Abstract
Capillary electrophoresis is a common technique used for glycosaminoglycan-derived disaccharide analysis because of its high resolving power, high separation efficiency, high sensitivity, short analysis time, and straightforward operation. CE coupled to laser-induced fluorescence (LIF) detection shows an approximately 100 times higher sensitivity than traditional UV detection at 232 nm. 2-Aminoacridone (AMAC) is a widely used fluorophore for labeling unsaturated disaccharides by deductive amination, which is one of the most important method of derivatization of disaccharides for CE-LIF detection. Outlined in this chapter is a protocol of analyzing glycosaminoglycan-derived disaccharides by CE-LIF with AMAC derivatization.
Keywords: Glycosaminoglycan, Disaccharide, 2-Aminoacridone, Capillary electrophoresis, Laser-induced fluorescence
1. Introduction
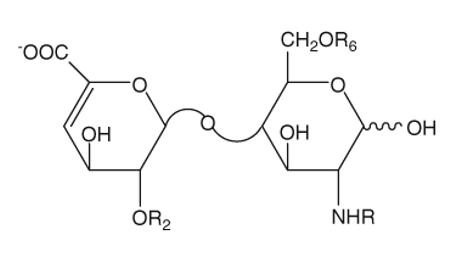
Heparin (HP), heparan sulfate (HS), chondroitin sulfate (CS), and dermatan sulfate (DS) are linear, highly charged polysaccharides that belong to the glycosaminoglycan (GAG) family (1), which are known to mediate in many life processes. HP is a widely used anticoagulant drug (2), and both HP and HS are implicated in cellular processes, such as regulation of enzymic catalysis, and cell–cell interaction (3, 4). CS/DS may be involved in participating and mediating cell–cell interaction and communication (5). The polysaccharide chains of HP/HS and CS/DS have closely related structures and consist of a repeating disaccharide structure, which consists of a hexosamine and a uronic acid. For HP/HS, the hexosamine could be either N-acetylated (GlcNAc), N-sulfonated (GlcNS), or unsubstituted (GlcNH), all of which can be 3- and/or 6-O-sulfonated; the uronic acid could be either glucuronic acid (GlcA) or iduronic acid (IdoA), both of which can be 2-O-sulfonated. For CS/DS, the hexosamine is N-acetylgalactosamine (GalNAc) and can be sulfated at 4- and/or 6-position, and the uronic acid could be sulfated at 2 position. Eight, commercially available, enzymatically prepared, HP/HS unsaturated disaccharide standards are described in Table 1, and CD/DS disaccharide standards are described in Table 2. Defining the structure of GAGs is an important factor in elucidating their structure–activity relationship. A common strategy for detailed structural analysis of GAGs involves either complete or partial depolymerization by either enzymatic or chemical means to obtain constituent disaccharides, for disaccharide analysis (6). Modern separation techniques, including high-performance liquid chromatography (HPLC) (7), gel permeation chromatography (GPC) (8, 9), polyacrylamide gel electrophoresis (PAGE) (10), and CE (11, 12), have been applied to HP/HS and CS/DS analysis to help solve many complex structures.
Table 1.
The structures of the eight Δ-disaccharide standards from HP/HS
| Reference number | Disaccharide | Formulas | R2 | R6 | R |
|---|---|---|---|---|---|
| 1 | TriS | ΔUA(2S)-GlcNS(6S) | SO3− | SO3− | SO3− |
| 2 | 2S6S | ΔUA(2S)-GlcNAc(6S) | SO3− | SO3− | Ac |
| 3 | 2SNS | ΔUA(2S)-GlcNS | SO3− | H | SO3− |
| 4 | NS6S | ΔUA-GlcNS(6S) | H | SO3− | SO3− |
| 5 | 2S | ΔUA(2S)-GlcNAc | SO3− | H | Ac |
| 6 | 6S | ΔUA-GlcNAc(6S) | H | SO3− | Ac |
| 7 | NS | ΔUA-GlcNS | H | H | SO3− |
| 8 | 0S | ΔUA-GlcNAc | H | H | Ac |
Table 2.
