel association between lung adenocarcinoma and the TP63 locus on 3q28 (Miki et al. 2010); however, when restricted to never-smoking males [odds ratios (OR) = 1.22, 95% confidence intervals (95% CI) = 0.96–1.56; p = 0.012] and never-smoking females [OR = 1.38, 95% CI = 1.20–1.57; p = 3.5 × 10−6 ], the association was less clear as it did not achieve genome-wide significance (p ≤ 10−7). To further explore the strength of this association with lung cancer risk in the absence of tobacco use, we expanded our initial pool of lung cancer studies conducted in East Asia (Hsiung et al. 2010) to include 10 studies conducted in Mainland China, Taiwan, South Korea, and Singapore and genotyped the TP63 SNPs (rsl0937405 and rs4488809) initially reported by Miki et al. (2010) in 3,467 never-smoking female cases and 3,787 never-smoking female controls.
Methods
Seven of the 10 lung cancer studies in East Asia pooled for this effort were described previously in our first lung cancer GWAS (Hsiung et al. 2010). These seven studies include the Genetic Epidemiological Study of Lung Adenocarcinoma (GELAC) (Jou et al. 2009), the Chinese Academy of Medical Sciences Cancer Hospital Study (CAMSCH) (Wu et al. 2009), the Seoul National University Study (SNU) (Kim et al. 2006), the Korea University Medical Center Study (KUMC) (Jung et al. 2008), the Kyungpook National University Hospital Study (KNUH) (Park et al. 2002), the Shanghai Women’s Health Cohort Study (SWHS) (Zhang et al. 2007; Zheng et al. 2005), and the Genes and Environment in Lung Cancer, Singapore Study (GEL-S) (Tang et al. 2010). For this effort, we pooled an additional three studies from Mainland China, including the Shenyang Lung Cancer Study (SLCS) (Yin et al. 2009), the Fudan Lung Cancer Study (FLCS), and the Tianjin Lung Cancer Study (TLCS) (Qian et al. 2011). All studies were case–control by design, except the SWHS, which was a prospective cohort study. Each study was approved by the local institutional review board and all study participants provided informed consent.
In the aggregate, we included 3,787 never-smoking female controls and 3,467 never-smoking female lung cancer cases, of which 2,557 (74%) were adenocarcinomas and 309 (9%) were squamous cell carcinomas. All cases and controls were genotyped for rs10937405 and rs4488809 using TaqMan assays designed by the National Cancer Institute Core Genotyping Facility (CGF). The TaqMan assays (Applied Biosystems Inc., Foster City, CA) were optimized on the ABI 7900HT detection system with high concordance with the sequence analysis of 102 individuals as listed on the SNP500Cancer website (http://www.snp500cancer.nci.nih.gov). SNU, KUMC, KNUH, and SWHS samples were all genotyped at the CGF. The GELAC and GEL-S studies were genotyped in Taiwan and Singapore, respectively. Genotyping for the CAMSCH, SLCS, TLCS and FLCS studies were conducted at their local institutions in Mainland China. All of the genotype frequencies showed fitness for Hardy–Weinberg proportion (p > 0.05) using a χ2 test. Genotype completion rates were greater than 97% across all studies.
Odds ratios (OR) and 95% confidence intervals (95% CI) were estimated by logistic regression models using the homozygote of the common allele as the reference group and adjusting for study and age as a continuous variable. Tests for trend were conducted by assigning the ordinal values (0,1,2) to the most prevalent genotypes in rank order of wild type, heterozygous, and variant homozygous genotypes, respectively. All statistical analyses were performed using SAS (Cary, NC).
Results
Ten lung cancer studies in East Asia included a total of 3,467 cases and 3,787 controls (Table 1). All cases and controls were never-smoking females. Cases and controls in each study were similar in age.
Table 1.
Region, study design, and number of cases and controls for each participating study
| Study center | Region | Cases (n = 3,467) | Controls (n = 3,787) | Age [mean (std)] |
Study design | |
|---|---|---|---|---|---|---|
| Cases | Controls | |||||
| GELAC | Taiwan | 1,193 | 1,145 | 59.6 (11.5) | 57.9 (12.6) | Case control |
