Abstract
Electrospun nanofiber meshes have emerged as a new generation of scaffold membranes possessing a number of features suitable for tissue regeneration. One of these features is the flexibility to modify their structure and composition to orchestrate specific cellular responses. In this study, we investigated the effects of nanofiber orientation and surface functionalization on human mesenchymal stem cell (hMSC) migration and osteogenic differentiation. We used an in vitro model to examine hMSC migration into a cell-free zone on nanofiber meshes and mitomycin C treatment to assess the contribution of proliferation to the observed migration. Poly (ɛ-caprolactone) meshes with oriented topography were created by electrospinning aligned nanofibers on a rotating mandrel, while randomly oriented controls were collected on a stationary collector. Both aligned and random meshes were coated with a triple-helical, type I collagen-mimetic peptide, containing the glycine-phenylalanine-hydroxyproline-glycine-glutamate-arginine (GFOGER) motif. Our results indicate that nanofiber GFOGER peptide functionalization and orientation modulate cellular behavior, individually, and in combination. GFOGER significantly enhanced the migration, proliferation, and osteogenic differentiation of hMSCs on nanofiber meshes. Aligned nanofiber meshes displayed increased cell migration along the direction of fiber orientation compared to random meshes; however, fiber alignment did not influence osteogenic differentiation. Compared to each other, GFOGER coating resulted in a higher proliferation-driven cell migration, whereas fiber orientation appeared to generate a larger direct migratory effect. This study demonstrates that peptide surface modification and topographical cues associated with fiber alignment can be used to direct cellular behavior on nanofiber mesh scaffolds, which may be exploited for tissue regeneration.
Introduction
Biomaterial-based implants offer a robust therapeutic strategy to improve tissue regeneration and construct integration.1 Acellular approaches for tissue regeneration, in which the implanted biomaterial recruits endogenous cells for repair, may be more readily translated into clinical practice than cell-based therapies.2–5 This is due to the technical challenges of cell delivery and survival, and the commercial difficulties associated with the manufacturing and storage of cells and obtaining regulatory approval. For purely biomaterial-based therapies to be effective, the biomaterial is usually required to function as both a scaffold and a biologically active agent to provide specific molecular signals for regulating cellular responses.6,7 The rational design of the material structure and composition is therefore essential for implant success.
Electrospun nanofiber meshes are a unique type of scaffold with structural features that, at least by scale, resemble the extracellular matrix (ECM). In addition, they exhibit large surface area and high porosity, making them suitable as a scaffold for guiding tissue regeneration by host cells.8–12 In a previous study, we demonstrated that nanofiber meshes made from a synthetic polymer are able to support the attachment, colonization, and osteogenic differentiation of progenitor cells.13 Synthetic polymers, however, lack biological ligands, and are not capable of directing intracellular signaling and response. Nanofiber meshes have also been fabricated from natural materials such as collagen and fibrinogen,14–16 but these are limited by poor mechanical strength and handling characteristics for in vivo applications.17 Approaches that incorporate bioactive molecules within a synthetic polymer backbone may provide an optimal combination of biological activity and mechanical integrity.
Tremendous advances have been made in imparting biofunctionality to synthetic materials by coating them with ECM components. These biomimetic material surfaces present adhesion motifs to engage the cell signal transduction machinery for directing cellular responses and tissue repair.6,18 Although adhesive proteins, such as type I collagen, fibronectin, and laminin, have been immobilized on material surfaces, these approaches are limited by protein purification and processing issues and a potential host immunogenic response.19–22 In addition, the multiple adhesion domains in a full-length protein may trigger conflicting intracellular signals, leading to suboptimal tissue repair. Therefore, there is a great need to develop peptides that mimic specific domains of natural proteins. These ECM-mimetic peptides can be synthesized and purified with relative ease, and further, can be designed to trigger a specific cellular response.23–27 One such peptide that has been recently investigated is a triple-helical, collagen-mimetic oligopeptide containing the glycine-phenylalanine-hydroxyproline-glycine-glutamate-arginine (GFOGER) domain from residues 502–507 of the α1(I) chain of type I collagen.28,29 It has been shown that the interaction of this adhesion motif with α2β1 integrin mediates osteoblast adhesion, differentiation, and matrix mineralization.30,31 This has been exploited to enhance the adhesion and osteogenic differentiation of progenitor cells and improve implant integration and bone formation by coating surfaces with the GFOGER peptide.32–34 This technique utilizes simple adsorption of the GFOGER peptide on implant surfaces in physiologic conditions, which may provide an additional advantage for clinical translation.
Another set of guidance strategies consists of topographical cues to influence cellular responses. It is now accepted that surface morphology, including roughness and texture, modulates cellular responses. For instance, titanium implants with rough microtopographies reduced the cell number and increased differentiation of osteoblast-like cells, thereby enhancing implant integration.35–38 The electrospinning process can be easily adapted to obtain fibrous matrices with varying structures. Fiber alignment, especially, has generated significant interest due to the fact that a number of native and regenerating tissues display an ordered architecture. Studies have shown that alignment of fibers along a particular direction affects cellular attachment and morphology as well as matrix deposition.39–41
The purpose of this study was to investigate the effects of nanofiber functionalization with the GFOGER peptide and orientation on hMSC function, to identify conditions that promote osteoprogenitor cell migration and differentiation. Nanofiber meshes were functionalized with the GFOGER peptide to improve cell migration and osteogenic differentiation. An oriented topography was obtained by electrospinning aligned nanofibers to enhance cellular migration. The individual and combined effects of nanofiber functionalization and orientation on hMSC function were investigated. We hypothesized that functionalizing nanofiber surfaces with the GFOGER peptide and aligning nanofiber orientation will modulate cell migration and differentiation in vitro.
Materials and Methods
Electrospinning and nanofiber mesh characterization
Nanofiber meshes were made by electrospinning, and characterized as described elsewhere.42 Briefly, a 12% (w/v) solution of poly (ɛ-caprolactone) (PCL) was made in a 90:10 volume ratio of hexafluoro-2-propanol:dimethylformamide (Sigma-Aldrich). A 3-mL syringe (Becton-Dickinson) was filled with the PCL solution and fitted with a 22-gauge blunt-end needle (Jensen Global, Inc.). The syringe was placed on a syringe pump (Harvard Apparatus), which was adjusted for a flow rate of 0.75 mL/h. To create a nanofiber mesh with random fiber alignment (random nanofiber mesh), a flat copper plate was placed at a distance of 20–23 cm from the needle tip. To obtain meshes with fibers aligned along the same direction (aligned nanofiber mesh), plates were collected on a mandrel rotating at ∼2500 rpm and placed 8–10 cm from the need tip. The syringe needle was attached to the positive end of a high voltage power supply (Gamma High Voltage Research), and the collector was grounded to create the electrostatic field required for electrospinning. After applying a voltage of 13–20 kV, the polymer solution was ejected from the needle toward the collector and deposited as nanoscaled fibers. The fibers were collected for 45–60 min to obtain meshes with sufficient thickness for cell culture experiments.
The nanofiber meshes were sputter coated with gold (Quorum Technologies) and their morphology visualized by scanning electron microscopy (SEM; Hitachi HTA). A custom MATLAB® (The MathWorks, Inc.) program was used to calculate the individual fiber diameter from the SEM images. The alignment of the fibers was quantified by measuring the fiber angle relative to the direction of rotation, using the Image-Pro software (Media Cybernetics, Inc.). In the case of random nanofiber meshes, the angles were measured with respect to an arbitrarily set line.
GFOGER peptide preparation and nanofiber surface coating
The peptide, GGYGGGPC(GPP)5GFOGER(GPP)5GPC, was synthesized by the Emory University Microchemical Facility as described previously.34 This peptide contains the GFOGER motif, where O refers to hydroxyproline. The purified peptide was lyophilized as a trifluoroacetic acid (TFA) salt. The peptide was reconstituted at a concentration of 10 mg/mL in a 0.1% TFA solution containing 0.01% sodium azide (NaN3). The stock solution was diluted to 50 μg/mL in phosphate-buffered saline (PBS; Mediatech, Inc.). Nanofiber mesh samples were sterilized by ethanol evaporation, wetted with 70% ethanol, and rinsed with excess PBS. The samples were then passively coated with GFOGER by submerging them in the dilute GFOGER solution for 2 h at room temperature or overnight at 4°C. For comparison, samples were coated with a 50 μg/mL purified rat type I collagen (Trevigen, Inc.) solution or left uncoated in PBS. The concentration for type I collagen (50 μg/mL) was chosen to provide saturating levels of the ligand. We have previously shown that cell adhesion reaches a saturation limit at collagen coating densities of 10 μg/mL.34 After rinsing again with PBS to remove any unbound peptide, the samples were ready for analysis or cell seeding.