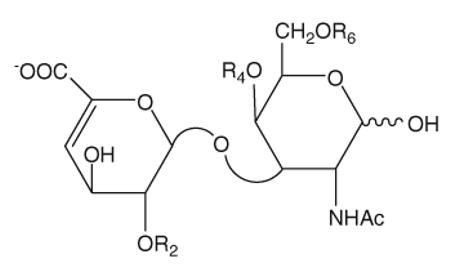
The structures of the eight Δ-disaccharide standards from CS/DS
| Reference number | Disaccharide | Formulas | R2 | R4 | R6 |
|---|---|---|---|---|---|
| 9 | TriS | ΔUA(2S)-GalNAc(4S)(6S) | SO3− | SO3− | SO3− |
| 10 | SD | ΔUA(2S)-GalNAc(6S) | SO3− | H | SO3− |
| 11 | SB | ΔUA(2S)-GalNAc(4S) | SO3− | SO3− | H |
| 12 | SE | ΔUA-GalNAc(4S)(6S) | H | SO3− | SO3− |
| 13 | 2S | ΔUA(2S)-GalNAc | SO3− | H | H |
| 14 | 6S | ΔUA-GalNAc(6S) | H | H | SO3− |
| 15 | 4S | ΔUA-GalNAc(4S) | H | SO3− | H |
| 16 | 0S | ΔUA-GalNAc | H | H | Ac |
Capillary electrophoresis (CE) is one of the most powerful techniques for GAG analysis because of its high resolving power, high separation efficiency, high sensitivity, short analysis time, straightforward operation (13), and compatibility with a variety of detection methods, including MS, NMR, and LIF (6). GAG-derived disaccharides can be detected by CE with UV detector at 232 nm created by the unsaturated bond in nonreducing uronic acid residue, which has an extinction coefficient of approximately 5,500 M−1 cm−1 (14). The addition of a fluorophore can greatly change the chromatographic properties of GAG-derived disaccharides and increase the sensitivity when detected by both UV and LIF detector (16). Reductive amination is one of the most frequently used derivatization method, and a number of labeling reagents have been applied to GAG analysis, such as 2-aminopyridine (2-AP), 7-aminonaphthalene-1,3-disulfonic acid (ANDS), 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS), and 2-aminoacridone (AMAC) (15). AMAC is a neutral fluorophore with λexc = 428 nm and λem = 525 nm, which has been previously used to HP/HS and CS/DS analysis (11, 12, 16–20). By using AMAC as labeling molecule, sensitivity for detection of GAG-derived disaccharides is greatly enhanced, and resolution is also improved (15).
2. Materials
Prepare all solutions with HPLC-grade water (purchased from Sigma-Aldrich) and analytical-grade reagents unless indicated otherwise. Prepare and store all solutions and reagents at room temperature unless indicated otherwise.
2.1. Separation of Eight HP/HS Unsaturated Disaccharide Standards and Eight CS/DS Disaccharide Standards by High-Performance Capillary Electrophoresis
2.1.1. Derivatization of Unsaturated HP/HS and CS/DS Disaccharides with AMAC
HP/HS and CS/DS unsaturated disaccharide standards (see Note 1).
AMAC (≥98%, Sigma-Aldrich). Store at −20°C.
Dimethyl sulfoxide (DMSO).
Labeling solution. 0.1 M AMAC in glacial acetic acid–DMSO (3:17, v/v). Store at 4°C.
Reducing agent. 1 M NaBH3 CN:82.84 mg sodium cyanoborohydride (NaBH3CN) in 1 mL water (see Note 2).
Reconstitution solution. DMSO–water (1:1, v/v). Store at 4°C.
Lyophilizer.
Centrifuge.
Water bath set to 45°C.
2.1.2. Separation and Calibration Curve of Eight HP/HS Disaccharide Standards and Eight CS/DS Disaccharide Standards
HPCE system (Agilent Technologies).
ZetaLif (Picometrics, France) detector (λexc = 488 nm).
