SUMMARY
Efforts to prevent human immunodeficiency virus (HIV-1) infection would benefit from understanding the factors that govern virus neutralization by antibodies. We present a mechanistic model for HIV-1 neutralization that includes both virus and antibody parameters. Variations in epitope integrity on the viral envelope glycoprotein (Env) trimer and Env reactivity to bound antibody influence neutralization susceptibility. In addition, we define an antibody-specific parameter, the perturbation factor (PF), that describes the degree of conformational change in the Env trimer required for a given antibody to bind. Minimally perturbing (low-PF) antibodies can efficiently neutralize viruses with a broad range of Env reactivities due to fast on-rates and high affinity for Env. Highly perturbing (high-PF) antibodies inhibit only viruses with reactive (perturbation-sensitive) Envs, often through irreversible mechanisms. Accounting for these quantifiable viral and antibody-associated parameters helps to predict the observed profiles of HIV-1 neutralization by antibodies with a wide range of potencies.
INTRODUCTION
The human immunodeficiency virus type 1 (HIV-1) envelope glycoprotein (Env) spike on the surface of virions binds host cell receptors (CD4 and CCR5) and mediates virus entry by fusing the viral and cell membranes (Wyatt et al., 1998). The unliganded Env trimer exists in a metastable, high-potential-energy state. During virus entry, this energy is channeled, through a series of receptor-induced conformational changes in Env, into the force required to fuse the viral and cell membranes (Blumenthal et al., 2012).
During persistent HIV-1 infection, the Env complex is a primary target for the antibody (Ab) response of the host. The HIV-1 Env surface is heavily glycosylated and exhibits variability among virus strains, minimizing the elicitation and efficacy of neutralizing Abs (Wei et al., 2003; Zwick and Burton, 2007). Neutralizing Abs generated by HIV-1-infected individuals vary tremendously in breadth and potency (Mascola, 2009). Although persistent HIV-1 variants typically escape these Abs, passive protection studies suggest that neutralizing Abs can potentially prevent acquisition of HIV-1 infection (reviewed in (Montefiori and Mascola, 2009) However, broadly neutralizing anti-HIV-1 Abs have been difficult to elicit in vaccinated animals or humans (Mascola et al., 1996).
A complete understanding of the mechanism of Ab-mediated neutralization of HIV-1 infection is lacking. Ab-mediated inhibition of HIV-1 infection depends upon the binding of Ab to the functional Env spike on the virus surface (Chen et al., 2009; Klasse and Sattentau, 2002; Parren et al., 1998; Sattentau and Moore, 1995; Tong et al., 2012; Yang et al., 2006). However, for a range of diverse HIV-1 variants and Abs, Ab binding to Env inconsistently predicts the potency of virus neutralization, suggesting that additional parameters contribute to virus inhibition. We recently identified a viral property, intrinsic Env reactivity (ER), which influences the susceptibility of HIV-1 variants to inactivation by Abs and other inhibitory ligands (Haim et al., 2011). ER describes the propensity of the high-potential-energy unliganded Env trimer to transition to lower-energy states upon perturbation. Viruses with high ER demonstrate global sensitivity to inhibition by multiple Abs that target different epitopes on the gp41 transmembrane and gp120 exterior Envs (Haim et al., 2011). In addition, viruses with high ER are more sensitive to cold-induced inactivation and more efficiently utilize low levels of CD4 for entry. Naturally-occurring HIV-1 variants exhibit a wide range of apparently continuous ER values, which can be estimated by measuring the sensitivity of virus entry to inhibition by a given level of bound soluble CD4 (sCD4). The increases in sensitivity of high-ER viruses to neutralization by multiple Abs do not arise from globally increased formation or exposure of the corresponding epitopes on Env (Haim et al., 2011). Thus, the efficiency of HIV-1 neutralization can be influenced not only by the affinity of Ab-Env binding but also by Env reactivity (ER) to Ab binding.
In our previous study (Haim et al., 2011), we made the unexpected discovery that the impact of ER on the efficiency of HIV-1 neutralization varied greatly for different Abs. This observation suggested that unappreciated properties of anti-Env Abs might limit the explanatory capabilities of current models of neutralization. Here we present a mechanistic model for HIV-1 neutralization that includes both viral and Ab parameters. We describe an Ab property that we designate the perturbation factor (PF). This property describes quantitatively the perturbation of Env conformation that is required for Ab binding. Using this parameter, we derive an expression that predicts with high accuracy the sensitivity of a given strain of HIV-1 to a given Ab, employing three input parameters: i) the efficiency of Ab binding to the trimeric, membrane-bound Env; ii) ER (a continuous property of Env); and iii) PF (a continuous property of Ab).