Analysis of GFOGER surface coating
The GFOGER adsorbed on the surface of the nanofibers was visualized and quantified using a biotinylated version of the GFOGER peptide. Biotin was conjugated to the carboxyl end of the peptide using the EZ-Link® Amine-PEG3-Biotin kit (Pierce Biotechnology), and unreacted biotin was removed by dialysis. To visualize the presence of the peptide on the nanofiber surface, the GFOGER-coated nanofiber mesh samples were incubated with 10 μg/mL of fluorescein-conjugated NeutrAvidin® (Molecular Probes) for 30 min at room temperature in the dark. After rinsing with excess PBS, images were taken on an inverted fluorescence microscope (Axio Observer.Z1; Carl Zeiss) using a fluorescein isothiocyanate filter. The saturation of the surface by GFOGER was investigated by incubating meshes with varying GFOGER concentrations of 0–50 μg/mL. To quantify the amount of the biotinylated GFOGER peptide adsorbed on the nanofiber surface, an enzyme-linked immunosorbent assay (ELISA) was performed using an anti-biotin antibody. Nonspecific adsorption of the antibody was first blocked by immersing the nanofiber meshes in 0.25% heat denatured serum albumin with 0.0005% Tween-20, 1 mM ethylenediaminetetraacetic acid (EDTA), and 0.025% NaN3 in PBS for 1 h at 37°C. The meshes were then incubated with an anti-biotin antibody (diluted 1:2000) conjugated to alkaline phosphatase (ALP; BN-34; Sigma-Aldrich) for 1 h at 37°C. An ALP substrate, 4-methylumbelliferyl phosphate, was used at a concentration of 60 μg/mL in the diethanolamine buffer (pH 9.5), to measure the amount of bound antibody. After incubating the meshes with the substrate solution for 1 h at 37°C, the fluorescence was read on a plate reader (HTS 7000; Perkins-Elmer) at an excitation of 360 nm and emission of 465 nm.
Human mesenchymal stem cell culture
The Center of Gene Therapy at Tulane University kindly provided the human mesenchymal stem cells (hMSCs). The isolation of the cells has been described previously by Sekiya and coworkers.43 Briefly, bone marrow aspirates were taken from the iliac crest of normal adult donors, the nucleated cells were isolated with a density gradient, and only the cells that adhered to the plate after 24 h were cultured further. Passage 1 cells frozen in 1-mL aliquots were shipped us. To expand a culture, the cells were thawed and plated at a density of 50 cells/cm2 in alpha minimum essential medium (Invitrogen), supplemented with 16% fetal bovine serum (Atlanta Biologicals), 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM L-glutamine (Invitrogen). This is termed the hMSC growth media. After the cells reached a confluency of ∼70%, they were harvested with 0.25% trypsin–EDTA (Invitrogen), counted, and either expanded again or seeded on nanofiber meshes. Passage 2–3 hMSCs were used in all experiments.
Investigating cell migration on nanofiber meshes in an in vitro model
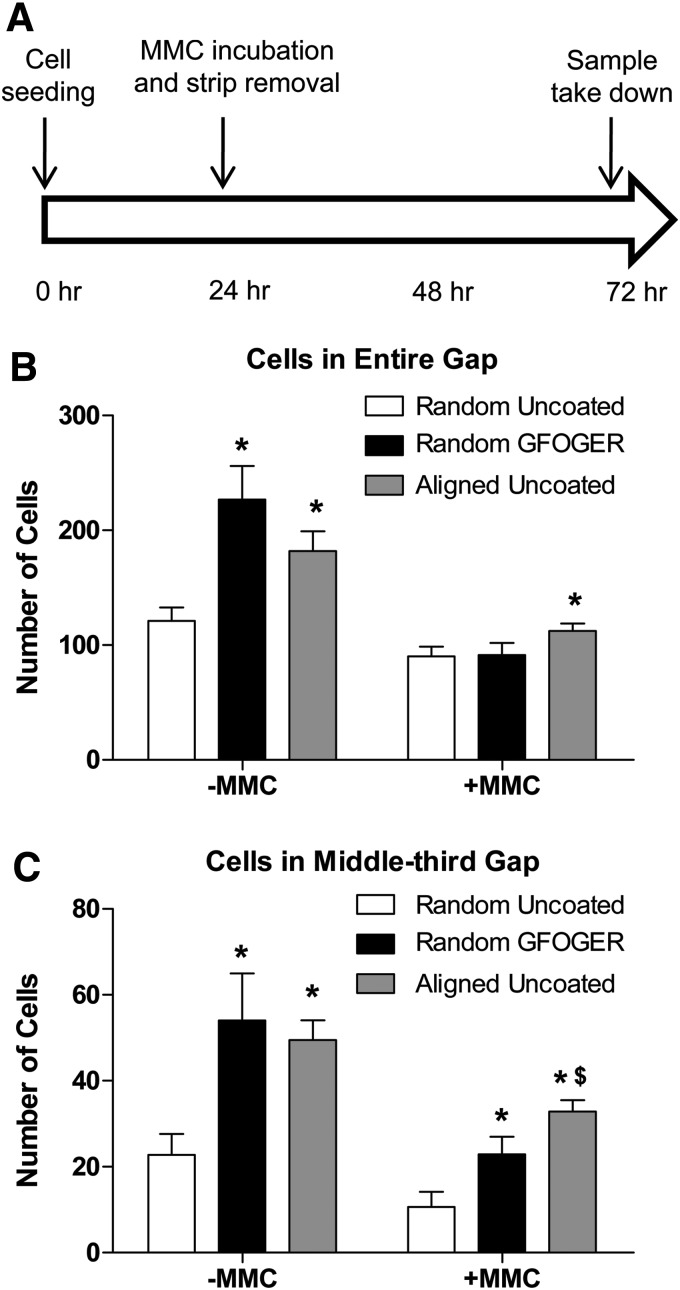
Rectangular samples (8 mm×12 mm) were cut from random and aligned nanofiber mesh sheets, sterilized, and coated with GFOGER or collagen, or left uncoated in 24-well tissue culture plates. A 0.9-mm-wide sterile stainless steel strip was then placed on the nanofiber mesh to create a region without cells (Fig. 5A). For aligned nanofiber meshes, the strip was placed perpendicular to the fiber orientation. Samples were submerged in 800 μL of hMSC growth media, after placement of a dead weight on the edges to prevent them from floating. hMSCs were then seeded on the nanofiber mesh samples at a density of 40,000 cells/cm2 in 200 μL of hMSC growth media. After 24 h, the strip was removed, and the cell migration into the gap was observed at various time points.
FIG. 5.
Role of cell proliferation in the cell migration model. (A) Time sequence of steps in this experiment. After seeding, cells were allowed to attach for 24 h, following which they were incubated with mitomycin C and the stainless strip was removed. Samples were taken down 48 h after strip removal. (B) The cell counts demonstrated that blocking proliferation resulted in reduced migration, indicating that both proliferation and migration contribute to cell migration. (C) In the middle-third gap, GFOGER coating and alignment resulted in a higher number of cells in the gap, even under mitomycin C incubation. * Indicates significantly greater than random uncoated with same MMC condition (p<0.05). $ Indicates significantly greater than random GFOGER with same MMC condition (p<0.05).
Cell migration was analyzed by staining the nuclei with 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes) and counting cells in the gap. Some samples were, in addition, stained with rhodaminephalloidin (Molecular Probes) to visualize the cell alignment. After samples were taken down, they were rinsed with PBS and fixed with 4% formaldehyde for 10 min. The samples were then incubated in 0.05% Triton-X (Sigma-Aldrich) for 5 min, rinsed in PBS, and incubated in 5 U/mL rhodaminephalloidin for 20 min. After a PBS rinse step, the samples were incubated in 5 μg/mL DAPI for 5 min. Samples were finally washed in excess PBS to remove any unbound dye, and images were taken on an inverted fluorescence microscope (Carl Zeiss). The DAPI images were further processed using ImageJ (NIH) to count the cell nuclei, and thereby quantify the cell number. Three fields of view along the gap were analyzed for each sample, and the mean cell number per field is presented, along with the standard error of the mean. To investigate the number of cells attached on day 0, four locations outside the gap were analyzed.