| SLCS | Mainland China | 450 | 450 | 56.6 (12.1) | 56.4(11.4) | Case control |
| FLCS | Mainland China | 424 | 404 | 59.2 (11.4) | 59.9(11.3) | Case control |
| CAMSCH | Mainland China | 321 | 321 | 56.9 (10.3) | 48.5 (13.7) | Case control |
| TLCS | Mainland China | 300 | 306 | 57.2 (9.8) | 57.2 (9.9) | Case control |
| SNU | South Korea | 225 | 258 | 61.6 (10.4) | 60.7 (10.2) | Case control |
| GEL-S | Singapore | 193 | 537 | 63.3 (12.0) | 64.8 (11.5) | Case control |
| SWHS | Mainland China | 193 | 200 | 59.0 (8.2) | 58.6 (8.4) | Cohort |
| KNUH | South Korea | 97 | 90 | 61.7 (9.3) | 61.1 (6.9) | Case control |
| KUMC | South Korea | 71 | 76 | 62.7 (10.8) | 60.9 (6.5) | Case control |
KUMC Korea University Study, KNUH Kyungpook National University Study, FLCS Fudan Lung Cancer Study, SLCS Shenyang Lung Cancer Study, SWHS Shanghai Women’s Health Cohort Study, SNU Seoul National University Study, CAMSCH Chinese Academy of Medical Sciences Cancer Hospital Study, GEL-S Genes and Environment in Lung Cancer, Singapore Study, GELAC Genetic Epidemiological Study of Lung Adenocarcinoma (in Taiwan), TLCS Tainjin Lung Cancer Study
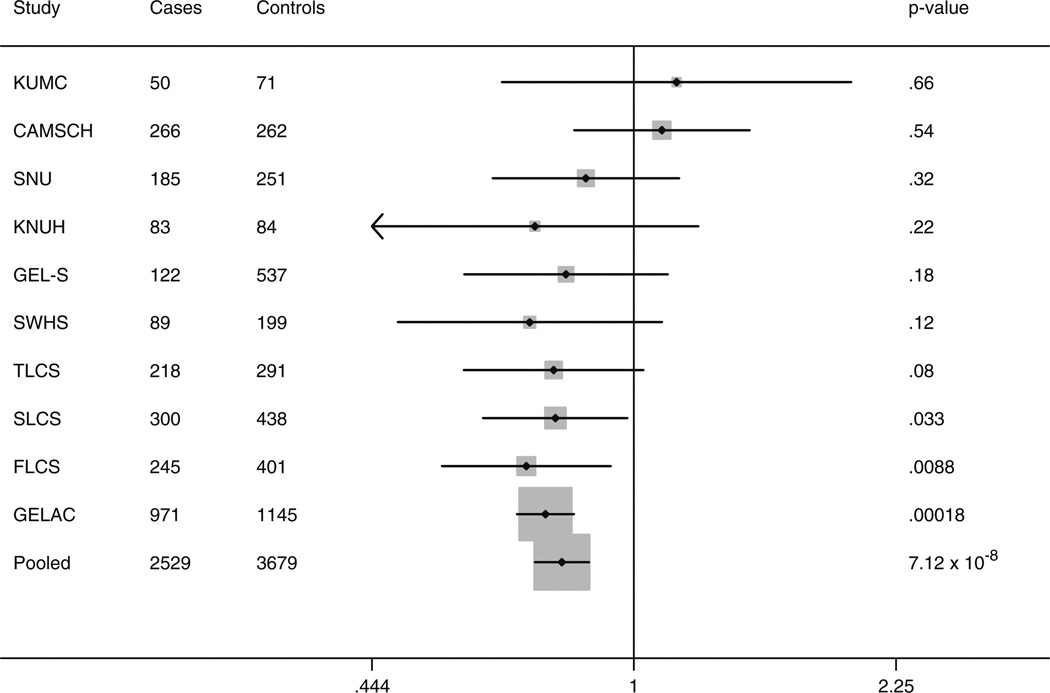
Of the two TP63 SNPs we genotyped, Table 2 shows that rs10937405 achieved genome-wide significance (p = 7.2 × 10−8; T allelic risk = 0.82, 95% CI = 0.76-0.88). The risk was predominately associated with the risk of lung adenocarcinoma (p = 7.1 × 10−8; allelic risk = 0.80, 95% CI = 0.74–0.87) (Table 2). The magnitude and direction of lung adenocarcinoma risk associated with rs10937405 were similar in most studies (heterogeneity p = 0.43) (Fig. 1; Supplemental Table 1). The T allele at rs10937405 was marginally associated with risk of squamous cell carcinoma (p = 0.037; allelic risk = 0.82, 95% CI = 0.67–0.99).
Table 2.
Lung cancer risk associated with TP63 rs 10937405 among never-smoking females from Asia, by histology
| rs number | Genotype | Controls |
Cases |
ORa | 95% CIa | p value | ||
|---|---|---|---|---|---|---|---|---|
| n | % | n | % | |||||
| rsl0937405 | All lung cancers | |||||||
| CC | 1,791 | 48.7 | 1,872 | 54.8 | ||||
| CT | 1,572 | 42.7 | 1,313 | 38.4 | 0.80 | 0.72–0.88 | 5.4 × 10−6 | |
| TT | 316 | 8.6 | 230 | 6.7 | 0.69 | 0.58–0.83 | 7.2 × 10−5 | |
| Trend | 0.82 | 0.76–0.88 | 7.2 × 10−8 | |||||
| Adenocarcinomas | ||||||||
| CC | 1,791 | 48.7 | 1,406 | 55.6 | ||||
| CT | 1,572 | 42.7 | 953 | 37.7 | 0.77 | 0.69–0.86 | 2.0 × 10−6 | |
| TT | 316 | 8.6 | 170 | 6.7 | 0.68 | 0.56–0.83 | 0.00015 | |
| Trend | 0.80 | 0.74–0.87 | 7.1 × 10−8 | |||||
| Squamous cell carcinomas | ||||||||
| CC | 1,791 | 48.7 | 163 | 54.0 | ||||
| CT | 1,572 | 42.7 | 121 | 40.1 | 0.84 | 0.66–1.07 | 0.17 | |
| TT | 316 | 8.6 | 18 | 6.0 | 0.63 | 0.38–1.03 | 0.068 | |
| Trend | 0.82 | 0.67–0.99 | 0.037 | |||||
Odds ratios (OR) and 95% confidence intervals (CI) adjusted for study and age
Fig. 1.
Lung adenocarcinoma risk associated with TP63 rs 10937405 among never-smoking females from Asia, by study. Odds ratios (OR) and 95% confidence intervals (CI) adjusted for age; KUMC Korea University Study, KNUH Kyungpook National University Study, FLCS Fudan Lung Cancer Study, SLCS Shenyang Lung Cancer Study, SWHS Shanghai Women’s Health Cohort Study, SNU Seoul National University Study, CAMSCH Chinese Academy of Medical Sciences Cancer Hospital Study, GELS Genes and Environment in Lung Cancer, Singapore Study, GELAC Genetic Epidemiological Study of Lung Adenocarcinoma (in Taiwan), TLCS Tainjin Lung Cancer Study
The associations between rs4488809 and lung cancer are provided for all lung cancer (p = 0.0011; allelic risk = 1.12, 95% CI = 1.05–1.19), adenocarcinoma (p = 7.4 × 10−5; allelic risk = 1.16, 95% CI = 1.08–1.24), and squamous cell carcinoma (p = 0.65; allelic risk = 0.96, 95% CI = 0.81–1.14) in Supplemental Table 2. Supplemental Table 3 presents the risk of rs4488809 associated with all lung cancers and lung adenocarcinomas by study. When both rsl0937405 (p = 0.0001; allelic risk = 0.82, 95% CI = 0.75–0.91) and rs4488809 (p = 0.31; allelic risk = 1.05, 95% CI = 0.96–1.14) were included in the same logistic regression model, only rs10937405 remained significant for adenocarcinoma.