Uncoated fused-silica capillary (50 μm i.d., 85 cm total length, 70 cm effective length).
Preconditioning solution. 1 M sodium hydroxide (NaOH) (see Note 3).
0.22 μm Steritop filters Millipore.
Running buffer. 50 mM phosphate buffer, pH 3.5 (see Note 4). Dissolve 0.6000 g monosodium phosphate (NaH2PO4) in about 90 mL of HPLC-grade water and titrate with 1 M HCl to pH 3.5. Make up volume to 100 mL.
2.2. Analysis of GAG-Derived Disaccharides in Biological Sample
2.2.1. Extract and Enzymic Degradation of GAGs
PBS buffer: Dissolve 8 g sodium chloride (NaCl), 0.20 g potassium chloride, 1.44 g disodium phosphate (Na2HPO4), and 0.24 g monopotassium phosphate (KH2PO4) in 800 mL of water. pH should be adjusted to 7.4 with dilute HCl. Make up volume with water to 1 L.
- Defatting Solutions
- 2:1 chloroform–methanol: Mix two parts chloroform to one part methanol (v/v)
- 1:1 chloroform–methanol: Mix equal parts of chloroform and methanol
- 1:2 chloroform–methanol: Mix one part chloroform with two parts methanol
Actinase E solution: 2 mg/mL solution of actinase E (Kaken Pharmaceutical, Japan). Add water to dry enzyme powder and let it dissolve slowly. This should be prepared immediately before use.
Water bath set to 55°C.
Tissue homogenizer: LabGEN 7 (Cole-Parmer, Illinois).
Strong anion-exchange column: Vivapure Q IEX H ion exchange columns (Sartorius Stedim, France)
Urea/chaps solution: Dissolve 480.48 g urea and 20 g chaps (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate) in 1 L of water.
200 mM NaCl solution: 11.69 g NaCl in 1 L water.
16% NaCl solution: 160 g NaCl in 1 L water.
Dialysis membrane (3, 10, 30 kDa MWCO Spectra/Por Dialysis Membrane, Spectrum Labs, California)
Centrifugal filters (3 and 3.5 kDa MWCO Amicon Ultra Centrifugal Filter Units, Millipore, Massachusetts)
Heparin lyase I (EC 4.2.2.7), heparin lyase II (no EC assigned), and heparin lyase III (EC 4.2.2.8). Cloning, E. coli expression, and purification of the recombinant heparin lyase I, II, and III from F. heparinum were performed in our laboratory as described (21–23). These enzymes are commercially available (Sigma Chemical, Missouri).
3. Methods
Carry out all procedures at room temperature unless otherwise specified.
3.1. Separation of Eight HP/HS Unsaturated Disaccharide Standards and Eight CS/DS Unsaturated Disaccharide Standards
3.1.1. Derivatization of Unsaturated HP/HS and CS/DS with AMAC
Lyophilize unsaturated disaccharide standards.
Add 5 μL (per 10 nmol of disaccharides) of 0.1 M AMAC in glacial acetic acid–DMSO (3:17, v/v) solution to the lyophilized disaccharides (see Note 5).
Add 5 μL (per 10 nmol of disaccharides) of reducing agent 1 M NaBH3CN to the reaction mixture (see Note 6).
Incubate the reaction mixture in water bath at 45°C for 4 h.
Make mixtures of HP/HS and CS/DS unsaturated disaccharide standards and make up the reaction mixture with re-constitution solution to desired volume (see Note 7).
3.1.2. Separation and Calibration Curve of Eight HP/HS Unsaturated Disaccharide Standards and Eight CS/DS Unsaturated Disaccharide Standards
- Precondition the capillary before each run (see Note 8).
- Flush with 1 M NaOH for 2 min
- Flush with water for 2 min
- Flush with running buffer for 3 min
Inject the sample by pressure mode for 50 mbar × 10 s at reversed polarity.
Separation is taken under 30 kV. The separation profiles of eight HP/HS unsaturated disaccharides and eight CS/DS unsaturated disaccharides are shown in Figs. 1 and 2.