Role of cell proliferation in the cell migration model
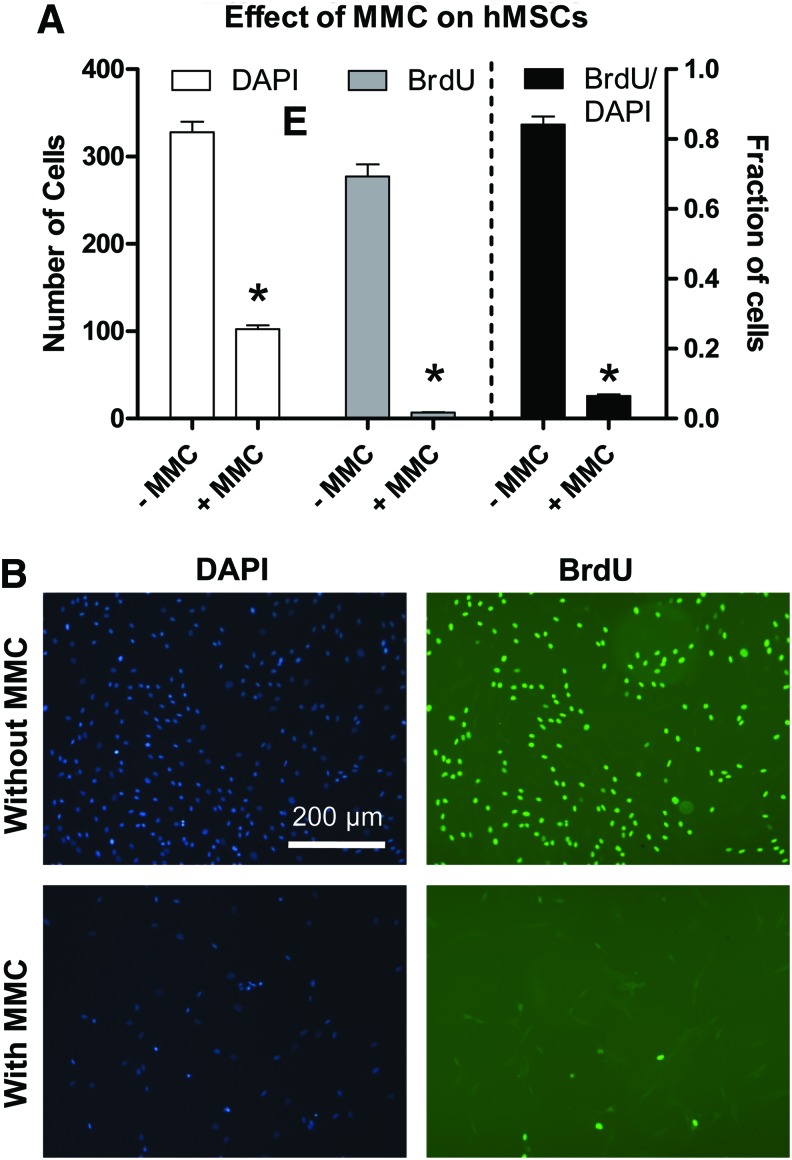
To investigate the influence of cell proliferation on the migration of cells on nanofiber meshes, mitomycin C was used to block proliferation. Mitomycin C, which is a known inhibitor of cell proliferation, crosslinks the strands of DNA, thereby inhibiting DNA replication.44,45 The effect of mitomycin C on hMSC proliferation was first studied on tissue culture plates. hMSCs were plated at a density of 20,000 cells/cm2, and after 24 h, they were incubated with 10 μg/mL mitomycin C for 60 min. Control samples remained in culture media. Cell proliferation was assessed using 5-bromo-2′-deoxyuridine (BrdU) labeling, according to the manufacturer's instructions (FLUOS; Roche Diagnostics). Forty-eight hours after cell seeding, the cells were incubated in hMSC growth media containing 10 μM BrdU for 24 h in the incubator. Cells were fixed with an ethanol-based fixative for 45 min, and denatured with 4 M HCl for 20 min to allow for antibody access. After neutralizing the pH with PBS, the cells were incubated with a monoclonal anti-BrdU antibody conjugated with fluorescein for 45 min at 37°C. The cells were finally stained with DAPI for the total number of cells and analyzed using fluorescence microscopy. Nine fields were examined in a 3×3 grid pattern, and the number of BrdU-positive cells along with the DAPI stained cells were counted. Data are presented as the BrdU and DAPI cell counts, as well as the proportion of BrdU-positive cells.
The effect of mitomycin C was next studied in the cell migration model on nanofiber meshes. Cells were seeded on rectangular nanofiber mesh samples at a density of 40,000 cells/cm2, as above. Twenty-four hours postseeding, mitomycin C was added to the media at a final concentration of 10 μg/mL, and incubated for 60 min. The media were slowly aspirated, fresh media were added, and the stainless strip was removed. After a further 48 h, the samples were taken down, stained with DAPI, and analyzed for cell migration into the gap.
Osteogenic differentiation and total DNA content on nanofiber meshes
Random and aligned nanofiber mesh samples were coated with GFOGER, collagen, or left uncoated. hMSCs were seeded on circular mesh samples (diameter: 12 mm), cut using biopsy punches (Acuderm), at a density of 20,000 cells/cm2. Four days after seeding, the hMSC growth media were replaced with osteogenic media, which consist of the hMSC growth media supplemented with 10 nM dexamethasone, 6 mM β-glycerol phosphate, 50 μg/mL ascorbic acid 2-phosphate, and 50 ng/mL L-thyroxine (Sigma-Aldrich). Media were changed every 3–4 days and the samples were cultured for 3 weeks. The samples were analyzed for DNA content, ALP activity, and calcium deposition, as described previously.13 Briefly, cells were lysed by freeze–thawing three times. The cell extract was used to measure the DNA amount by the PicoGreen®dsDNA Quantitation Kit (Molecular Probes) and the ALP activity by the use of p-nitrophenyl phosphate. The calcium content of a separate set of mesh samples was determined by using the dye Arsenazo III, after overnight incubation in 1 N acetic acid.
Data analysis
Data were analyzed using analysis of variance (ANOVA) and Tukey's tests for pairwise comparisons. Whenever required, the raw data were transformed using a natural logarithmic transformation to make the data normal and the variance independent of the mean.46 The Student's t-test was used for two-sample comparison, while analyzing cell adhesion efficiency (Fig. 5C). Other statistical tests that were performed for specific comparisons are mentioned in the Results section. The significance level for the above analyses was set at p<0.05. Minitab® 15 (Minitab, Inc.) was used for all statistical analysis.
Results
Nanofiber mesh morphology and alignment
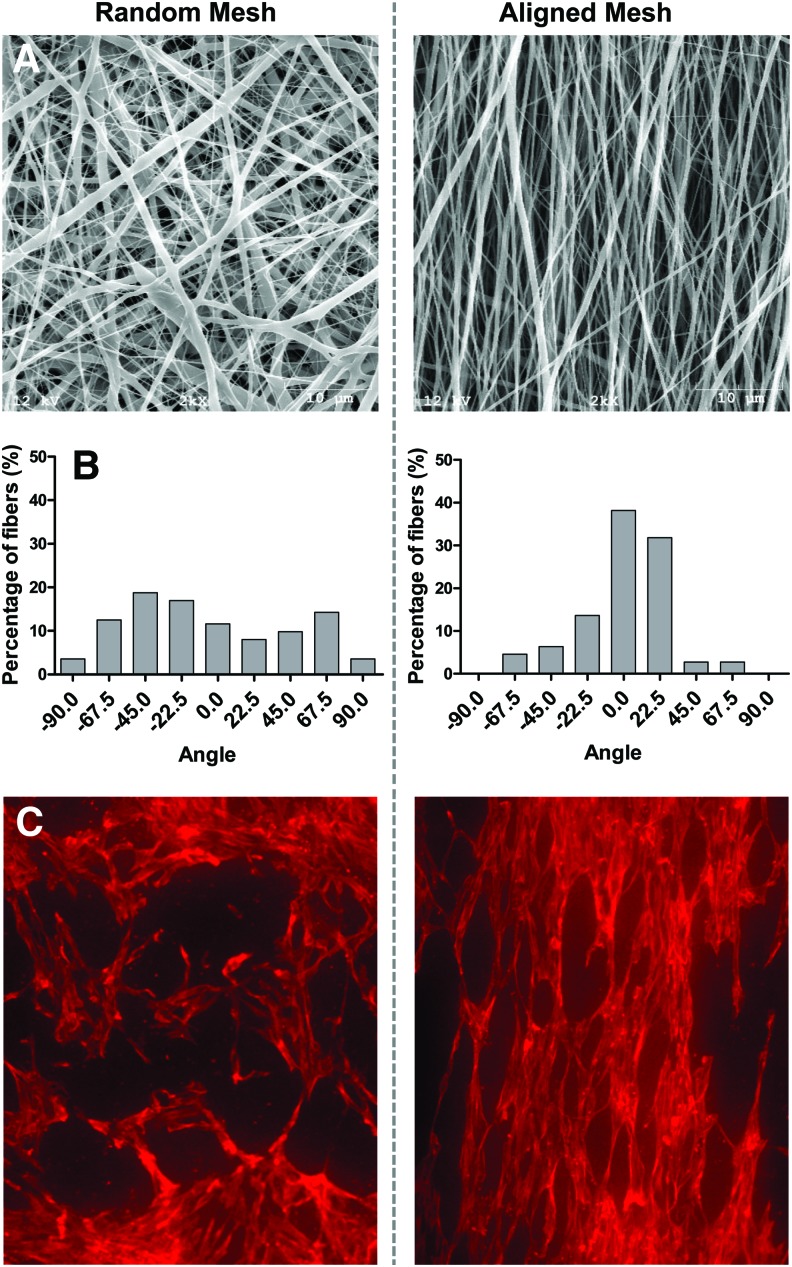
PCL nanofiber meshes were produced by electrospinning and characterized by analyzing SEM images. Interconnected, nonwoven fibers that were mostly bead free were obtained (Fig. 1A). The resulting nanofiber mesh is a porous structure, but due to the multiple fiber layers, the effective pore size appeared to be <2 μm, much less than hMSC dimensions. A flat stationary collector was used to obtain random nanofiber meshes with no dominant fiber orientation. The mean and median fiber diameters for random meshes were found to be 168 and 123 nm, respectively. The distribution of fiber diameters indicated that the highest frequency occurred for fibers between 75 and 125 nm, with 90% of the fibers between 50 and 300 nm. To obtain aligned nanofiber meshes, a rotating mandrel was used to orient the fibers along the direction of rotation of the mandrel surface. In this case, the mean and median fiber diameters were observed to be 256.4 and 227.4 nm, respectively. The fibers of the aligned meshes were found to be significantly larger in diameter than those in the random meshes (Mann–Whitney test; p<0.001; nonparametric Mann–Whitney test chosen due to non-normality of fiber diameter data). The angle of the fibers with respect to an arbitrary line was measured to quantify fiber alignment (Fig. 1B). Although there is a moderate spread in the orientation of the aligned fibers, a preferred fiber direction was observed, with 89% of the fibers between −45° and +45°. For random meshes, this metric was calculated to be only 52%. Using a z-test for proportions to compare the percentages of aligned fibers, alignment was found to be significantly higher in the case of aligned meshes (p<0.0001). hMSCs seeded on aligned nanofiber meshes exhibited a polarized morphology along the preferred fiber direction, while in the case of random nanofiber meshes, the cells did not display any regular orientation (Fig. 1C).