RESULTS
Relationship between Ab-Env Binding and Neutralization of Primary HIV-1
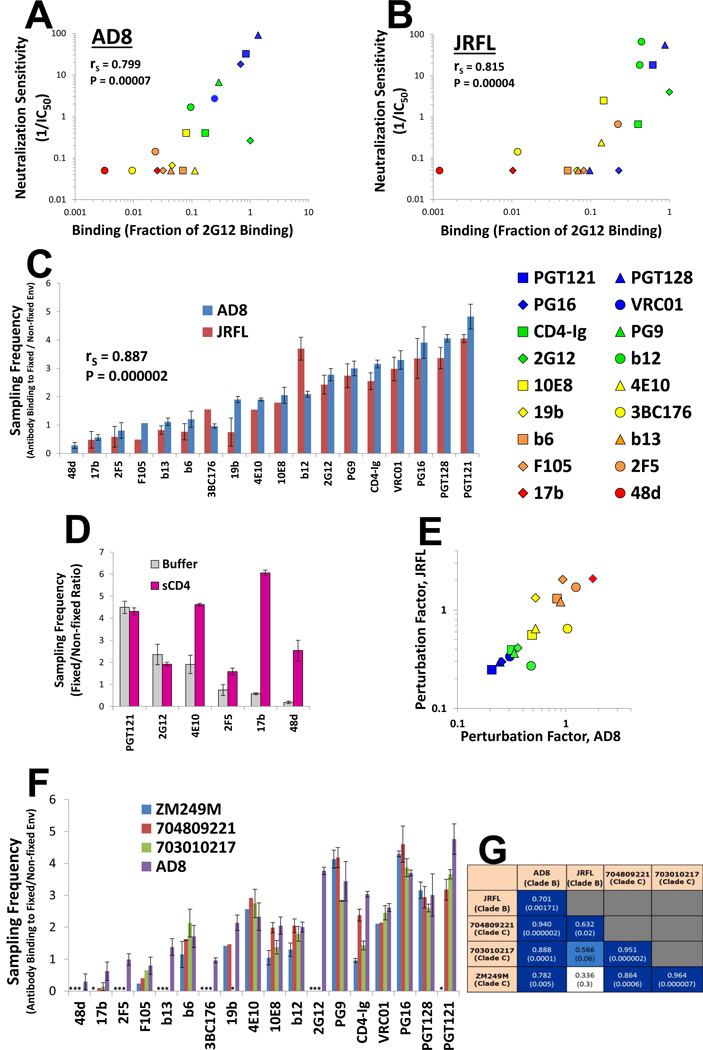
Ab inhibition of HIV-1 infection is affected by the efficiency of Ab binding to the Env spike on the virus surface (Chen et al., 2009; Klasse and Sattentau, 2002; Parren et al., 1998; Sattentau and Moore, 1995; Tong et al., 2012; Yang et al., 2006). We studied the binding of Abs to the trimeric, membrane-anchored form of Env from two primary HIV-1 strains, AD8 and JRFL, expressed on the surface of HOS cells. The AD8 and JRFL Envs exhibit near-complete proteolytic maturation in HOS cells and thus better reflect the antigenicity of the functional Env trimer (Haim et al., 2013; Pancera and Wyatt, 2005). A panel of monoclonal Abs was tested for the efficiency of Env binding and, in parallel, for the ability to neutralize viruses pseudotyped with the AD8 and JRFL Envs. A strong correlation was observed between the binding of each Ab to Env expressed on the surface of HOS cells and the capacity of the Ab to inhibit infection by viruses containing that Env (Figure 1, A and B). These correlations were significantly better when binding was measured in HOS cells rather than in COS-1 or 293T cells (Figure S1A), reflecting more efficient Env cleavage in HOS cells (Haim et al., 2013). Therefore, for the primary AD8 and JRFL HIV-1 strains, Ab binding efficiency to the trimeric, membrane-bound and cleaved Env is directly related to neutralization potency.
Figure 1. Ab Interaction with Primary HIV-1 Env Trimers.