Discussion
We have replicated the association between lung adeno-carcinoma and a SNP marker, rs10937405 that localizes to the TP63 gene. This marker was previously reported by a GWAS conducted in Japan and South Korea (Miki et al. 2010). We have been able to expand the initial findings by evaluating this risk in women who have never smoked tobacco. Contrary to Miki et al. (2010), TP63 rsl0937405 showed a stronger association with lung cancer in our population than did TP63 rs4488809. A recent study among Han Chinese also found that genetic variation in the TP63 region was associated with lung cancer risk, with the effects being more pronounced for rs4488809, although the results were not reported for never-smoking females (Hu et al. 2011).
The location of the SNP rs10937405 maps to the TP63 gene, whose product, p63, is an important component of the p53 family of genes. The p53 pathway plays a critical role in cell-cycle regulation by functioning as a tumor suppressor in numerous cancers, and p53 mutations occur in about two-thirds of all human cancer (Hernandez-Boussard et al. 1999). Further, p63 has been found to play an important role in cancer development and progression through its interaction with mutant p53 (Melino 2011). An isoform of TP63 has been proposed to have oncogenic properties on the basis of its dominant negative effects on p53, and TP63 genomic gains have been identified as potential indicators of pre-invasive lung lesions and early lung cancer diagnosis (Massion et al. 2009).
It is notable that lung cancer risk was not associated with TP63 variants at the genome-wide significance level in previous GWAS conducted in subjects of European descent (Amos et al. 2008; Hung et al. 2008; McKay et al. 2008; Wang et al. 2008; Landi et al. 2009). A GWAS carried out in Europe reported a suggestive but weak association between lung cancer and TP63 rs4488809 (Hung et al. 2008). Given that the allele frequencies are similar between the Asian and Caucasian HapMap populations for TP63 rs10937405 (T allele: Asian = 30%, Caucasian = 36%) and TP63 rs4488809 (T allele: Asian = 48%, Caucasian = 49%), and that the numbers of cases and controls in the Caucasian studies were larger than in the East Asian studies, these efforts had similar power to detect an association. The weaker association found in Caucasians compared with East Asians may be due to the different environmental exposures in North America and Europe compared to those in East Asia. In particular, the subjects in the European descent lung cancer GWAS were primarily tobacco smokers, whereas our Asian study populations were restricted to never-smokers. Alternatively, genomic coverage with the current platforms of the causal, functional variant in this region may be higher for Asian compared with Caucasian populations; therefore, the association between the tested SNPs and the causal variant may be lower in Caucasians as compared to Asians.
Globally, cigarette smoking is the primary risk factor for lung cancer, with 85% of tumors in men and 47% in women attributed to smoking tobacco (Parkin et al. 2005). Although only 10–15% of lung cancers occur in non-smokers, this still accounts for about 200,000 incident cancer cases worldwide per year. Extensive research has been conducted on the environmental and genetic determinants of nonsmoking lung cancer throughout Asia (Lan et al. 2000, 2002, 2004; Hosgood III et al. 2007, 2008; Yang et al. 2004; Xu et al. 1989; Lubin et al. 2005), where lung cancer rates among nonsmoking women in some regions are among the highest in the world (Mumford et al. 1987; Lam 2005). The relatively high rates of lung cancer among nonsmoking females in Asia are partially attributed to known environmental risk factors, such as exposure to combustion products of indoor heating and cooking solid fuel and cooking oil fumes (Lubin et al. 2005; Lan et al. 2002; Hosgood III et al. 2011). In-home coal used for heating and cooking has been classified as carcinogenic in humans (Straif et al. 2006), and genetic variation has been shown to influence this association (Hosgood III et al. 2007, 2008; Lan et al. 2000). North American and Western European populations do not typically experience these high levels of indoor air pollution exposures attributed to indoor heating and cooking.
In conclusion, our study of never-smoking lung cancer among Asian females has reported an association with genetic variants in the TP63 region. Since it is unlikely that genetic variation alone will explain the lung cancer burden among never-smoking females in Asia, further research is needed to identify environmental exposures and gene–environment interactions in these populations. Further, the functional genetic variants and mechanisms underpinning this association will require additional studies including fine mapping and laboratory studies.
Supplementary Material
Footnotes
Electronic supplementary material The online version of this article (doi: 10.1007/s00439-012-1144-8) contains supplementary material, which is available to authorized users.
Conflicts of interest The authors have no conflicts of interest to report.
Contributor Information
H. Dean Hosgood, III, Email: hosgoodd@mail.nih.gov, Division of Cancer Epidemiology and Genetics, National Cancer Institute, 6120 Executive Blvd., EPS 8118, MCS 7240, Bethesda, MD 20892-7240, USA.
Wen-Chang Wang, Division of Biostatistics and Bioinformatics, Institute of Population Health Sciences, National Health Research Institutes, Zhunan, Taiwan.
Yun-Chul Hong, Department of Preventive Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea; Institute of Environmental Medicine, Seoul National University Medical Research Center, Seoul, Republic of Korea.
Jiu-Cun Wang, Ministry of Education Key Laboratory of Contemporary Anthropology, State Key Laboratory of Genetic Engineering, School of Life Sciences, Fudan University, Shanghai 200433, China.
Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Medical, University Cancer Hospital and Institute, Tianjin 300060, People’s Republic of China.
I-Shou Chang, National Institute of Cancer Research National Health Research, Institutes, Zhunan, Taiwan.
Chien-Jen Chen, Genomics Research Center, Academia Sinica, Taipei, Taiwan; Graduate Institute of Epidemiology, College of Public Health, National Taiwan University, Taipei, Taiwan.
Daru Lu, Ministry of Education Key Laboratory of Contemporary Anthropology, State Key Laboratory of Genetic Engineering, School of Life Sciences, Fudan University, Shanghai 200433, China.
Zhihua Yin, Department of Epidemiology, School of Public Health, China Medical University, No 92 North Second Road, Heping District, Shenyang 110001, People’s Republic of China.
Chen Wu, Departments of Etiology and Carcinogenesis and State Key Laboratory of Molecular Oncology, Cancer Institute and Hospital, Chinese Academy of Medical Science and Peking, Union Medical College, Beijing, China.