FIG. 1.
Fiber and cell alignment on nanofiber meshes. (A) Scanning electron microscopy (SEM) images of random and nanofiber meshes at 2000× magnification. (B) The fiber angle was measured from the SEM images and the distribution plotted to assess the fiber alignment. Fibers had a preferential orientation in the aligned mesh only. (C) Human mesenchymal stem cells (hMSCs) were seeded on random and aligned nanofiber meshes, and the cell alignment was observed by staining with rhodaminephalloidin. Images are at 10× magnification. hMSCs aligned along the fiber direction in the case of the aligned nanofiber mesh. Color images available online at www.liebertpub.com/tea
GFOGER coating of nanofiber meshes
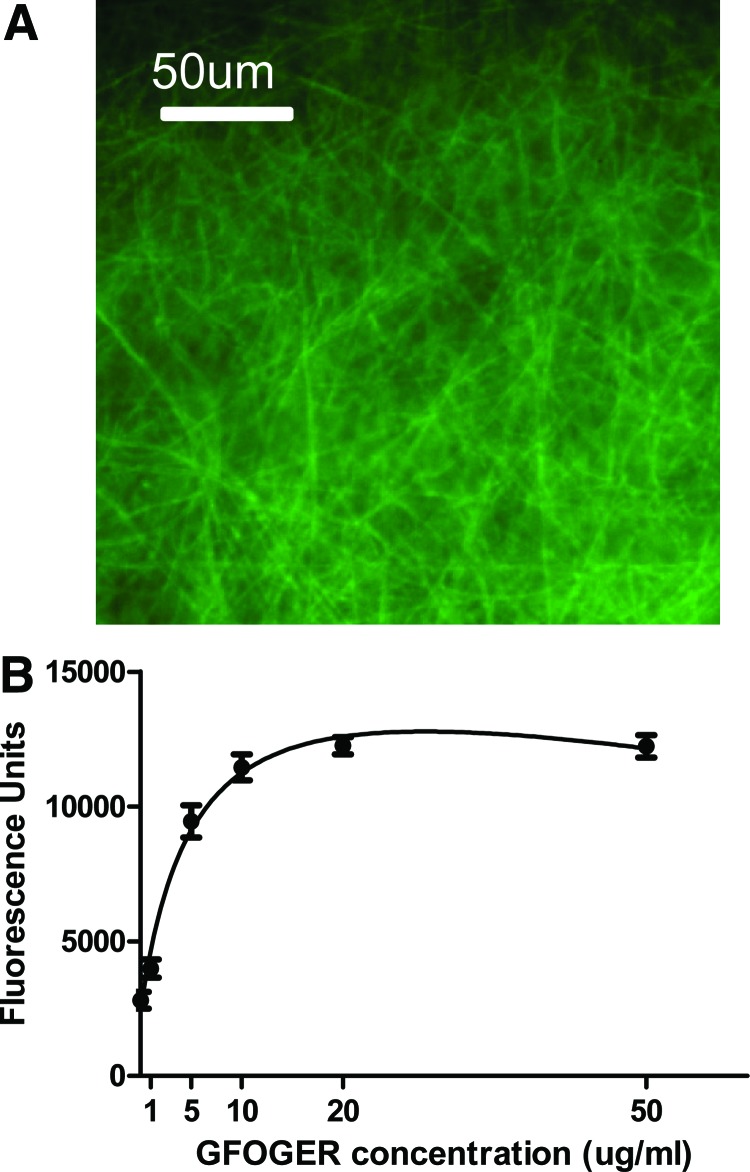
The collagen-mimetic peptide, GGYGGGPC(GPP)5GFOGER(GPP)5GPC, containing the GFOGER motif, was synthesized by stepwise solid-phase procedures.28,47,48 The GFOGER peptide was passively adsorbed on the surface of nanofiber meshes at a concentration of 50 μg/mL. The adsorbed peptide was visualized by coating the meshes with a biotinylated GFOGER peptide and incubating in a fluorescein-conjugated NeutrAvidin. The images revealed that the peptide coated the individual fibers uniformly over the entire mesh area (Fig. 2A). An ELISA was performed to quantify the amount of the GFOGER peptide adsorbed on the nanofibers with varying peptide concentrations. The saturation curve indicated that the relative surface density increased with increasing peptide concentrations, with the surface being saturated at a concentration of ∼20 μg/mL (Fig. 2B). The peptide concentration, at which the surface is 50% saturated, was calculated to be 3.5 μg/mL. For all further experiments, a peptide concentration of 50 μg/mL was used to ensure saturation of the surface for maximal biological responses.
FIG. 2.
Glycine-phenylalanine-hydroxyproline-glycine-glutamate-arginine (GFOGER) coating of nanofiber meshes. (A) Biotinylated GFOGER was passively adsorbed on nanofiber meshes, and the coating visualized using fluorescein-conjugated NeutrAvidin®. (B) The amount of GFOGER adsorbed on nanofiber meshes with varying GFOGER concentrations was quantified by an enzyme-linked immunosorbent assay. Color images available online at www.liebertpub.com/tea
Cellular migration on nanofiber meshes
An in vitro model was developed to study the effect of GFOGER coating and fiber alignment on cellular migration on top of nanofiber meshes. A cell-free region was created on the mesh, and the migration of hMSCs into this gap was examined by analyzing the DAPI stained images. To observe baseline migration, we first performed the experiments with uncoated, random nanofiber meshes. We verified that a cell-free region was generated in day 0 samples. The mean gap width was found to be 0.87±0.02 mm, consistent with the width of the stainless steel strip (0.9 mm). The cell count in the gap was found to be negligible on day 0. The ability of GFOGER coating and fiber alignment to enhance cellular migration in this model was studied on day 2 after strip removal. Some samples were coated with collagen to compare to the collagen-mimetic GFOGER peptide. The DAPI images revealed that the gaps on the GFOGER-coated meshes were completely confluent with cells (Fig. 3B). Aligned and collagen-coated meshes displayed moderately more cell migration than random and uncoated meshes, respectively. The cell numbers were determined in two regions: the entire gap and the middle-third of the gap. The middle-third region of the gap was analyzed as a more stringent measure of cellular migration. Analysis of the cell counts indicates that both coating and fiber alignment had a significant effect on cell migration in both the regions (Fig. 3C, D). In the entire gap region on random meshes, GFOGER-coated samples displayed higher cell numbers, compared to both uncoated and collagen samples. On aligned meshes, both GFOGER- and collagen-coated samples had higher cell counts than the uncoated samples. Fiber alignment enhanced cell migration on uncoated and collagen-coated samples. However, this effect was not seen on the GFOGER-coated meshes, probably due to the fact that the gap was saturated with cells, even on the GFOGER-coated random meshes. Analysis of the middle-third region of the gap revealed that the cell counts displayed the following order for both random and aligned meshes: GFOGER>Collagen>Uncoated. Fiber alignment enhanced the cell migration on both uncoated and collagen-coated samples, similar to the observation in the entire gap.
FIG. 3.