(A,B) Correlation between binding of monoclonal Abs and CD4-Ig (at 2 µ;g/ml) to the indicated cell-surface Envs and the reciprocal of the Ab concentration (in µ;g/ml) required for half-maximal inhibition of the infection of viruses with the same Envs. Binding of each Ab is expressed as a fraction of the binding of the 2G12 Ab (at 2 µ;g/ml) to Env. (C) Sampling frequency of the Env conformations competent for binding the indicated monoclonal Abs and CD4-Ig. Error bars, SEM. (D) Effect of sCD4 binding (15 µ;g/ml) on the AD8 Env sampling frequency associated with the indicated Abs. For the absolute binding values, see Figure S1C. (E) Correlation between the PF values measured for each Ab on the AD8 and JRFL Envs. The 48d Ab is excluded from this correlation due to the low binding values for the JRFL Env. The symbols are color coded based on PF (blue = low, red = high). Spearman rank-order correlation coefficient, rS; P-value, two-tailed T-test. (F) PF values for the panel of Abs measured on the AD8 and three clade C HIV-1 Envs. Asterisks represent sampling frequencies that could not be calculated due to undetectable Ab binding to that Env. (G) Spearman rank-order coefficients for correlations between PF values of the Ab panel, measured on clade B and C HIV-1 Envs. P values are indicated in parentheses and the boxes are color coded according to the strength of the correlation. Data are represented as mean +/− SEM. Figure 1 is related to Figure S1.
Measurement of the Sampling Frequency of the Ab Binding-competent States of Env
Hypothetically, in its metastable unliganded state, the HIV-1 Env trimer may sample different conformations, only some of which allow the binding of a particular Ab. We define the sampling frequency as the fraction of total Env molecules that is in a conformation competent for binding the Ab. To investigate the sampling frequency associated with different Abs, we compared the binding of each Ab to glutaraldehyde (GA)-fixed and untreated Env on the surface of transfected HOS cells. Binding efficiency of an Ab to fixed Env relative to its binding to unfixed Env represents the relative occupancy of those conformations competent for Ab binding at a given time point (Yuan et al., 2006). To study Env conformational sampling (and not the intactness of the epitope), we tested only Abs that achieved measurable levels of binding. The observed pattern of sampling frequencies associated with the Abs was very similar for the AD8 and JRFL Envs (Figure 1C and Figure S1B). The 48d and 17b Abs, which recognize CD4-induced epitopes, bound very poorly to Env and their binding was further reduced upon GA fixation. By contrast, the binding of several Abs increased after GA treatment. Apparently, in these cases, limiting Env conformational change improves Ab binding, suggesting that the unliganded Env trimer frequently samples conformations recognized by these Abs. After CD4 binding, the sampling frequency associated with the CD4-induced Abs and the gp41 MPER-directed 2F5 and 4E10 Abs increased (Figure 1D and Figure S1C). For other Abs, such as PGT121 and 2G12, the sampling frequency was similar in the unliganded and CD4-bound states of Env.
That CD4 binding to Env followed by GA treatment significantly increased the binding of CD4-induced and anti-gp41 Abs suggested that GA fixation measures the frequency with which the Env trimer samples conformational states compatible with Ab binding, rather than reflecting an alteration of the target epitopes through, for example, lysine modification. Similarly, we previously observed that GA fixation of the gp120 Env core, which lacks the V1, V2 and V3 loops, does not decrease the binding of the Abs used in our study (Kwon et al., 2012). Therefore, for these Abs, the ratio of Ab binding to fixed versus unfixed Env reflects the frequency with which Env conformational states competent for Ab binding are sampled.
The sampling frequency associated with each Ab epitope (i.e., the relative occupancy of those conformations that can bind the Ab) is determined by the sum of the free energy of these conformations relative to that of all sampled conformational states (Guggenheim, 1955). Higher energy conformations (i.e., conformations that are more structurally perturbed relative to the basal conformation of the molecule) are less frequently sampled. Therefore, the greater the magnitude of structural change required for Ab binding (i.e., the higher the energy state), the lower its spontaneous sampling frequency. The degree of perturbation required for Ab binding can be derived by calculating the reciprocal of the measured sampling frequency, and is herein designated the perturbation factor (PF). Our measured sampling frequency and calculated PF provide indications of the Env structural changes that are required for Ab binding, which are often related to, but not identical with, the Env structural changes that are induced by Ab binding (Hammes et al., 2009). Thus, for example, although CD4 binding can induce dramatic structural changes in Env, the high sampling frequency of its bindingcompetent state indicates that minimal changes in Env are required for binding of this ligand (Figure 1C).