Wei Zheng, Department of Medicine, Vanderbilt Epidemiology Center, Vanderbilt-Ingram Cancer Center, Institute for Medicine and Public Health, Vanderbilt University School of Medicine, Nashville, TN, USA.
Biyun Qian, Department of Epidemiology and Biostatistics, Tianjin Medical, University Cancer Hospital and Institute, Tianjin 300060, People’s Republic of China.
Jae Yong Park, Department of Internal Medicine, School of Medicine, Kyungpook National University, Daegu, Republic of Korea; Department of Biochemistry, School of Medicine, Kyungpook National University, Daegu, Republic of Korea; Cancer Research Center, Kyungpook National University Hospital, Daegu, Republic of Korea.
Yeul Hong Kim, Genomic Research Center for Lung and Breast/Ovarian Cancers, Korea University Anam Hospital, Seoul, Republic of Korea; Division of Brain, Department of Internal Medicine, Korea University College of Medicine, Seoul, Republic of Korea; Division of Oncology/Hematology, Department of Internal Medicine, Korea University College of Medicine, Seoul, Republic of Korea.
Nilanjan Chatterjee, Division of Cancer Epidemiology and Genetics, National Cancer Institute, 6120 Executive Blvd., EPS 8118, MCS 7240, Bethesda, MD 20892-7240, USA.
Ying Chen, Department of Epidemiology and Public Health, National University of Singapore, Singapore, Singapore.
Gee-Chen Chang, Division of Chest Medicine, Department of Internal Medicine, Taichung Veterans General Hospital, Taichung, Taiwan; Institute of Biomedical Sciences, National Chung-Hsing, University, Taichung, Taiwan; School of Medicine, China Medical University, Taichung, Taiwan; School of Medicine, National Yang-Ming University, Taipei, Taiwan.
Chin-Fu Hsiao, Division of Biostatistics and Bioinformatics, Institute of Population Health Sciences, National Health Research Institutes, Zhunan, Taiwan.
Meredith Yeager, Division of Cancer Epidemiology and Genetics, National Cancer Institute, 6120 Executive Blvd., EPS 8118, MCS 7240, Bethesda, MD 20892-7240, USA; Department of Health and Human Services, Core Genotyping Facility, Advanced Technology Program, Science Applications International Corporation-Frederick, National Cancer Institute, National Institutes of Health, Frederick, MD, USA.
Ying-Huang Tsai, Department of Pulmonary and Critical Care, Chang Gung Memorial Hospital, Chia Yi, Taiwan; Department of Respiratory Care, Chang Gung University, Taoyuan, Taiwan.
Hu Wei, Division of Cancer Epidemiology and Genetics, National Cancer Institute, 6120 Executive Blvd., EPS 8118, MCS 7240, Bethesda, MD 20892-7240, USA.
Young Tae Kim, Cancer Research Institute, Seoul National University College of Medicine, Seoul, Republic of Korea; Department of Thoracic and Cardiovascular Surgery, Clinical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea.
Wei Wu, Department of Epidemiology, School of Public Health, China Medical University, No 92 North Second Road, Heping District, Shenyang 110001, People’s Republic of China.
Zhenhong Zhao, Ministry of Education Key Laboratory of Contemporary Anthropology, State Key Laboratory of Genetic Engineering, School of Life Sciences, Fudan University, Shanghai 200433, China.
Wong-Ho Chow, Division of Cancer Epidemiology and Genetics, National Cancer Institute, 6120 Executive Blvd., EPS 8118, MCS 7240, Bethesda, MD 20892-7240, USA.
Xiaoling Zhu, Department of Epidemiology and Biostatistics, Tianjin Medical, University Cancer Hospital and Institute, Tianjin 300060, People’s Republic of China.
Yen-Li Lo, Division of Biostatistics and Bioinformatics, Institute of Population Health Sciences, National Health Research Institutes, Zhunan, Taiwan.
Sook Whan Sung, Department of Thoracic and Cardiovascular Surgery, Seoul National University College of Medicine, Seoul, Republic of Korea; Department of Thoracic and Cardiovascular Surgery, Seoul National University Bundang Hospital, Geongi-do, Republic of Korea.
Kuan-Yu Chen, Department of Internal Medicine, National Taiwan University Hospital and National Taiwan University College of Medicine, Taipei, Taiwan.
Jeff Yuenger, Division of Cancer Epidemiology and Genetics, National Cancer Institute, 6120 Executive Blvd., EPS 8118, MCS 7240, Bethesda, MD 20892-7240, USA; Department of Health and Human Services, Core Genotyping Facility, Advanced Technology Program, Science Applications International Corporation-Frederick, National Cancer Institute, National Institutes of Health, Frederick, MD, USA.
Joo Hyun Kim, Department of Preventive Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea; Institute of Environmental Medicine, Seoul National University Medical Research Center, Seoul, Republic of Korea; Department of Thoracic and Cardiovascular Surgery, Clinical Research Institute, Seoul National University Hospital, Seoul, Republic of Korea.
Liming Huang, Departments of Etiology and Carcinogenesis and State Key Laboratory of Molecular Oncology, Cancer Institute and Hospital, Chinese Academy of Medical Science and Peking, Union Medical College, Beijing, China.
Ying-Hsiang Chen, Division of Biostatistics and Bioinformatics, Institute of Population Health Sciences, National Health Research Institutes, Zhunan, Taiwan.
Yu-Tang Gao, Department of Epidemiology, Shanghai Cancer Institute, Shanghai, China.
Ming-Shyan Huang, School of Medicine, Kaohsiung Medical University, Kaohsiung, Taiwan; Department of Internal Medicine, Kaohsiung Medical University Hospital, Kaohsiung, Taiwan.
Tae Hoon Jung, Department of Internal Medicine, School of Medicine, Kyungpook National University, Daegu, Republic of Korea.
Neil Caporaso, Division of Cancer Epidemiology and Genetics, National Cancer Institute, 6120 Executive Blvd., EPS 8118, MCS 7240, Bethesda, MD 20892-7240, USA.