Effect of coating and fiber alignment on cell migration. (A) Schematic of cell migration model. A stainless steel strip is placed on the nanofiber mesh sample before seeding to create a cell-free region. After 24 h, the strip is removed and cell migration into the gap is observed by fluorescence microscopy. The larger rectangle covers the entire gap created by the strip, whereas the narrower rectangle represents the middle-third of the gap. (B) Images of 4′,6-diamidino-2-phenylindole stained nanofiber mesh samples illustrating the differences in the degree of cell migration. Scale bar is 1 mm and applies to all images. (C, D) The number of cells present in both entire gap as well as the middle-third of the gap was counted to quantify cell migration. Analysis of variance revealed that both GFOGER coating and fiber alignment enhance cell migration. (E) Effect of GFOGER on initial cell adhesion. The cell adhesion efficiency was investigated by quantifying the number of cells that attached outside the gap on day 0. There was no effect of GFOGER coating on cell adhesion efficiency. * Indicates significantly greater than uncoated with same fiber orientation. ** Indicates significantly greater than collagen with same fiber orientation. $ Indicates greater than random orientation with same coating. Significance was set at p<0.05. Color images available online at www.liebertpub.com/tea
One potential reason for the observed enhancement in cell migration due to GFOGER could be due to higher number of cells having attached on GFOGER-coated samples on day 0. To investigate whether GFOGER coating had a significant effect on cell adhesion efficiency, we counted the cells that attached outside the gap on day 0. The cell counts were observed to be equivalent in both the uncoated and GFOGER-coated groups, indicating that GFOGER coating did not modify the cell adhesion efficiency (Fig. 3E). This result shows that both sets of samples start with a comparable number of cells on the gap border.
Effect of cell proliferation on the migration of cells on nanofiber meshes
To assess the contribution of cell proliferation to the cell migration observed on nanofiber meshes, we blocked proliferation by treating the cells with mitomycin C, a known inhibitor of cell proliferation, 24 h after seeding. We verified that mitomycin C was able to inhibit proliferation of hMSCs on tissue culture plates by staining with BrdU, a marker of cell proliferation (Fig. 4). The results demonstrate that 48 h after mitomycin incubation, the number of proliferating cells (BrdU positive) decreased, thereby reducing the total number of cells (seen with DAPI staining) at this time point. In the absence of mitomycin C, the proportion of proliferating cells during the 24-h period was 84.1%. This proportion decreased significantly to 6.4% in the presence of mitomycin C, indicating that mitomycin C effectively blocked proliferation in hMSCs.
FIG. 4.
Effect of mitomycin C (MMC) on hMSC proliferation on tissue culture plastic. Cells were incubated with mitomycin C 24 h postseeding to block proliferation, and proliferation was assessed by 5-bromo-2′-deoxyuridine (BrdU) staining that ended 48 h after mitomycin C incubation. (A) The number of BrdU and 4′,6-diamidino-2-phenylindole (DAPI) stained cells were counted in a 3×3 grid pattern. Data are presented as the BrdU and DAPI cell counts, as well as the proportion of BrdU-positive cells. * Indicates significantly less than without MMC (p<0.05). (B) BrdU and DAPI images illustrating that cell proliferation was inhibited by mitomycin C. Scale bar is 200 μm and applies to all images. Color images available online at www.liebertpub.com/tea
The effect of inhibiting cell proliferation on the migration of hMSCs on nanofiber meshes was next investigated. hMSCs were seeded on nanofiber meshes, with a stainless steel strip placed on top, and allowed to attach for 24 h (Fig. 5A). The cells were incubated with mitomycin C for 60 min, and the strip was removed. After 48 h of strip removal, the samples were stained with DAPI and analyzed for cell migration before (Fig. 5B, C). The analysis of both the entire gap and the middle-third gap regions revealed that in the absence of mitomycin C, both GFOGER coating and fiber alignment enhanced cell migration, as observed previously. With mitomycin C incubation, the overall cell migration decreased, indicating that cell proliferation contributed significantly to the observed cell number. In the presence of mitomycin C, only the aligned, uncoated mesh samples had a significantly higher cell count than the random, uncoated mesh samples, in the entire gap region. In contrast, in the middle-third of the gap, the random, GFOGER-coated samples demonstrated a significantly higher migration than the random, uncoated samples. Further, the aligned, uncoated group displayed a higher cell count than the random, GFOGER-coated group. These results suggest that cell proliferation contributed to the observed cell migration on meshes, and that proliferation has a larger influence on migration due to GFOGER coating, compared to fiber alignment.
Influence of nanofiber coating and alignment on total DNA content and osteogenic differentiation
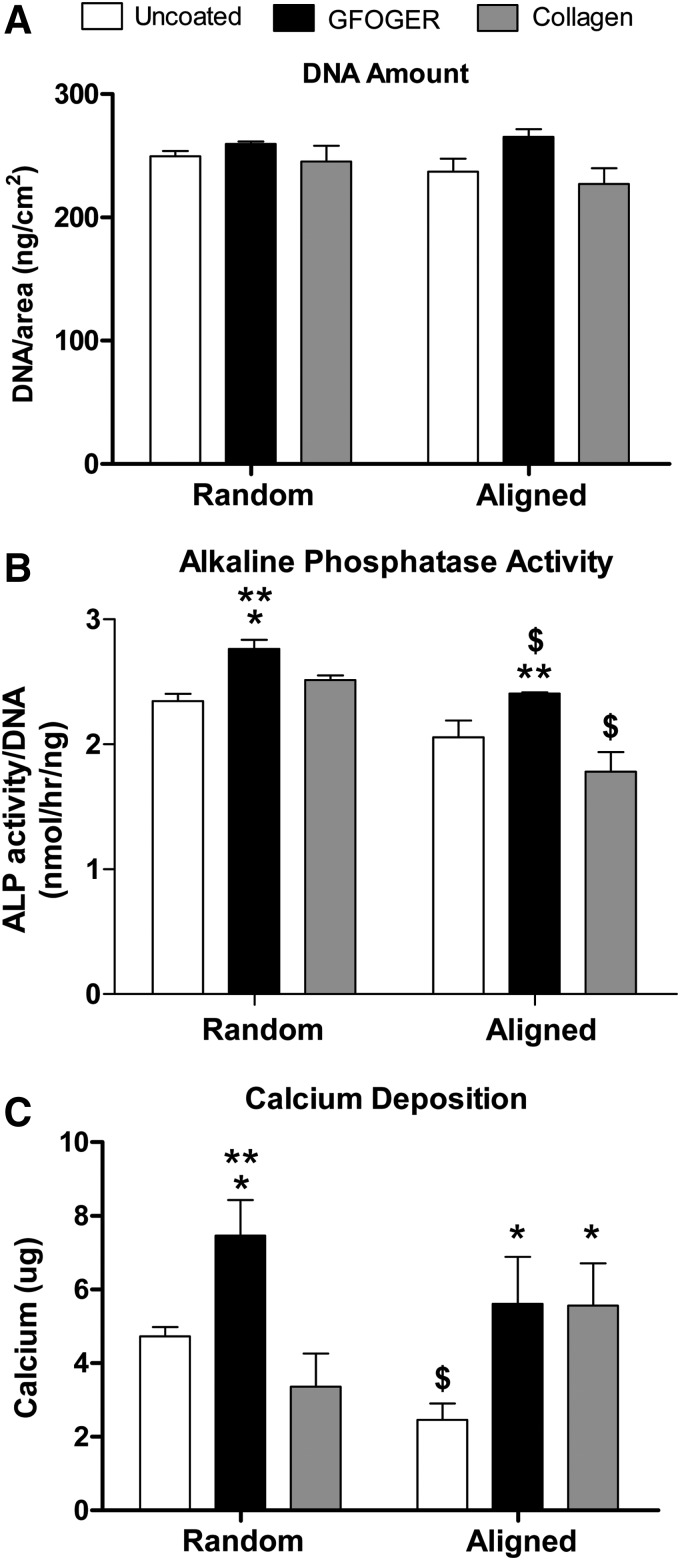
Cells were seeded on circular nanofiber meshes and cultured in either hMSC growth or osteogenic media for 21 days. This experiment did not involve the use of a stainless steel strip, and the cells were uniformly distributed on the mesh. The effect of coating and fiber alignment on DNA content in osteogenic media was first investigated. ANOVA revealed that GFOGER coating had an overall significant effect on the amount of DNA, but there were no significant individual differences between the groups (Fig. 6A).
FIG. 6.
Effect of coating and fiber alignment on cell number and osteogenic differentiation of hMSCs in osteogenic media. Cells were seeded on circular nanofiber mesh samples and cultured in osteogenic media for 21 days. (A) DNA amount. No significant differences were seen in the amount of DNA between groups. (B) Alkaline phosphatase (ALP) activity. Samples coated with GFOGER demonstrated increased ALP activity, whereas those with aligned fibers displayed a reduction in ALP activity. (C) Calcium deposition. GFOGER coating enhanced calcium deposition by hMSCs on nanofiber meshes. Fiber alignment did not have a significant overall effect.
The influence of nanofiber coating and alignment on the osteogenic differentiation of hMSCs in osteogenic media was assessed by measuring the ALP activity and calcium deposition. Analysis of ALP activity data revealed that both coating and fiber alignment had a significant effect on the ALP activity, and that the GFOGER-coated samples displayed a higher ALP activity than the uncoated and collagen-coated groups (Fig. 6B). In the case of random nanofiber meshes, the GFOGER group also demonstrated a significantly higher ALP activity than the uncoated and collagen groups, whereas, on aligned meshes, the GFOGER group displayed a significantly higher activity than the collagen group only. Fiber alignment was found to reduce the ALP activity in the case of GFOGER- and collagen-coated samples.