Although HIV-1 strain-dependent Env variation slightly influenced the absolute PF values associated with each Ab, very strong rank order correlations were observed among Ab PFs calculated using different HIV-1 Env strains (Figure 1E and see below). Apparently, the clade B Envs studied herein are sufficiently similar that the PF is primarily a property of the Ab. Thus, anti-Env Abs can be classified according to their PF. The Ab symbols are color-coded in Figure 1E and throughout the manuscript according to the PF (low PF = blue, high PF = red).
Comparison of Ab PFs Using Envs from Divergent Clade B and Clade C HIV-1
To determine whether the PF values measured for the above clade B strains are maintained in genetically diverse strains of HIV-1, we examined the Envs of three clade C transmitted/founder HIV-1 isolates from geographically distinct regions: isolate ZM249M from Zambia, isolate 704809221 from South Africa and isolate 703010217 from Malawi (Figure 1F and Figure S1D). These clade C Envs were antigenically distinct from the clade B Envs, and were not recognized by several of the Abs that recognized the AD8 and JRFL Envs. For the remainder of the Abs, there were very strong correlations among the PF values measured both within the clade C group and with the clade B AD8 Env (Figure 1G). The correlation between JRFL Env and the clade C Envs was not as good as that of AD8, suggesting that a limited level of variation in the PFs may exist. Interestingly, the clade C isolates demonstrated significantly different PF values for CD4-Ig, indicating that the Env state competent for binding CD4 is differentially sampled in HIV-1 isolates. Overall, despite limited variation, PF values are largely maintained across diverse HIV-1 strains from clades B and C.
Effect of Ab PF on Binding and Inhibition of Primary HIV-1 Envs
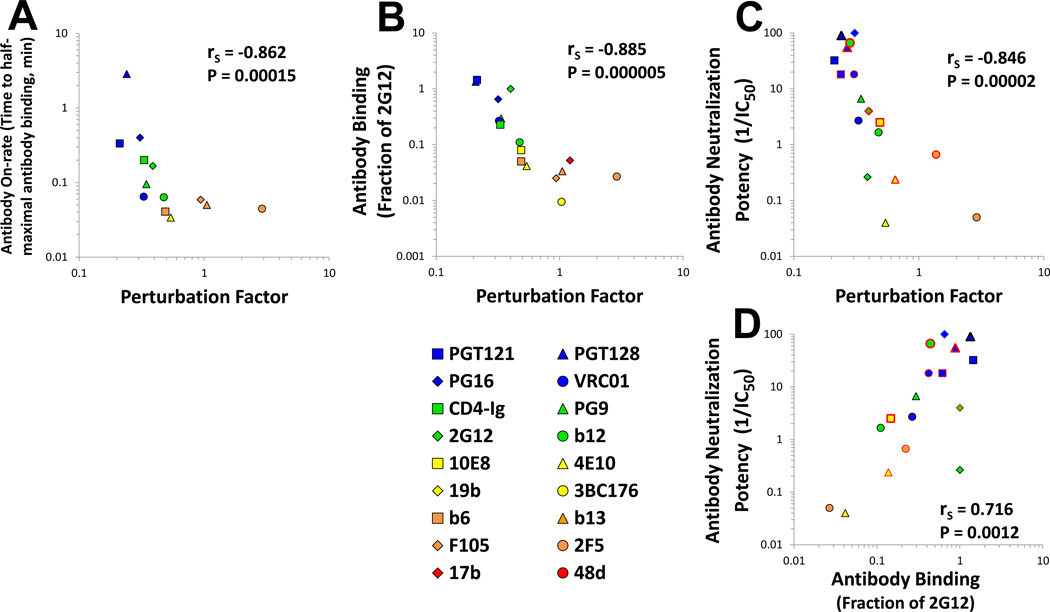
The on-rate of binding of some anti-Env Abs to the gp120 monomer correlates with neutralization efficiency (Sattentau and Moore, 1995; Steckbeck et al., 2005). To examine the relationships among PF, Env binding and HIV-1 neutralization, we measured the on-rate of Ab binding to cell-surface Env. Strong inverse correlations were observed between the PF of the Ab and its on-rate (Figure 2A), and between the PF and the steady-state binding of the Ab to Env (Figure 2B). We then compared the PF of each Ab with the efficiency with which it inhibited HIV-1 infection. Analysis of the group of highly efficient Abs that can achieve 90% or higher inhibition of the AD8 and JRFL primary isolates revealed a very strong inverse correlation between the PF of each Ab and its virus-neutralizing potency (Figure 2C). Indeed, the inverse correlation between Ab PF and neutralization potency was at least as strong as the positive correlation between Ab-Env binding and neutralization efficiency (Figure 2D). Analysis of the entire group of Abs showed that the level of correlation between PF and neutralization potency (rS = −0.855, P-value<0.000001) was similar to that between binding and neutralization potency (rS = 0.857, P-value<0.000001). Thus, Ab-mediated neutralization depends upon the intactness of the epitope (specific to the Env) and its degree of exposure (a property of the epitope or the Ab). Given the integrity of the relevant epitope, the degree of conformational perturbation of the unliganded trimer that is required for the Ab to bind is inversely related to the Ab on-rate and steady-state level of binding to Env, which influence neutralization potency.