Run a series of disaccharide standard solutions of different concentrations. Draw calibration curve for HP/HS and CS/DS (see Table 3).
Fig. 1.
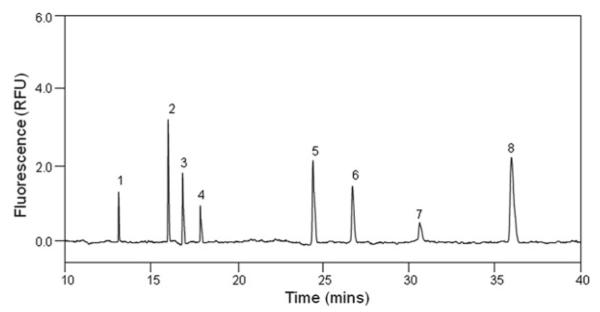
Electrophoregram of eight HP/HS Δ-disaccharides. Analysis was performed at 25°C, pressure injection of 50 mbar × 10 s, using 50 mM phosphate buffer, pH 3.5, under 30 kV with reversed polarity.
Fig. 2.
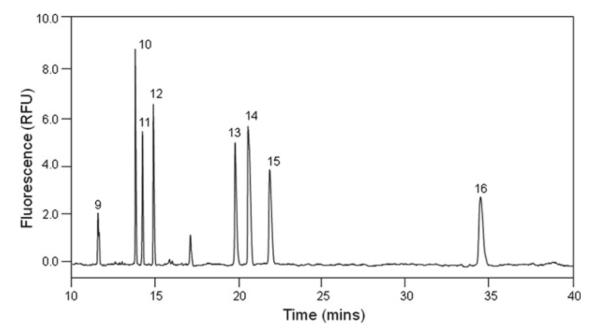
Electrophoregram of eight CS/DS Δ-disaccharides. Analysis was performed at 25°C, pressure injection of 50 mbar × 10 s, using 50 mM phosphate buffer, pH 3.5, under 30 kV with reversed polarity.
Table 3.
Linearity equations for AMAC-derivatized HP/HS and CS/D Δ-disaccharides
| Disaccharide type | Reference number | Disaccharide | Linearity equations |
|---|---|---|---|
| HP/HSa | 1 | TriS | Υ = 0.72718 X + 0.06179, R2 = 0.98859 |
| 2 | 2S6S | Υ = 2.34914 X + 0.28360, R2 = 0.99523 | |
| 3 | NS2S | Υ = 1.25743 X + 0.30320, R2 = 0.98814 | |
| 4 | NS6S | Υ = 0.83603 X + 0.17811, R2 = 0.98841 | |
| 5 | 2S | Υ = 6.05419 X + 0.55524, R2 = 0.99884 | |
| 6 | 6S | Υ = 4.33948 X + 0.34730, R2 = 0.99313 | |
| 7 | NS | Υ = 5.02764 X + 0.75179, R2 = 0.98820 | |
| 8 | 0S | Υ = 7.66238 X + 1.03039, R2 = 0.99517 | |
| CS/DSb | 9 | TriS | Υ = 1.86003 X + 1.87668, R2 = 0.99517 |
| 10 | SD | Υ = 1.68848 X + 8.25444, R2 = 0.97362 | |
| 11 | SB | Υ = 1.01521 X + 3.28328, R2 = 0.97650 | |
| 12 | SE | Υ = 1.63450 X + 1.91876, R2 = 0.99964 | |
| 13 | 2S | Υ = 3.03737 X + 7.05318, R2 = 0.99149 | |
| 14 | 6S | Υ = 3.14119 X + 6.78632, R2 = 0.99826 | |
| 15 | 4S | Υ = 2.69588 X + 6.61650, R2 = 0.99273 | |
| 16 | 0S | Υ = 1.55614 X + 8.42892, R2 = 0.98708 |
Tested range is from 0.2 to 5 ng/μL
Tested range is from 0.3 to 25 ng/μL
3.2. Analysis of GAG-Derivatized Disaccharides in Biological Sample
3.2.1. Extract and Enzymic Degradation of GAGs
Cut tissues into 2–3 cm square pieces. Wash tissues in PBS buffer to remove excess blood. Freeze-dry to remove excess water.