Under osteogenic stimulation, hMSCs deposited calcium on the nanofiber meshes, indicative of an osteoblast phenotype (Fig. 6C). GFOGER coating significantly enhanced calcium deposition on both random and aligned meshes, compared to uncoated meshes. In addition, on random meshes, GFOGER-coated samples demonstrated a significantly higher calcium deposition than collagen-coated samples. Collagen coating increased the calcium levels only in the case of aligned meshes. Fiber alignment did not have an overall significant effect on calcium deposition, although a reduction was observed in the case of uncoated meshes.
Discussion
In this study, we developed bioactive nanofiber meshes with ordered topography for enhancing tissue repair. Nanofibers were functionalized by passive adsorption of the collagen-mimetic GFOGER peptide, and the nanotopography was patterned by aligning nanofiber orientation. The effects of these parameters on the migration, proliferation, and osteogenic differentiation of hMSCs were studied. Our results indicate that nanofiber surface functionalization and orientation modulate cellular behavior, individually and in combination. GFOGER coating and fiber alignment enhanced hMSC migration on nanofiber meshes. Both cellular proliferation and individual cell migration contributed to the observed migration, with proliferation exerting a larger influence on migration due to GFOGER coating, compared to fiber alignment. In contrast, only GFOGER coating enhanced the osteogenic differentiation of hMSCs.
The migration of hMSCs into a gap region on top of nanofiber meshes was studied in vitro using a modified version of the scratch wound healing assay, which is performed frequently to study migration on tissue culture plastic.45,49 Cell migration was investigated because it is an important initial step in recruiting endogenous progenitor cells for tissue repair in vivo. In addition to analyzing the entire gap region, we also measured cell migration in the middle-third area of the gap to isolate cells that infiltrated farther as a more stringent measure of migration. It is possible that some of the cells located just inside the gap boundary may be there simply due to the division of cells outside the gap, rather than due to an active response to nanofiber surface modification.
The functionalization of nanofiber surface with GFOGER resulted in the largest increase of hMSC migration (as much as a 16-fold increase), higher than both collagen coating and fiber alignment. Whereas cells did not completely occupy the gap region on uncoated meshes even after 5 days (results not shown), the gap in the GFOGER-coated samples was confluent with cells, after just 2 days. To investigate whether the groups started with comparable number of cells at the gap boundary, we measured the cell adhesion efficiency at 24 h, the time of strip removal. It was observed that this was indeed the case, with no difference detected between the number of cells outside the gap. This result is different from previous published work,32,34 where GFOGER improved adhesion efficiency on tissue culture plastic/glass after a 1-h attachment period. However, the comparable adhesion efficiency of uncoated nanofiber meshes to GFOGER-coated samples is likely due to the highly textured surface and a longer cell attachment period. It should also be noted that GFOGER coating displayed a significantly larger effect on cell migration compared to collagen coating, even though their effects on cell adhesion have been shown to be equivalent previously.34
The alignment of fibers enhanced hMSC migration into the gap along the alignment direction on uncoated and collagen-coated meshes. Fiber alignment did not appear to influence migration for the GFOGER-coated groups, however, this may be due to the time period (48-h poststrip removal) utilized in the experimental protocol. We hypothesize that the lack of alignment effect on GFOGER-coated samples was due to the fact that the gap in these samples was already confluent with cells, even on random meshes, and there was no additional room for cells to infiltrate on aligned meshes. This hypothesis remains to be tested in future experiments, where shorter time periods will be studied. However, our current data clearly support the enhanced migration effect of aligned fibers on uncoated and collagen fibers. Although individual fibers were not perfectly oriented along a single direction, the cells that attached to the meshes aligned themselves along the fiber direction. This contact guidance may explain the improved migration of cells into the gap. Contact guidance due to fiber alignment has been reported previously by Yang et al. and Bashur et al., where they observed an increase in cell spreading and aspect ratio on aligned meshes.41,50 The fiber diameter in the case of aligned meshes was found to be higher than that in random meshes (256 nm vs. 168 nm). This was probably due to differences in the electrostatic fields between the stationary and rotating collector setups. Although different configurations of the rotating mandrel were attempted, we did not find it feasible to get similar fiber diameters, while maintaining good fiber alignment. It is possible that the difference in fiber diameter may contribute to the observed differences in cell behavior on aligned nanofiber meshes. However, the fibers are still in a narrow submicron range, and the cells may not be able to sense the subtle differences in fiber diameter. This hypothesis is supported by studies that have reported insignificant differences in cell function with varying fiber diameters within the submicron range.50,51
The observed migration of cells on nanofiber meshes occurs either due to the physical migration of individual cells, or due to cell proliferation, or a combination of both. To isolate the individual contributions, we blocked proliferation by incubating the cells with mitomycin C. In the absence of proliferation, the migration was reduced in all groups, indicating that the migration on nanofiber meshes is dependent, in part, on cell proliferation. It is possible that part of the reduction in total cell numbers may be due to a decrease in the viability of hMSCs due to mitomycin C; however, previous reports suggest that this is not a significant factor.52 The reduction in migration was especially high for the GFOGER-coated samples, in which the improvement in migration was abolished, when measured in the entire gap region. In the middle-third gap region, however, GFOGER coating still enhanced cell migration, indicating that at least some of the positive effects of GFOGER coating on migration are due to cell migration. This also suggests that the cell counts in the middle-third gap region are a more sensitive measure of cell migration than those in the entire gap. Compared to the GFOGER samples, the aligned meshes were less affected by the inhibition of proliferation, and demonstrated more migration in both the gap regions. Overall, these results suggest that proliferation contributed to the hMSC migration that was observed on nanofiber meshes. Kark et al. reported similarly that the gap closure on tissue culture plates, by rat MSCs in response to platelet releasate, was due to the combined effects of individual cell migration and proliferation.45 The mode of migration on GFOGER-coated samples appears to be more dependent on proliferation, whereas it is more dependent on direct migration in the case of aligned meshes.
The mechanisms for the improved cellular migration on GFOGER-coated nanofiber meshes are likely related to the adhesive properties of the collagen-mimetic GFOGER peptide. It is now known that the GFOGER hexapeptide, which is a sequence in the α1(I) chain of type I collagen, is a major binding site for the α2β1 integrin.28,48 The α2β1 integrin is abundantly expressed in a wide variety of cells, including MSCs.53 Since the engagement of integrins with ECM molecules regulate the adhesion-related function of cells, which include migration and proliferation, it is likely that the GFOGER-α2β1 integrin interaction may be driving the migration on nanofiber meshes.54–57 Senger et al. demonstrated that blocking the α2β1 integrin by a soluble antibody resulted in ∼40% inhibition of endothelial cell migration toward type I collagen.58 In addition, Reyes and coworkers have reported that cell adhesion was greater and specific to the α2β1 integrin, when titanium surfaces were coated with GFOGER, compared to uncoated, RGD-coated, and even serum-coated samples.33 Finally, the GFOGER peptide has been shown to promote the formation of mature integrin-mediated focal adhesions, an important event for postadhesion intracellular signaling.34
The effects of the mesh fiber orientation and GFOGER coating on the proliferation and differentiation of hMSCs on nanofiber meshes were also investigated. Proliferation was studied indirectly by determining the DNA content as a measure of the cell number. However, this provides only a snapshot of the number of cells at any time point, and any changes in the DNA content could be due to the differences in cell attachment, viability, and proliferation rate. Since the cell attachment and viability were not significantly affected by the mesh design, the DNA amount may be interpreted to be largely determined by the proliferation rate. We have quantified the DNA amount in the first week of culture and observed a higher DNA content on the GFOGER-coated meshes in osteogenic media, suggesting an early enhancement of proliferation by GFOGER (data not shown). However, by 21 days, there was no significant effect of GFOGER coating on the DNA content.
The osteogenic differentiation of hMSCs on nanofiber meshes was evaluated by measuring the ALP activity and the calcium deposition. ALP is a membrane-bound enzyme that hydrolyzes phosphate esters during mineralization, and is considered a marker of early osteogenic differentiation.59 On the other hand, calcium deposition is an end point measure of matrix mineralization. GFOGER coating enhanced both the ALP activity and calcium deposition, indicating that the peptide is able to promote differentiation of hMSCs to an osteoblast-like phenotype. The GFOGER-α2β1 integrin interaction has been implicated in mediating osteoblast adhesion, differentiation, and matrix mineralization.30,31 Reyes and coworkers have previously reported the osteogenic differentiation of immature osteoblasts and rat MSCs when cultured on GFOGER-coated surfaces.32,33 They demonstrated that the peptide triggers signaling pathways that result in the upregulation of Runx2/Cbfa1, a transcriptional activator essential for osteogenic differentiation.32,60–62 In contrast to GFOGER coating, fiber alignment did not have an overall positive effect on osteogenic differentiation, and in some groups resulted in a reduction in ALP activity and calcium deposition. This may be due to the cells being more migratory on aligned meshes.