Figure 2. Relationship between the Perturbation Factor (PF) and Ab On-Rate, Binding Efficiency and Neutralization Potency.
(A,B) Inverse correlation between the PF associated with each Ab and on-rate and efficiency of Ab binding to AD8 Env. (C,D) Correlation between Ab PF or binding measured on the AD8 and JRFL Envs and Ab neutralization of virus containing these Envs. The AD8 (black-bordered symbols) and JRFL (red-bordered symbols) data are pooled. Data points are colored according to PF. Spearman rank-order correlation coefficient, rS; P value, two-tailed T-test.
Ab Inhibition of Reactive and Non-reactive HIV-1 Env Variants
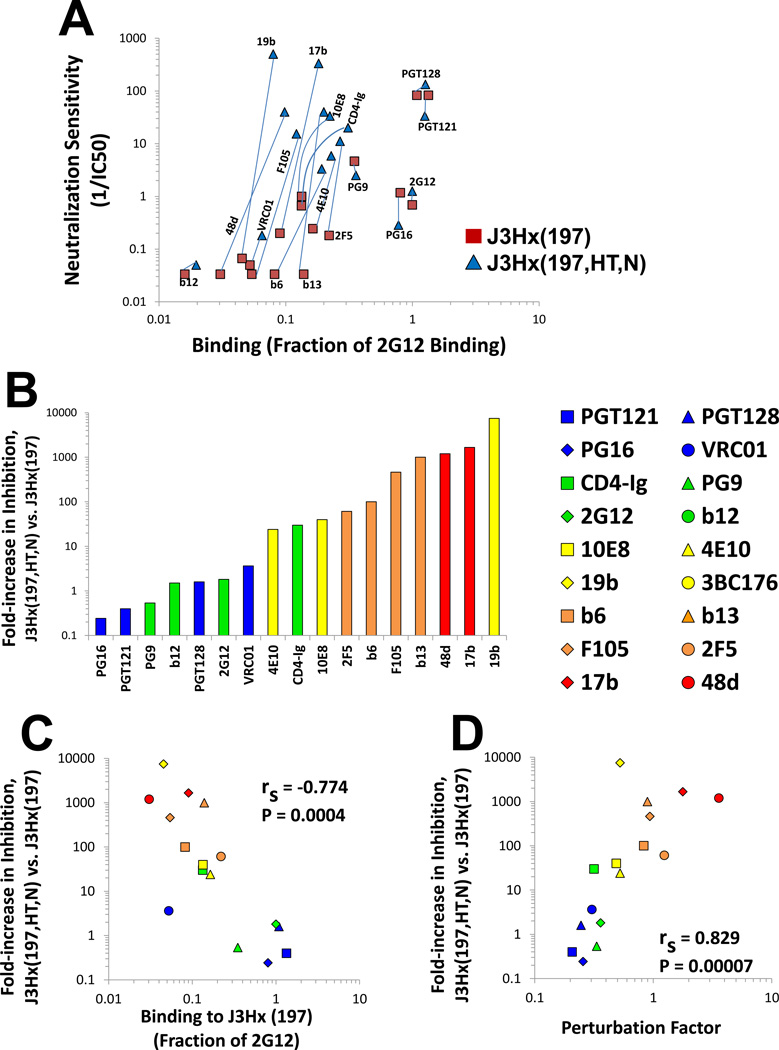
For the AD8 and JRFL primary HIV-1 isolates, the efficiency with which each Ab binds the Env trimer correlated strongly with its capacity to inhibit infection by the corresponding virus. However, many primary HIV-1 strains are more sensitive to Ab neutralization (i.e., have higher envelope reactivity (ER) values) than AD8 and JRFL (Haim et al., 2011). We examined the binding and neutralization profiles of a panel of Abs and CD4-Ig to a pair of closely-matched HIV-1 Envs that differ in ER. The J3Hx(197) is a variant of the AD8 Env that lacks a potential N-linked glycosylation site at asparagine 197, which increases the exposure of the coreceptor-binding (CoR-BS) site (Kolchinsky et al., 2001a). Although this change enhances the binding of Abs that target epitopes overlapping the CoR-BS, sensitivity to these Abs is only minimally affected. Thus, like AD8 from which it is derived, J3Hx(197) is a low-ER Env and is relatively resistant to Ab neutralization and cold-induced inactivation (Haim et al., 2011). As was seen for the low-ER AD8 and JRFL viruses, the neutralization of the J3Hx(197) virus by each Ab strongly correlated with the binding of the Ab to J3Hx(197) Env trimers (rS = 0.844, P-value= 0.00004) (Figure 3A). These Env binding and virus neutralization profiles were compared with those of a J3Hx(197) variant, J3Hx(197,HT,N), that contains two changes in gp41, NM625/626HT and D674N. These gp41 changes minimally affect the binding of most Abs to the Env trimer (Figure 3A), but have been shown to significantly increase ER (Haim et al., 2011). A detailed description of these changes is given in the Extended Experimental Procedure Section.