Xueying Zhao, Ministry of Education Key Laboratory of Contemporary Anthropology, State Key Laboratory of Genetic Engineering, School of Life Sciences, Fudan University, Shanghai 200433, China.
Zhang Huan, Department of Epidemiology and Biostatistics, Tianjin Medical, University Cancer Hospital and Institute, Tianjin 300060, People’s Republic of China.
Dianke Yu, Departments of Etiology and Carcinogenesis and State Key Laboratory of Molecular Oncology, Cancer Institute and Hospital, Chinese Academy of Medical Science and Peking, Union Medical College, Beijing, China.
Chang Ho Kim, Department of Internal Medicine, School of Medicine, Kyungpook National University, Daegu, Republic of Korea.
Wu-Chou Su, Department of Internal Medicine, National Cheng Kung University Hospital and College of Medicine, Tainan, Taiwan.
Xiao-Ou Shu, Department of Medicine, Vanderbilt Epidemiology Center, Vanderbilt-Ingram Cancer Center, Institute for Medicine and Public Health, Vanderbilt University School of Medicine, Nashville, TN, USA.
In-San Kim, Department of Biochemistry, School of Medicine, Kyungpook National University, Daegu, Republic of Korea.
Bryan Bassig, Division of Cancer Epidemiology and Genetics, National Cancer Institute, 6120 Executive Blvd., EPS 8118, MCS 7240, Bethesda, MD 20892-7240, USA.
Yuh-Min Chen, School of Medicine, National Yang-Ming University, Taipei, Taiwan; Chest Department, Taipei Veterans General Hospital, Taipei, Taiwan.
Sung Ick Cha, Department of Internal Medicine, School of Medicine, Kyungpook National University, Daegu, Republic of Korea.
Wen Tan, Departments of Etiology and Carcinogenesis and State Key Laboratory of Molecular Oncology, Cancer Institute and Hospital, Chinese Academy of Medical Science and Peking, Union Medical College, Beijing, China.
Hongyan Chen, Ministry of Education Key Laboratory of Contemporary Anthropology, State Key Laboratory of Genetic Engineering, School of Life Sciences, Fudan University, Shanghai 200433, China.
Tsung-Ying Yang, Division of Chest Medicine, Department of Internal Medicine, Taichung Veterans General Hospital, Taichung, Taiwan.
Jae Sook Sung, Genomic Research Center for Lung and Breast/Ovarian Cancers, Korea University Anam Hospital, Seoul, Republic of Korea; Division of Brain, Department of Internal Medicine, Korea University College of Medicine, Seoul, Republic of Korea.
Chih-Liang Wang, Department of Pulmonary and Critical Care, Chang Gung Memorial Hospital, Chia Yi, Taiwan.
Xuelian Li, Department of Epidemiology, School of Public Health, China Medical University, No 92 North Second Road, Heping District, Shenyang 110001, People’s Republic of China.
Kyong Hwa Park, Genomic Research Center for Lung and Breast/Ovarian Cancers, Korea University Anam Hospital, Seoul, Republic of Korea; Division of Oncology/Hematology, Department of Internal Medicine, Korea University College of Medicine, Seoul, Republic of Korea.
Chong-Jen Yu, Department of Internal Medicine, National Taiwan University Hospital and National Taiwan University College of Medicine, Taipei, Taiwan.
Jeong-Seon Ryu, Department of Internal Medicine, Inha University College of Medicine, Incheon, Republic of Korea.
Yongbing Xiang, Department of Epidemiology, Shanghai Cancer Institute, Shanghai, China.
Amy Hutchinson, Division of Cancer Epidemiology and Genetics, National Cancer Institute, 6120 Executive Blvd., EPS 8118, MCS 7240, Bethesda, MD 20892-7240, USA; Department of Health and Human Services, Core Genotyping Facility, Advanced Technology Program, Science Applications International Corporation-Frederick, National Cancer Institute, National Institutes of Health, Frederick, MD, USA.
Jun Suk Kim, Genomic Research Center for Lung and Breast/Ovarian Cancers, Korea University Anam Hospital, Seoul, Republic of Korea; Division of Brain, Department of Internal Medicine, Korea University College of Medicine, Seoul, Republic of Korea; Division of Oncology/Hematology, Department of Internal Medicine, Korea University College of Medicine, Seoul, Republic of Korea.
Qiuyin Cai, Department of Medicine, Vanderbilt Epidemiology Center, Vanderbilt-Ingram Cancer Center, Institute for Medicine and Public Health, Vanderbilt University School of Medicine, Nashville, TN, USA.
Maria Teresa Landi, Division of Cancer Epidemiology and Genetics, National Cancer Institute, 6120 Executive Blvd., EPS 8118, MCS 7240, Bethesda, MD 20892-7240, USA.
Kyoung-Mu Lee, Department of Preventive Medicine, Seoul National University College of Medicine, Seoul, Republic of Korea.
Jen-Yu Hung, School of Medicine, Kaohsiung Medical University, Kaohsiung, Taiwan; Department of Internal Medicine, Kaohsiung Medical University Hospital, Kaohsiung, Taiwan.
Ju-Yeon Park, Cancer Research Institute, Seoul National University College of Medicine, Seoul, Republic of Korea.
Margaret Tucker, Division of Cancer Epidemiology and Genetics, National Cancer Institute, 6120 Executive Blvd., EPS 8118, MCS 7240, Bethesda, MD 20892-7240, USA.
Chien-Chung Lin, Department of Internal Medicine, National Cheng Kung University Hospital and College of Medicine, Tainan, Taiwan.
Yangwu Ren, Department of Epidemiology, School of Public Health, China Medical University, No 92 North Second Road, Heping District, Shenyang 110001, People’s Republic of China.
Reury-Perng Perng, Chest Department, Taipei Veterans General Hospital, Taipei, Taiwan.
Chih-Yi Chen, Cancer Center, China Medical University Hospital, Taipei, Taiwan.
Li Jin, Ministry of Education Key Laboratory of Contemporary Anthropology, State Key Laboratory of Genetic Engineering, School of Life Sciences, Fudan University, Shanghai 200433, China.