Defat tissues using defatting solutions A–C. Immerse tissue successively in each solution, starting with A, for 12–24 h. Pour off solution (see Note 9). Allow tissue to fully dry (to remove all organic solvent) before continuing.
Submerse dry, defatted tissue in actinase E solution and incubate at 55°C for at least 12 h. Use 5–10 mL solution for every 1 g of tissue. After the first 12 h, homogenize tissue with tissue homogenizer. If undigested tissue remains, additional enzyme may be added and incubate for additional time as needed.
Filter digested material through 0.22 μm filter to remove unsolubilized material. Add urea and chaps to make the resulting filtered solution 8 M urea and 2 wt.% chaps (0.48 g urea and 0.02 g chaps for every mL of solution).
Isolate GAGs from tissue using strong anion-exchange (SAX) column. GAGs were isolated following a modified version of the manufacturer’s protocol. The following should be briefly done: Wash column 2–3 times with 1 column volume (c.v.) of water. Wash column with 1 c.v. of urea/chaps solution. Load filtered, digested sample (see Note 10). Wash sample 3 times with 1 c.v. of 200 mM NaCl. Elute sample with 2 washes of 0.5 c.v. 16% NaCl.
For every 1 mL of purified GAGs isolated above, add 4 mL methanol (methanol to make 80% of total solution). Mix well, place in 4°C fridge overnight. Centrifuge at 5,000 × g for 30 min and pour off supernatant to isolate precipitated GAG.
Isolated GAG is then desalted by dissolving in a minimal amount of water and dialyzed (against 1 L water for every 10 mg of GAG) or loaded onto a spin column (3 kDa MWCO) and washed with 5 column volumes of water.
GAG samples (5 μg) were incubated with the chondroitinase ABC (5 mU) and chondroitinase ACII (2 mU) at 37°C for 10 h. The enzymatic products were recovered by centrifugal filtration (30 kDa MWCO). CS/DS disaccharides, passed through the filter, were freeze-dried and ready for CE-LIF analysis.
The heparinase I, II, and III (5 mU each) were added into the remainder and incubated at 37°C for 10 h. The products were again recovered by centrifugal filtration (10 kDa MWCO), and the HP/HS disaccharides were similarly collected and freeze-dried and ready for CE-LIF analysis.
3.2.2. Derivatization of GAG-Derived Disaccharides with AMAC
Add 5 μL (per estimated 10 nmol of disaccharides) of 0.1 M AMAC in glacial acetic acid–DMSO (3:17, v/v) solution to the lyophilized disaccharides.
Add 5 μL (per estimated 10 nmol of disaccharides) of reducing agent 1 M NaBH3CN to the reaction mixture.
Incubate the reaction mixture at 45°C for 4 h.
Make up the reaction mixture with reconstitution solution to desired volume.
3.2.3. Determination of GAG-Derived Disaccharides
Precondition the capillary and inject the sample with the same procedure as suggested in Subheading 3.1.2 . Electrophoregrams are shown in Figs. 3 and 4; peaks were identified by either coinjection or comparing with standard unsaturated disaccharide profiles. Presence of GAG-derivatized disaccharides was also proven by mass spectrometry.
Fig. 3.
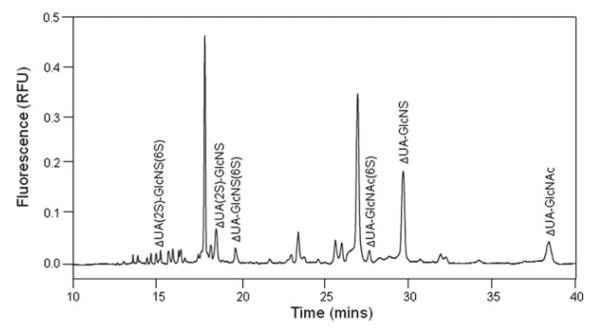
Electrophoregram of HP/HS-derivatized disaccharides from camel liver. Analysis was performed at 25°C, pressure injection of 50 mbar × 10 s, using 50 mM phosphate buffer, pH 3.5, under 30 kV with reversed polarity. Unlabeled small and large peaks are fluorescent compounds not corresponding to the eight HP/HS-derivatized disaccharide standards.