Figure 3. Ab-Env Binding, PF and Neutralization of AD8 Variants that Differ in Env Reactivity (ER).
(A) Relationship between Ab binding and inhibition of two AD8 Env variants that differ in the level of ER. Light blue lines are drawn between data points representing the same Ab tested with the two Envs. Abs with an IC50 value greater than 20 µ;g/ml were assigned a neutralization value of 0.05. (B) Fold increase in neutralization sensitivity (1/IC50) of J3Hx(197,HT,N) relative to J3Hx(197) for the indicated Abs and CD4-Ig. The colors of the bars indicate the PF values of the Abs, measured on the AD8 Env. (C,D) Correlations between binding efficiency or PF of Abs measured on the J3Hx(197) Env and fold increase in neutralizing potency (1/IC50) for J3Hx(197,HT,N) relative to J3HX(197). Spearman rank-order correlation coefficient, rS; P value, two-tailed T test.
Due to its high ER, the J3Hx(197,HT,N) virus was neutralized very efficiently by many Abs, even those with low or modest levels of binding to the J3Hx(197,HT,N) Env trimer (Figure 3A). Compared with the neutralization of J3Hx(197), the IC50 values of some Abs decreased up to 4 orders of magnitude for inhibition of the J3Hx(197,HT,N) virus. In contrast to J3Hx(197), no correlation was observed between Env binding and virus neutralization for J3Hx(197,HT,N) (rS = 0.066) (Figure 3A). Furthermore, no correlation was observed between the measured change in binding efficiency and the change in neutralization of the two Env variants (data not shown).
The binding-neutralization profiles in Figure 3A suggest that ER exerts varying effects on the virus-neutralizing potency of different Abs, as previously observed (Haim et al., 2011). The effect of ER on virus inhibition by each Ab can be measured by the fold-decrease in IC50 of the J3Hx(197,HT,N) virus relative to that of the J3Hx(197) virus (Figure 3B). The impact of ER on virus inhibition by each Ab was strongly associated with the efficiency of binding to the Env trimer; the lower the binding efficiency of the Ab, the larger the effect of ER on inhibition (Figure 3C). As expected from the relationship of Env trimer binding and PF (Figure 2B), there was a very strong correlation between the PF of each Ab and the impact of changes in ER on Ab neutralization potency (Figure 3D). Thus, the more conformational perturbation of Env required for Ab binding, the stronger the inhibition of viruses with the perturbation-sensitive (high-ER) Envs, relative to the inhibition of viruses with the perturbation-resistant (low-ER) Envs. In summary, if an Ab can bind to Env, the PF of the Ab modulates the contribution of the ER to virus neutralization.