Kun-Chieh Chen, Division of Chest Medicine, Department of Internal Medicine, Taichung Veterans General Hospital, Taichung, Taiwan.
Yao-Jen Li, Graduate Institute of Epidemiology, College of Public Health, National Taiwan University, Taipei, Taiwan.
Yu-Fang Chiu, Division of Biostatistics and Bioinformatics, Institute of Population Health Sciences, National Health Research Institutes, Zhunan, Taiwan.
Fang-Yu Tsai, National Institute of Cancer Research National Health Research, Institutes, Zhunan, Taiwan.
Pan-Chyr Yang, Department of Internal Medicine, National Taiwan University Hospital and National Taiwan University College of Medicine, Taipei, Taiwan.
Joseph F. Fraumeni, Jr., Division of Cancer Epidemiology and Genetics, National Cancer Institute, 6120 Executive Blvd., EPS 8118, MCS 7240, Bethesda, MD 20892-7240, USA
Adeline Seow, Department of Epidemiology and Public Health, National University of Singapore, Singapore, Singapore.
Dongxin Lin, Departments of Etiology and Carcinogenesis and State Key Laboratory of Molecular Oncology, Cancer Institute and Hospital, Chinese Academy of Medical Science and Peking, Union Medical College, Beijing, China.
Baosen Zhou, Department of Epidemiology, School of Public Health, China Medical University, No 92 North Second Road, Heping District, Shenyang 110001, People’s Republic of China.
Stephen Chanock, Division of Cancer Epidemiology and Genetics, National Cancer Institute, 6120 Executive Blvd., EPS 8118, MCS 7240, Bethesda, MD 20892-7240, USA; Department of Health and Human Services, Core Genotyping Facility, Advanced Technology Program, Science Applications International Corporation-Frederick, National Cancer Institute, National Institutes of Health, Frederick, MD, USA.
Chao Agnes Hsiung, Division of Biostatistics and Bioinformatics, Institute of Population Health Sciences, National Health Research Institutes, Zhunan, Taiwan.
Nathaniel Rothman, Division of Cancer Epidemiology and Genetics, National Cancer Institute, 6120 Executive Blvd., EPS 8118, MCS 7240, Bethesda, MD 20892-7240, USA.
Qing Lan, Division of Cancer Epidemiology and Genetics, National Cancer Institute, 6120 Executive Blvd., EPS 8118, MCS 7240, Bethesda, MD 20892-7240, USA.
References
- Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, Dong Q, Zhang Q, Gu X, Vijayakrishnan J, Sullivan K, Matakidou A, Wang Y, Mills G, Doheny K, Tsai YY, Chen WV, Shete S, Spitz MR, Houlston RS. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Boussard T, Rodriguez-Tome P, Montesano R, Hainaut P. IARC p53 mutation database: a relational database to compile and analyze p53 mutations in human tumors and cell lines. International Agency for Research on Cancer. Hum Mutat. 1999;14:1–8. doi: 10.1002/(SICI)1098-1004(1999)14:1<1::AID-HUMU1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Hosgood HD, III, Wei H, Sapkota A, Choudhury I, Bruce N, Smith KR, Rothman N, Lan Q. Household coal use and lung cancer: systematic review and meta-analysis of case–control studies, with an emphasis on geographic variation. Int J Epidemiol. 2011;40(3):719–728. doi: 10.1093/ije/dyq259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosgood HD, III, Berndt SI, Lan Q. GST genotypes and lung cancer susceptibility in Asian populations with indoor air pollution exposures: a meta-analysis. Mutat Res. 2007;636:134–143. doi: 10.1016/j.mrrev.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosgood HD, III, Menashe I, Shen M, Yeager M, Yuenger J, Rajaraman P, He X, Chatterjee N, Caporaso N, Zhu Y, Chanock S, Zheng T, Lan Q. Pathway-based evaluation of 380 candidate genes and lung cancer susceptibility suggests the importance of the cell cycle pathway. Carcinogenesis. 2008;29:1938–1943. doi: 10.1093/carcin/bgn178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiung CA, Lan Q, Hong YC, Chen CJ, Hosgood HD, Chang IS, Chatterjee N, Brennan P, Wu C, Zheng W, Chang GC, Wu T, Park JY, Hsiao CF, Kim YH, Shen H, Seow A, Yeager M, Tsai YH, Kim YT, Chow WH, Guo H, Wang WC, Sung SW, Hu Z, Chen KY, Kim JH, Chen Y, Huang L, Lee KM, Lo YL, Gao YT, Kim JH, Liu L, Huang MS, Jung TH, Jin G, Caporaso N, Yu D, Kim CH, Su WC, Shu XO, Xu P, Kim IS, Chen YM, Ma H, Shen M, Cha SI, Tan W, Chang CH, Sung JS, Zhang M, Yang TY, Park KH, Yuenger J, Wang CL, Ryu JS, Xiang Y, Deng Q, Hutchinson A, Kim JS, Cai Q, Landi MT, Yu CJ, Park JY, Tucker M, Hung JY, Lin CC, Perng RP, Boffetta P, Chen CY, Chen KC, Yang SY, Hu CY, Chang CK, Fraumeni JF, Chanock S, Yang PC, Rothman N, Lin D. The 5pl5.33 locus is associated with risk of lung adenocarcinoma in never-smoking females in Asia. PLoS Genet. 2010;6:el001051. doi: 10.1371/journal.pgen.1001051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Wu C, Shi Y, Guo H, Zhao X, Yin Z, Yang L, Dai J, Hu L, Tan W, Li Z, Deng Q, Wang J, Wu W, Jin G, Jiang Y, Yu D, Zhou G, Chen H, Guan P, Chen Y, Shu Y, Xu L, Liu X, Liu L, Xu P, Han B, Bai C, Zhao Y, Zhang H, Yan Y, Ma H, Chen J, Chu M, Lu F, Zhang Z, Chen F, Wang X, Jin L, Lu J, Zhou B, Lu D, Wu T, Lin D, Shen H. A genome-wide association study identifies two new lung cancer susceptibility loci at 13ql2.12 and 22ql2.2 in Han Chinese. Nat Genet. 2011;43(8):792–796. doi: 10.1038/ng.875. [DOI] [PubMed] [Google Scholar]
- Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, Chen C, Goodman G, Field JK, Liloglou T, Xinarianos G, Cassidy A, McLaughlin J, Liu G, Narod S, Krokan HE, Skorpen F, Elvestad MB, Hveem K, Vatten L, Linseisen J, Clavel-Chapelon F, Vineis P, Bueno-de-Mesquita HB, Lund E, Martinez C, Bingham S, Rasmuson T, Hainaut P, Riboli E, Ahrens W, Benhamou S, Lagiou P, Trichopoulos D, Holcatova I, Merletti F, Kjaerheim K, Agudo A, Macfarlane G, Talamini R, Simonato L, Lowry R, Conway DI, Znaor A, Healy C, Zelenika D, Boland A, Delepine M, Foglio M, Lechner D, Matsuda F, Blanche H, Gut I, Heath S, Lathrop M, Brennan P. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature. 2008;452:633–637. doi: 10.1038/nature06885. [DOI] [PubMed] [Google Scholar]
- Jou YS, Lo YL, Hsiao CF, Chang GC, Tsai YH, Su WC, Chen YM, Huang MS, Chen HL, Chen CJ, Yang PC, Hsiung CA. Association of an EGFR intron 1 SNP with never-smoking female lung adenocarcinoma patients. Lung cancer (Amsterdam, Netherlands) 2009;64:251–256. doi: 10.1016/j.lungcan.2008.09.014. [DOI] [PubMed] [Google Scholar]
- Jung HY, Whang YM, Sung JS, Shin HD, Park BL, Kim JS, Shin SW, Seo HY, Seo JH, Kim YH. Association study of TP53 polymorphisms with lung cancer in a Korean population. J Hum Genet. 2008;53:508–514. doi: 10.1007/s10038-008-0278-y. [DOI] [PubMed] [Google Scholar]
- Kim JH, Kim H, Lee KY, Choe KH, Ryu JS, Yoon HI, Sung SW, Yoo KY, Hong YC. Genetic polymorphisms of ataxia telangiectasia mutated affect lung cancer risk. Hum Mol Genet. 2006;15:1181–1186. doi: 10.1093/hmg/ddl033. [DOI] [PubMed] [Google Scholar]
- Lam WK. Lung cancer in Asian women-the environment and genes. Respirology. 2005;10:408–17. doi: 10.1111/j.1440-1843.2005.00723.x. [DOI] [PubMed] [Google Scholar]
- Lan Q, He X, Costa DJ, Tian L, Rothman N, Hu G, Mumford JL. Indoor coal combustion emissions, GSTM1 and GSTT1 genotypes, and lung cancer risk: a case-control study in Xuan Wei, China. Cancer Epidemiol Biomarkers Prev. 2000;9:605–608. [PubMed] [Google Scholar]
- Lan Q, Chapman RS, Schreinemachers DM, Tian L, He X. Household stove improvement and risk of lung cancer in Xuanwei, China. J Natl Cancer Inst. 2002;94:826–835. doi: 10.1093/jnci/94.11.826. [DOI] [PubMed] [Google Scholar]
- Lan Q, Mumford JL, Shen M, DeMarini DM, Bonner MR, He X, Yeager M, Welch R, Chanock S, Tian L, Chapman RS, Zheng T, Keohavong P, Caporaso N, Rothman N. Oxidative damage-related genes AKR1C3 and OGG1 modulate risks for lung cancer due to exposure to PAH-rich coal combustion emissions. Carcinogenesis. 2004;25:2177–2181. doi: 10.1093/carcin/bgh240. [DOI] [PubMed] [Google Scholar]
- Landi MT, Chatterjee N, Yu K, Goldin LR, Goldstein AM, Rotunno M, Mirabello L, Jacobs K, Wheeler W, Yeager M, Bergen AW, Li Q, Consonni D, Pesatori AC, Wacholder S, Thun M, Diver R, Oken M, Virtamo J, Albanes D, Wang Z, Burdette L, Doheny KF, Pugh EW, Laurie C, Brennan P, Hung R, Gaborieau V, McKay JD, Lathrop M, McLaughlin J, Wang Y, Tsao MS, Spitz MR, Wang Y, Krokan H, Vatten L, Skorpen F, Arnesen E, Benhamou S, Bouchard C, Metsapalu A, Vooder T, Nelis M, Valk K, Field JK, Chen C, Goodman G, Sulem P, Thorleifsson G, Rafnar T, Eisen T, Sauter W, Rosenberger A, Bickeboller H, Risch A, Chang-Claude J, Wichmann HE, Stefansson K, Houlston R, Amos CI, Fraumeni JF, Jr, Savage SA, Bertazzi PA, Tucker MA, Chanock S, Caporaso NE. A genome-wide association study of lung cancer identifies a region of chromosome 5pl5 associated with risk for adenocarcinoma. Am J Hum Genet. 2009;85:679–691. doi: 10.1016/j.ajhg.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin JH, Wang ZY, Wang LD, Boice JD, Jr, Cui HX, Zhang SR, Conrath S, Xia Y, Shang B, Cao JS, Kleinerman RA. Adjusting lung cancer risks for temporal and spatial variations in radon concentration in dwellings in Gansu Province, China. Radiat Res. 2005;163:571–579. doi: 10.1667/rr3109. [DOI] [PubMed] [Google Scholar]