In conclusion, this study demonstrates that nanofiber orientation and surface functionalization modulate hMSC migration and osteogenic differentiation. Coating nanofibers with the collagen-mimetic peptide GFOGER enhanced the migration, proliferation, and osteogenic differentiation of hMSCs, likely by engaging the α2β1 integrin receptor. Fiber alignment increased cell migration on nanofiber meshes along the direction of fiber orientation due to contact guidance, but did not demonstrate a positive effect on osteogenic differentiation. Overall, these results indicate that modulating nanofiber mesh design parameters related to fiber orientation and surface functionalization represent promising biomaterial-based strategies for improving cell function and tissue regeneration.
Acknowledgments
This study was supported with funding from NIH R37 DE013033, NIH R01 AR051336, the Armed Forces Institute for Regenerative Medicine (AFIRM), and the Center for Advanced Engineering and Soldier Survivability (CABSS). The authors would like to thank Dr. Ravi Bellamkonda for helpful discussions and Dr. Vivek Mukhatyar for technical assistance with nanofiber mesh electrospinning and analyses, and Dr. Ayona Chatterjee for statistical consulting.
Disclosure Statement
The authors do not have any competing financial interests.
References
- 1.Peppas N.A., and Langer R.New challenges in biomaterials. Science 263, 1715, 1994 [DOI] [PubMed] [Google Scholar]
- 2.Griffin T.J., Cheung W.S., and Hirayama H.Hard and soft tissue augmentation in implant therapy using acellular dermal matrix. Int J Periodontics Restorative Dent 24, 352, 2004 [PubMed] [Google Scholar]
- 3.Badylak S.F.The extracellular matrix as a scaffold for tissue reconstruction. Semin Cell Dev Biol 13, 377, 2002 [DOI] [PubMed] [Google Scholar]
- 4.Swiontkowski M.F., Aro H.T., Donell S., Esterhai J.L., Goulet J., Jones A., et al. Recombinant human bone morphogenetic protein-2 in open tibial fractures A subgroup analysis of data combined from two prospective randomized studies. J Bone Joint Surg Am 88, 1258, 2006 [DOI] [PubMed] [Google Scholar]
- 5.Athanasiou K.A., Niederauer G.G., and Agrawal C.M.Sterilization, toxicity, biocompatibility and clinical applications of polylactic acid/polyglycolic acid copolymers. Biomaterials 17, 93, 1996 [DOI] [PubMed] [Google Scholar]
- 6.Sakiyama-Elbert S., and Hubbell J.Functional biomaterials: design of novel biomaterials. Annu Rev Mater Res 31, 183, 2001 [Google Scholar]
- 7.Drury J.L., and Mooney D.J.Hydrogels for tissue engineering: scaffold design variables and applications. Biomaterials 24, 4337, 2003 [DOI] [PubMed] [Google Scholar]
- 8.Xin X., Hussain M., and Mao J.J.Continuing differentiation of human mesenchymal stem cells and induced chondrogenic and osteogenic lineages in electrospun PLGA nanofiber scaffold. Biomaterials 28, 316, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoshimoto H., Shin Y.M., Terai H., and Vacanti J.P.A biodegradable nanofiber scaffold by electrospinning and its potential for bone tissue engineering. Biomaterials 24, 2077, 2003 [DOI] [PubMed] [Google Scholar]
- 10.Pham Q.P., Sharma U., and Mikos A.G.Electrospinning of polymeric nanofibers for tissue engineering applications: a review. Tissue Eng 12, 1197, 2006 [DOI] [PubMed] [Google Scholar]
- 11.Brown T.D., Dalton P.D., and Hutmacher D.W.Direct writing by way of melt electrospinning. Adv Mater 23, 5651, 2011 [DOI] [PubMed] [Google Scholar]
- 12.Brown T.D., Slotosch A., Thibaudeau L., Taubenberger A., Loessner D., Vaquette C., et al. Design and fabrication of tubular scaffolds via direct writing in a melt electrospinning mode. Biointerphases 7, 13, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolambkar Y.M., Peister A., Ekaputra A.K., Hutmacher D.W., and Guldberg R.E.Colonization and osteogenic differentiation of different stem cell sources on electrospun nanofiber meshes. Tissue Eng Part A 16, 3219, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McManus M.C., Boland E.D., Simpson D.G., Barnes C.P., and Bowlin G.L.Electrospun fibrinogen: feasibility as a tissue engineering scaffold in a rat cell culture model. J Biomed Mater Res A 81, 299, 2007 [DOI] [PubMed] [Google Scholar]
- 15.Barnes C.P., Sell S.A., Boland E.D., Simpson D.G., and Bowlin G.L.Nanofiber technology: designing the next generation of tissue engineering scaffolds. Adv Drug Deliv Rev 59, 1413, 2007 [DOI] [PubMed] [Google Scholar]
- 16.Buttafoco L., Kolkman N.G., Engbers-Buijtenhuijs P., Poot A.A., Dijkstra P.J., Vermes I., et al. Electrospinning of collagen and elastin for tissue engineering applications. Biomaterials 27, 724, 2006 [DOI] [PubMed] [Google Scholar]
- 17.Barnes C.P., Pemble C.W., Brand D.D., Simpson D.G., and Bowlin G.L.Cross-linking electrospun type II collagen tissue engineering scaffolds with carbodiimide in ethanol. Tissue Eng 13, 1593, 2007 [DOI] [PubMed] [Google Scholar]
- 18.Garcia A.J., and Reyes C.D.Bio-adhesive surfaces to promote osteoblast differentiation and bone formation. J Dent Res 84, 407, 2005 [DOI] [PubMed] [Google Scholar]
- 19.Muhonen V., Fauveaux C., Olivera G., Vigneron P., Danilov A., Nagel M.D., et al. Fibronectin modulates osteoblast behavior on Nitinol. J Biomed Mater Res A 88, 787, 2009 [DOI] [PubMed] [Google Scholar]
- 20.Bernhardt R., van den Dolder J., Bierbaum S., Beutner R., Scharnweber D., Jansen J., et al. Osteoconductive modifications of Ti-implants in a goat defect model: characterization of bone growth with SR muCT and histology. Biomaterials 26, 3009, 2005 [DOI] [PubMed] [Google Scholar]
- 21.Lutolf M.P., and Hubbell J.A.Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat Biotechnol 23, 47, 2005 [DOI] [PubMed] [Google Scholar]
- 22.Hilbig H., Kirsten M., Rupietta R., Graf H.L., Thalhammer S., Strasser S., et al. Implant surface coatings with bone sialoprotein, collagen, and fibronectin and their effects on cells derived from human maxillar bone. Eur J Med Res 12, 6, 2007 [PubMed] [Google Scholar]
- 23.Bellamkonda R., Ranieri J.P., and Aebischer P.Laminin oligopeptide derivatized agarose gels allow three-dimensional neurite extension in vitro. J Neurosci Res 41, 501, 1995 [DOI] [PubMed] [Google Scholar]
- 24.Pierschbacher M.D., and Ruoslahti E.Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature 309, 30, 1984 [DOI] [PubMed] [Google Scholar]
- 25.Kao W.J., Lee D., Schense J.C., and Hubbell J.A.Fibronectin modulates macrophage adhesion and FBGC formation: the role of RGD, PHSRN, and PRRARV domains. J Biomed Mater Res 55, 79, 2001 [DOI] [PubMed] [Google Scholar]
- 26.Petrie T.A., Capadona J.R., Reyes C.D., and Garcia A.J.Integrin specificity and enhanced cellular activities associated with surfaces presenting a recombinant fibronectin fragment compared to RGD supports. Biomaterials 27, 5459, 2006 [DOI] [PubMed] [Google Scholar]
- 27.Massia S.P., Rao S.S., and Hubbell J.A.Covalently immobilized laminin peptide Tyr-Ile-Gly-Ser-Arg (YIGSR) supports cell spreading and co-localization of the 67-kilodalton laminin receptor with alpha-actinin and vinculin. J Biol Chem 268, 8053, 1993 [PubMed] [Google Scholar]
- 28.Knight C.G., Morton L.F., Peachey A.R., Tuckwell D.S., Farndale R.W., and Barnes M.J.The collagen-binding A-domains of integrins alpha(1)beta(1) and alpha(2)beta(1) recognize the same specific amino acid sequence, GFOGER, in native (triple-helical) collagens. J Biol Chem 275, 35, 2000 [DOI] [PubMed] [Google Scholar]
- 29.Knight C.G., Morton L.F., Onley D.J., Peachey A.R., Messent A.J., Smethurst P.A., et al. Identification in collagen type I of an integrin alpha2 beta1-binding site containing an essential GER sequence. J Biol Chem 273, 33287, 1998 [DOI] [PubMed] [Google Scholar]