Binding and Neutralization Profiles of the High-ER HXBc2 Env and a Low-ER Variant
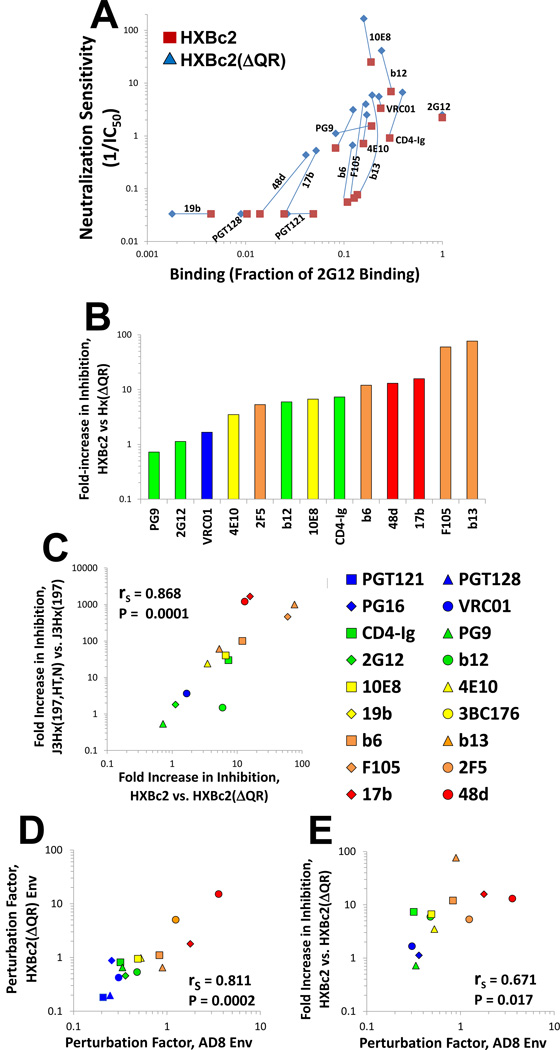
Laboratory adaptation of HIV-1 is often associated with acquisition of increased sensitivity to neutralizing Abs (Mascola et al., 1996; Moore et al., 1995; Pugach et al., 2004; Wrin et al., 1995). HXBc2 is a laboratory-adapted strain of HIV-1 that has a high-ER Env and therefore is relatively sensitive to Ab neutralization (Haim et al., 2011). We found that deletion of a pair of residues (Gln-Arg) in the gp120 V3 variable region rendered the resulting virus (HXBc2(AQR)) more resistant to neutralization by most Abs (Figure 4A). These changes in sensitivity were not caused by a general decrease in the binding of the Abs to Env trimers (Figure 4A); moreover, relative to HXBc2, the HXBc2(AQR) virus exhibited decreased sensitivity to cold-induced inactivation (data not shown). Therefore, the HXBc2 and HXBc2(AQR) variants are closely matched viruses that apparently differ in ER. Presumably, the presence of the Gln-Arg residues in the HXBc2 V3 region influences the conformation of the trimer association domain (TAD) (Mao et al., 2013), which is located at the apex of the Env trimer, is composed of the V1 /V2 and V3 regions of gp120, and can influence the level of ER (Haim et al., 2011).
Figure 4. Ab-Env Binding, PF and Neutralization of HXBc2 Variants with Different ER Values.
(A) Relationship between Ab binding and inhibition of the high-ER HXBc2 Env and the low-ER HXBc2(AQR) Env. Abs that had a measured IC50 value greater than 30 µ;g/ml were assigned a neutralization value of 0.03. (B) Fold increase in neutralization sensitivity (1/IC50) of virus containing the HXBc2 Env relative to the HXBc2(AQR) Env for the indicated Abs and CD4-Ig. The colors of the bars indicate the PF values of the Abs, measured on the AD8 Env. (C) Correlation between the effects of ER on inhibition by each Ab for the J3Hx(197,HT,N)-J3Hx(197) pair and the HXBc2-HXBc2(AQR) pair. (D) Correlation between the PF values measured on the AD8 and HXBc2(AQR) Envs. (E) Correlation between PF of Abs measured on the HXBc2(AQR) Env and fold increase in neutralizing potency (1/IC50) for HXBc2(AQR). Abs are colored according to PF values, measured on the AD8 Env. Spearman rank-order correlation coefficient, rS; P value, two-tailed T-test. Figure 4 is related to Figure S2.
Similar to the pattern observed with the J3Hx(197,HT,N) and J3Hx(197) viruses, neutralization by some Abs was dramatically affected by changes in ER, as indicated by the difference between the IC50 values for the HXBc2 and HXBc2(AQR) variants (Figure 4, A and B). Furthermore, there was a striking correlation between the J3Hx(197)-J3Hx(197,HT,N) pair and the HXBc2-HXBc2(ΔQR) pair with respect to the effect of ER changes on inhibition by each Ab (Figure 4C). Therefore, some Abs are unaffected by the level of Env reactivity and inhibit both high-ER and low-ER Envs similarly, whereas other Abs are significantly affected by the level of Env reactivity and primarily inhibit high-ER Envs. This quantitative relationship between Ab and ER is maintained in the context of different Envs, supporting the suggestion that a property of the Ab is involved.