Fig. 4.
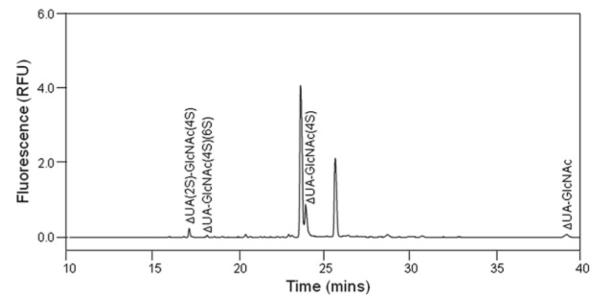
Electrophoregram of CS/DS-derivatized disaccharides from camel liver. Analysis was performed at 25°C, pressure injection of 50 mbar × 10 s, using 50 mM phosphate buffer, pH 3.5, under 30 kV with reversed polarity. Unlabeled small and large peaks are fluorescent compounds not corresponding to the eight CS/DS-derivatized disaccharide standards.
4. Notes
Unsaturated disaccharide standards of CS/DS (ΔDi-0S: ΔUA-GalNAc, ΔDi-4S: ΔUA-GalNAc4S, ΔDi-6S: ΔUAGalNAc6S, ΔDi-2S: ΔUA2S-GalNAc, ΔDi-diSB: ΔUA2S-GalNAc4S, ΔDi-diSD: ΔUA2S-GalNAc6S, ΔDi-diSE: ΔUA-GalNAc4S6S, ΔDi-triS: ΔUA2SGalNAc4S6S). Unsaturated disaccharide standards of heparin/HS (ΔDi-0S: ΔUA-GlcNAc, ΔDi-NS: ΔUA-GlcNS, ΔDi-6S: ΔUA-GlcNAc6S, ΔDi-2S: ΔUA2S-GlcNAc, ΔDi-2SNS: ΔUA2S-GlcNS, ΔDi-NS6S: ΔUA-GlcNS6S, ΔDi-2S6S: ΔUA2S-GlcNAc6S, ΔDi-triS: ΔUA2S-GlcNS6S) were obtained from SEIKAGAKU CORPORATION (Japan). Make 1 μg/μL standard solution with HPLC-grade water and store at −20°C.
The reducing agent solution must be made fresh prior to use.
All solutions used for capillary electrophoresis must be filtered through a 0.22 μm membrane filter and degassed before use.
Be aware that pH will change with temperature and buffer additives. This separation buffer is suggested by Militsopoulou et al. (11). It is worth noting that below pH 4.0, the ionization of silanols is low and the electroosmotic flow mobility is insignificant. So when conventional electrode polarity is reversed, negatively charged disaccharides are drawn to the anode only under the influence of electrophoresis.
The coupling reaction between unsaturated disaccharides and the fluorophore proceeds through reductive amination, which involves the reducing end of disaccharide and amine group of AMAC. 100–500 times of excess free AMAC tag is used to produce high derivatization yield.
The purpose to add NaBH3CN to reaction mixture is to stabilize the conjugate formed from reductive amination between unsaturated disaccharides and AMAC.
Under the suggested separation conditions, excess AMAC receives positive charge, which means that AMAC will not enter the capillary at reversed polarity mode. This is proven by injecting AMAC solution only into the capillary and applying the same separation condition. No peak is found in the electrophoregram.
Condition of capillary is critical to migration time of analytes and peak shape. Generally, new capillary will be washed with MeOH, 1 M HCl, 1 M NaOH, and operating buffer, with water flushing at each interval, until the baseline is good enough for analysis. After each day, the capillary should be washed with 1 M NaOH and water for 5 min and dried with air.