- Massion PP, Zou Y, Uner H, Kiatsimkul P, Wolf HJ, Baron AE, Byers T, Jonsson S, Lam S, Hirsch FR, Miller YE, Franklin WA, Varella-Garcia M. Recurrent genomic gains in preinvasive lesions as a biomarker of risk for lung cancer. PLoS ONE. 2009;4:e5611. doi: 10.1371/journal.pone.0005611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay JD, Hung RJ, Gaborieau V, Boffetta P, Chabrier A, Byrnes G, Zaridze D, Mukeria A, Szeszenia-Dabrowska N, Lissowska J, Rudnai P, Fabianova E, Mates D, Bencko V, Foretova L, Janout V, McLaughlin J, Shepherd F, Montpetit A, Narod S, Krokan HE, Skorpen F, Elvestad MB, Vatten L, Njolstad I, Axelsson T, Chen C, Goodman G, Barnett M, Loomis MM, Lubinski J, Matyjasik J, Lener M, Oszutowska D, Field J, Liloglou T, Xinarianos G, Cassidy A, Vineis P, Clavel-Chapelon F, Palli D, Tumino R, Krogh V, Panico S, Gonzalez CA, Ramon QJ, Martinez C, Navarro C, Ardanaz E, Larranaga N, Kham KT, Key T, Bueno-de-Mesquita HB, Peeters PH, Trichopoulou A, Linseisen J, Boeing H, Hallmans G, Overvad K, Tjonneland A, Kumle M, Riboli E, Zelenika D, Boland A, Delepine M, Foglio M, Lechner D, Matsuda F, Blanche H, Gut I, Heath S, Lathrop M, Brennan P. Lung cancer susceptibility locus at 5pl5.33. Nat Genet. 2008;40:1404–1406. doi: 10.1038/ng.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melino G. p63 is a suppressor of tumorigenesis and metastasis interacting with mutant p53. Cell Death Differ. 2011;18:1487–1499. doi: 10.1038/cdd.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki D, Kubo M, Takahashi A, Yoon KA, Kim J, Lee GK, Zo JI, Lee JS, Hosono N, Morizono T, Tsunoda T, Kamatani N, Chayama K, Takahashi T, Inazawa J, Nakamura Y, Daigo Y. Variation in TP63 is associated with lung adenocarcinoma susceptibility in Japanese and Korean populations. Nat Genet. 2010;42:893–896. doi: 10.1038/ng.667. [DOI] [PubMed] [Google Scholar]
- Mumford JL, He XZ, Chapman RS, Cao SR, Harris DB, Li XM, Xian YL, Jiang WZ, Xu CW, Chuang JC. Lung cancer and indoor air pollution in Xuan Wei, China. Science. 1987;235:217–220. doi: 10.1126/science.3798109. [DOI] [PubMed] [Google Scholar]
- Park JY, Lee SY, Jeon HS, Bae NC, Chae SC, Joo S, Kim CH, Park JH, Kam S, Kim IS, Jung TH. Polymorphism of the DNA repair gene XRCC1 and risk of primary lung cancer. Cancer Epidemiol Biomarkers Prev. 2002;11:23–27. [PubMed] [Google Scholar]
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- Qian B, Zhang H, Zhang L, Zhou X, Yu H, Chen K. Association of genetic polymorphisms in DNA repair pathway genes with non-small cell lung cancer risk. Lung Cancer. 2011;73(2):138–146. doi: 10.1016/j.lungcan.2010.11.018. [DOI] [PubMed] [Google Scholar]
- Straif K, Baan R, Grosse Y, Secretan B, El F, Ghissassi F, Cogliano V. Carcinogenicity of household solid fuel combustion and of high-temperature frying. Lancet Oncol. 2006;7:977–978. doi: 10.1016/s1470-2045(06)70969-x. [DOI] [PubMed] [Google Scholar]
- Tang L, Lim WY, Eng P, Leong SS, Lim TK, Ng AW, Tee A, Seow A. Lung cancer in Chinese women: evidence for an interaction between tobacco smoking and exposure to inhalants in the indoor environment. Environ Health Perspect. 2010;118:1257–1260. doi: 10.1289/ehp.0901587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Broderick P, Webb E, Wu X, Vijayakrishnan J, Matakidou A, Qureshi M, Dong Q, Gu X, Chen WV, Spitz MR, Eisen T, Amos CI, Houlston RS. Common 5pl5.33 and 6p21.33 variants influence lung cancer risk. Nat Genet. 2008;40:1407–1409. doi: 10.1038/ng.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Hu Z, Yu D, Huang L, Jin G, Liang J, Guo H, Tan W, Zhang M, Qian J, Lu D, Wu T, Lin D, Shen H. Genetic variants on chromosome 15q25 associated with lung cancer risk in Chinese populations. Cancer Res. 2009;69:5065–5072. doi: 10.1158/0008-5472.CAN-09-0081. [DOI] [PubMed] [Google Scholar]
- Xu ZY, Blot WJ, Xiao HP, Wu A, Feng YP, Stone BJ, Sun J, Ershow AG, Henderson BE, Fraumeni JF., Jr Smoking, air pollution, and the high rates of lung cancer in Shenyang, China. J Natl Cancer Inst. 1989;81:1800–1806. doi: 10.1093/jnci/81.23.1800. [DOI] [PubMed] [Google Scholar]
- Yang XR, Wacholder S, Xu Z, Dean M, Clark V, Gold B, Brown LM, Stone BJ, Fraumeni JF, Jr, Caporaso NE. CYP1A1 and GSTM1 polymorphisms in relation to lung cancer risk in Chinese women. Cancer Lett. 2004;214:197–204. doi: 10.1016/j.canlet.2004.06.040. [DOI] [PubMed] [Google Scholar]
- Yin Z, Su M, Li X, Li M, Ma R, He Q, Zhou B. ERCC2, ERCC1 polymorphisms and haplotypes, cooking oil fume and lung adenocarcinoma risk in Chinese non-smoking females. J Exp Clin Cancer Res. 2009;28:153. doi: 10.1186/1756-9966-28-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Shu XO, Gao YT, Ji BT, Yang G, Li HL, Kilfoy B, Rothman N, Zheng W, Chow WH. Family history of cancer and risk of lung cancer among nonsmoking Chinese women. Cancer Epidemiol Biomarkers Prev. 2007;16:2432–2435. doi: 10.1158/1055-9965.EPI-07-0398. [DOI] [PubMed] [Google Scholar]
- Zheng W, Chow WH, Yang G, Jin F, Rothman N, Blair A, Li HL, Wen W, Ji BT, Li Q, Shu XO, Gao YT. The Shanghai Women’s Health Study: rationale, study design, and baseline characteristics. Am J Epidemiol. 2005;162:1123–1131. doi: 10.1093/aje/kwi322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.