- 30.Mizuno M., Fujisawa R., and Kuboki Y.Type I collagen-induced osteoblastic differentiation of bone-marrow cells mediated by collagen-alpha2beta1 integrin interaction. J Cell Physiol 184, 207, 2000 [DOI] [PubMed] [Google Scholar]
- 31.Mizuno M., and Kuboki Y.Osteoblast-related gene expression of bone marrow cells during the osteoblastic differentiation induced by type I collagen. J Biochem (Tokyo) 129, 133, 2001 [DOI] [PubMed] [Google Scholar]
- 32.Reyes C.D., and Garcia A.J.Alpha2beta1 integrin-specific collagen-mimetic surfaces supporting osteoblastic differentiation. J Biomed Mater Res A 69, 591, 2004 [DOI] [PubMed] [Google Scholar]
- 33.Reyes C.D., Petrie T.A., Burns K.L., Schwartz Z., and Garcia A.J.Biomolecular surface coating to enhance orthopaedic tissue healing and integration. Biomaterials 28, 3228, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reyes C.D., and Garcia A.J.Engineering integrin-specific surfaces with a triple-helical collagen-mimetic peptide. J Biomed Mater Res A 65, 511, 2003 [DOI] [PubMed] [Google Scholar]
- 35.Carlsson L., Rostlund T., Albrektsson B., and Albrektsson T.Removal torques for polished and rough titanium implants. Int J Oral Maxillofac Implants 3, 21, 1988 [PubMed] [Google Scholar]
- 36.Martin J.Y., Schwartz Z., Hummert T.W., Schraub D.M., Simpson J., Lankford J. Jr., et al. Effect of titanium surface roughness on proliferation, differentiation, and protein synthesis of human osteoblast-like cells (MG63). J Biomed Mater Res 29, 389, 1995 [DOI] [PubMed] [Google Scholar]
- 37.Buser D., Schenk R.K., Steinemann S., Fiorellini J.P., Fox C.H., and Stich H.Influence of surface characteristics on bone integration of titanium implants. A histomorphometric study in miniature pigs. J Biomed Mater Res 25, 889, 1991 [DOI] [PubMed] [Google Scholar]
- 38.Lincks J., Boyan B.D., Blanchard C.R., Lohmann C.H., Liu Y., Cochran D.L., et al. Response of MG63 osteoblast-like cells to titanium and titanium alloy is dependent on surface roughness and composition. Biomaterials 19, 2219, 1998 [DOI] [PubMed] [Google Scholar]
- 39.Zhong S., Teo W.E., Zhu X., Beuerman R.W., Ramakrishna S., and Yung L.Y.An aligned nanofibrous collagen scaffold by electrospinning and its effects on in vitro fibroblast culture. J Biomed Mater Res A 79, 456, 2006 [DOI] [PubMed] [Google Scholar]
- 40.Baker B.M., and Mauck R.L.The effect of nanofiber alignment on the maturation of engineered meniscus constructs. Biomaterials 28, 1967, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang F., Murugan R., Wang S., and Ramakrishna S.Electrospinning of nano/micro scale poly(L-lactic acid) aligned fibers and their potential in neural tissue engineering. Biomaterials 26, 2603, 2005 [DOI] [PubMed] [Google Scholar]
- 42.Kolambkar Y.M., Dupont K.M., Boerckel J.D., Huebsch N., Mooney D.J., Hutmacher D.W., et al. An alginate-based hybrid system for growth factor delivery in the functional repair of large bone defects. Biomaterials 32, 65, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sekiya I., Larson B.L., Smith J.R., Pochampally R., Cui J.G., and Prockop D.J.Expansion of human adult stem cells from bone marrow stroma: conditions that maximize the yields of early progenitors and evaluate their quality. Stem Cells 20, 530, 2002 [DOI] [PubMed] [Google Scholar]
- 44.Tomasz M.Mitomycin C: small, fast and deadly (but very selective). Chem Biol 2, 575, 1995 [DOI] [PubMed] [Google Scholar]
- 45.Kark L.R., Karp J.M., and Davies J.E.Platelet releasate increases the proliferation and migration of bone marrow-derived cells cultured under osteogenic conditions. Clin Oral Implants Res 17, 321, 2006 [DOI] [PubMed] [Google Scholar]
- 46.Kutner M.H., Nachtsheim C.J., Neter J., and Li W.Applied Linear Statistical Models. 5th ed. New York, NY: McGraw-Hill, 2005 [Google Scholar]
- 47.Reyes C.D., and Garcia A.J.A centrifugation cell adhesion assay for high-throughput screening of biomaterial surfaces. J Biomed Mater Res A 67, 328, 2003 [DOI] [PubMed] [Google Scholar]
- 48.Morton L.F., Peachey A.R., Zijenah L.S., Goodall A.H., Humphries M.J., and Barnes M.J.Conformation-dependent platelet adhesion to collagen involving integrin alpha 2 beta 1-mediated and other mechanisms: multiple alpha 2 beta 1-recognition sites in collagen type I. Biochem J 299 (Pt 3), 791, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shang Y.C., Wang S.H., Xiong F., Zhao C.P., Peng F.N., Feng S.W., et al. Wnt3a signaling promotes proliferation, myogenic differentiation, and migration of rat bone marrow mesenchymal stem cells. Acta Pharmacol Sin 28, 1761, 2007 [DOI] [PubMed] [Google Scholar]
- 50.Bashur C.A., Dahlgren L.A., and Goldstein A.S.Effect of fiber diameter and orientation on fibroblast morphology and proliferation on electrospun poly(D,L-lactic-co-glycolic acid) meshes. Biomaterials 27, 5681, 2006 [DOI] [PubMed] [Google Scholar]
- 51.Badami A.S., Kreke M.R., Thompson M.S., Riffle J.S., and Goldstein A.S.Effect of fiber diameter on spreading, proliferation, and differentiation of osteoblastic cells on electrospun poly(lactic acid) substrates. Biomaterials 27, 596, 2006 [DOI] [PubMed] [Google Scholar]
- 52.McCarroll J.A., Phillips P.A., Kumar R.K., Park S., Pirola R.C., Wilson J.S., et al. Pancreatic stellate cell migration: role of the phosphatidylinositol 3-kinase(PI3-kinase) pathway. Biochem Pharmacol 67, 1215, 2004 [DOI] [PubMed] [Google Scholar]
- 53.Heino J.The collagen receptor integrins have distinct ligand recognition and signaling functions. Matrix Biol 19, 319, 2000 [DOI] [PubMed] [Google Scholar]
- 54.Kim H.K., Oh D.S., Lee S.B., Ha J.M., and Joe Y.A.Antimigratory effect of TK1–2 is mediated in part by interfering with integrin alpha2beta1. Mol Cancer Ther 7, 2133, 2008 [DOI] [PubMed] [Google Scholar]
- 55.Ferdous Z., Peterson S.B., Tseng H., Anderson D.K., Iozzo R.V., and Grande-Allen K.J.A role for decorin in controlling proliferation, adhesion, and migration of murine embryonic fibroblasts. J Biomed Mater Res A 93, 419, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Michael K.E., Dumbauld D.W., Burns K.L., Hanks S.K., and Garcia A.J.Focal adhesion kinase modulates cell adhesion strengthening via integrin activation. Mol Biol Cell 20, 2508, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Clark E.A., and Brugge J.S.Integrins and signal transduction pathways: the road taken. Science 268, 233, 1995 [DOI] [PubMed] [Google Scholar]
- 58.Senger D.R., Perruzzi C.A., Streit M., Koteliansky V.E., de Fougerolles A.R., and Detmar M.The alpha(1)beta(1) and alpha(2)beta(1) integrins provide critical support for vascular endothelial growth factor signaling, endothelial cell migration, and tumor angiogenesis. Am J Pathol 160, 195, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seibel M.J.Molecular markers of bone turnover: biochemical, technical and analytical aspects. Osteoporos Int 11 Suppl 6, S18, 2000 [DOI] [PubMed] [Google Scholar]
- 60.Takeuchi Y., Suzawa M., Kikuchi T., Nishida E., Fujita T., and Matsumoto T.Differentiation and transforming growth factor-beta receptor down-regulation by collagen-alpha2beta1 integrin interaction is mediated by focal adhesion kinase and its downstream signals in murine osteoblastic cells. J Biol Chem 272, 29309, 1997 [DOI] [PubMed] [Google Scholar]
- 61.Byers B.A., Pavlath G.K., Murphy T.J., Karsenty G., and Garcia A.J.Cell-type-dependent up-regulation of in vitro mineralization after overexpression of the osteoblast-specific transcription factor Runx2/Cbfal. J Bone Miner Res 17, 1931, 2002 [DOI] [PubMed] [Google Scholar]
- 62.Xiao G., Jiang D., Thomas P., Benson M.D., Guan K., Karsenty G., et al. MAPK pathways activate and phosphorylate the osteoblast-specific transcription factor, Cbfa1. J Biol Chem 275, 4453, 2000 [DOI] [PubMed] [Google Scholar]