If necessary, defatting solutions may need to be filtered as they are removed to retain small tissue particles. As a general rule of thumb, use about 10–50 mL of each defatting solution for every 1 g of dry tissue weight; more can be used for fattier tissues.
Each column will have a max binding capacity, which will be the absolute amount of GAG the column can isolate; overloading the column will result in sample loss. The amount of each digested, filtered tissue to load onto each column should be based on the anticipated GAG content of the tissue which can be found in literature (typically on the order of 10–50 mg/g dry tissue).
References
- 1.Volpi N, Maccari F, Linhardt RJ. Capillary electrophoresis of complex natural polysaccharides. Electrophoresis. 2008;29:3095–3106. doi: 10.1002/elps.200800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Linhardt RJ. Heparin: an important drug enters its seventh decade. Chem Ind. 1991;2:45–50. [Google Scholar]
- 3.Grag HG, Linhardt RJ, Hales CA. Chemistry and Biology of Heparin and Heparan Sulfate. Elsevier B. V; New York, NY: 2005. [Google Scholar]
- 4.Linhardt RJ, Turnbull JE, Wang HM, Loganathan D, Gallagher JT. Examination of the substrate specificity of heparin and heparan sulfate lyases. Biochemistry. 1990;29:2611–2617. doi: 10.1021/bi00462a026. [DOI] [PubMed] [Google Scholar]
- 5.Alicia M, Hitchcock AM, Yates KE, Shortkroff S, Costello CE, Zaia J. Optimized extraction of glycosaminoglycans from normal and osteoarthritic cartilage for glycomics profiling. Glycobiology. 2007;17:25–35. doi: 10.1093/glycob/cwl046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang B, Solakyildirim K, Chang Y, Linhardt RJ. Hyphenated techniques for the analysis of heparin and heparan sulfate. Anal Bioanal Chem. 2011;399:541–557. doi: 10.1007/s00216-010-4117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rice KG, Kim YS, Grant AC, Merchant ZM, Linhardt RJ. High-performance liquid chromatographic separation of heparin derived oligosaccharides. Anal Biochem. 1985;150:325–331. doi: 10.1016/0003-2697(85)90518-4. [DOI] [PubMed] [Google Scholar]
- 8.Hileman RE, Smith AE, Toida T, Linhardt RJ. Preparation and structure of heparin lyase-derived heparan sulfate oligosaccharides. Glycobiology. 1997;7:231–239. doi: 10.1093/glycob/7.2.231. [DOI] [PubMed] [Google Scholar]
- 9.Chuang WL, McAllister H, Rabenstein L. Chromatographic methods for product-profile analysis and isolation of oligosaccharides produced by heparinase-catalyzed depolymerization of heparin. J Chromatogr A. 2001;932:65–74. doi: 10.1016/s0021-9673(01)01241-9. [DOI] [PubMed] [Google Scholar]
- 10.Rice KG, Rottink MK, Linhardt RJ. Fractionation of heparin-derived oligosaccharides by gradient polyacrylamide-gel electrophoresis. Biochem J. 1987;244:515–522. doi: 10.1042/bj2440515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Militsopoulou M, Lamari FN, Hjerpe A, Karamanos NK. Determination of twelve heparin- and heparan sulfate-derived disaccharides as 2-aminoacridone derivatives by capillary zone electrophoresis using ultraviolet and laser-induced fluorescence detection. Electrophoresis. 2002;23:1104–1109. doi: 10.1002/1522-2683(200204)23:7/8<1104::AID-ELPS1104>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 12.Mitropoulou TN, Lamari F, Syrokou A, Hjerpe A, Karamanos NK. Identification of oligomeric domains within dermatan sulfate chains using differential enzymic treatments, derivatization with 2-aminoacridone and capillary electrophoresis. Electrophoresis. 2001;22:2458–2463. doi: 10.1002/1522-2683(200107)22:12<2458::AID-ELPS2458>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 13.Mao W, Thanawiroon C, Linhardt RJ. Capillary electrophoresis for the analysis of glycosaminoglycans and glycosaminoglycan-derived oligosaccharides. Biomed Chromatogr. 2002;16:77–94. doi: 10.1002/bmc.153. [DOI] [PubMed] [Google Scholar]
- 14.Skidmore MA, Guimond SE, Dumax-Vorzet AF, Atrih A, Yates EA, Turnbull JE. High sensitivity separation and detection of heparan sulfate disaccharides. J Chromatogr A. 2006;1135:52–56. doi: 10.1016/j.chroma.2006.09.064. [DOI] [PubMed] [Google Scholar]
- 15.Lamari FN, Kuhn R, Karamanos NK. Derivatization of carbohydrates for chromatographic, electrophoretic and mass spectrometric structure analysis. J Chromatogr B. 2003;793:15–36. doi: 10.1016/s1570-0232(03)00362-3. [DOI] [PubMed] [Google Scholar]
- 16.Militsopoulou M, Lecomte C, Bayle C, Couderc F, Karamanos NK. Laser-induced fluorescence as a powerful detection tool for capillary electrophoretic analysis of heparin/heparan sulfate disaccharides. Biomed Chromatogr. 2003;17:39–41. doi: 10.1002/bmc.207. [DOI] [PubMed] [Google Scholar]
- 17.Mastrogianni O, Lamari F, Syrokou A, Militsopoulou M, Hjerpe A, Karamanos NK. Microemulsion electrokinetic capillary chromatography of sulfated disaccharides derived from glycosaminoglycans. Electrophoresis. 2001;22:2743–2745. doi: 10.1002/1522-2683(200108)22:13<2743::AID-ELPS2743>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 18.Hitchcock AM, Bowman MJ, Staples GO, Zaia J. Improved workup for glycosaminoglycan disaccharide analysis using CE with LIF detection. Electrophoresis. 2008;29:4538–4548. doi: 10.1002/elps.200800335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zinellu A, Pisanu S, Zinellu E, Lepedda AJ, Cherchi GM, Sotgia S, Carru C, Deiana L, Formato M. A novel LIF-CE method for the separation of hyaluronan- and chondroitin sulfate-derived disaccharides: application to structural and quantitative analyses of human plasma low- and high-charged chondroitin sulfate isomers. Electrophoresis. 2007;28:2439–2447. doi: 10.1002/elps.200600668. [DOI] [PubMed] [Google Scholar]
- 20.Viola M, Vigetti D, Karousou E, Bartolini B, Genasetti A, Rizzi M, Clerici M, Pallotti F, Luca GD, Passi A. New electrophoretic and chromatographic techniques for analysis of heparin and heparan sulfate. Electrophoresis. 2008;29:3168–3174. doi: 10.1002/elps.200700855. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida E, Arakawa S, Matsunaga T, Toriumi S, Tokuyama S, Morikawa K, Tahara Y. Cloning, sequencing, and expression of the gene from bacillus circulans that codes for a heparinase that degrades both heparin and heparan sulfate. Biosci Biotechnol Biochem. 2002;66:1873–1879. doi: 10.1271/bbb.66.1873. [DOI] [PubMed] [Google Scholar]
- 22.Shaya D, Tocilj A, Li Y, Myette J, Venkataraman G, Sasisekharan R, Cygler M. Crystal structure of heparinase II from Pedobacter heparinus and its complex with a disaccharide product. J Biol Chem. 2006;281:15525–15535. doi: 10.1074/jbc.M512055200. [DOI] [PubMed] [Google Scholar]
- 23.Godavarti R, Davis M, Venkataraman G, Cooney C, Langer R, Sasisekharan R. Heparinase III from Flavobacterium heparinum: cloning and recombinant expression in Escherichia coli. Biochem Biophys Res Commun. 1996;225:751–758. doi: 10.1006/bbrc.1996.1246. [DOI] [PubMed] [Google Scholar]