We measured the sampling frequency associated with the binding of different Abs to the HXBc2(AQR) Env and calculated the PF values for each Ab. As expected, the Ab PF values measured with the HXBc2(AQR) Env correlated well with those calculated using the AD8 and JRFL Envs (Figure 4D). Furthermore, the PF of each Ab correlated well with the difference in IC50 values for neutralization of the HXBc2 versus HXBc2(AQR) viruses (Figure 4E). Interestingly, the rank order of the calculated PF values was maintained despite very different efficiencies of binding of some Abs. For example, although the PGT121 and PGT128 Abs bound very poorly to HXBc2 and HXBc2(AQR) Envs, and therefore did not inhibit these viruses, their PFs were identical to those measured using other Envs that bound these Abs with high efficiency (Figure S2). That the PF values are consistent in the context of different Envs despite very different binding efficiencies further supports the hypotheses that: i) the PF is primarily a property of the Ab, and ii) ‘epitope integrity’ and PF are two factors that independently affect binding and thus neutralization.
A General Expression that Describes HIV-1 Inhibition by Ab
We derived an expression that accounts for the above observations and predicts the neutralization of a range of HIV-1 isolates by diverse Abs. Neutralization can be thought of as a result of Ab binding to the HIV-1 Env trimer as well as the consequences of Ab binding:
| (1) |
The consequences of Ab binding to the HIV-1 Env trimer depend upon ER, which we have found to be modulated by the PF. Thus,
| (2) |
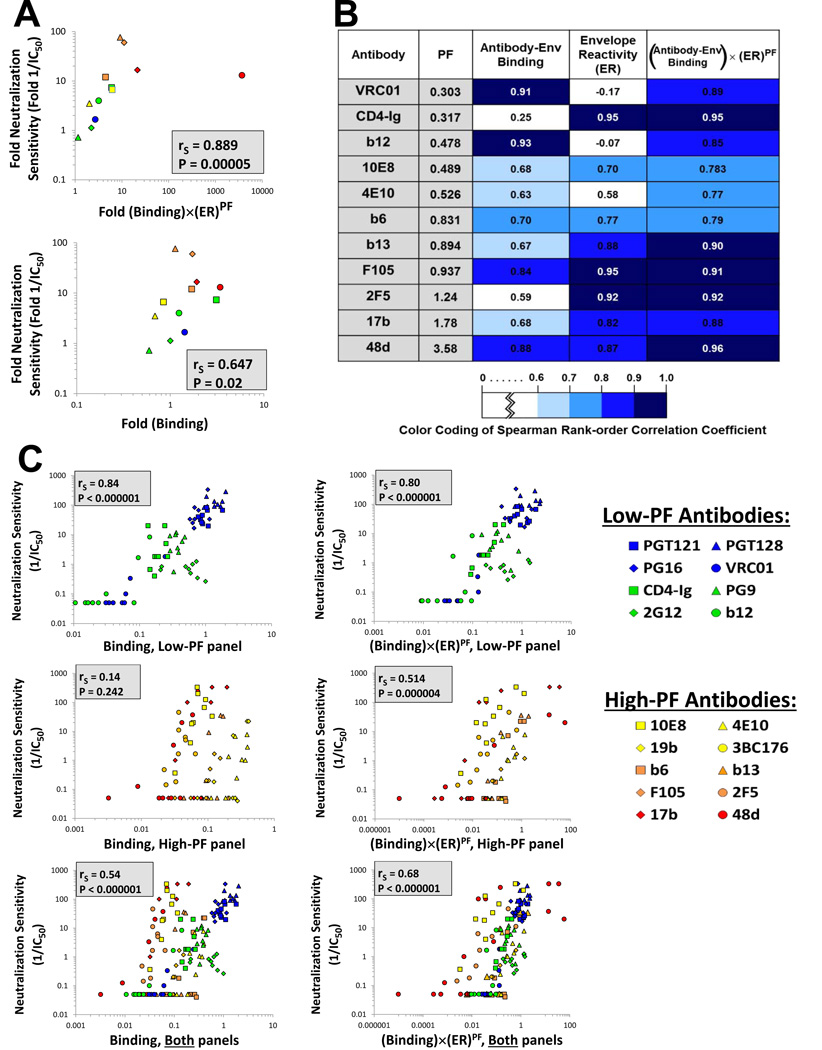
To test the validity of the above expression, we first studied the neutralization of the HXBc2 and HXBc2(ΔQR) variants described above, which differ in ER, by the panel of Abs. We examined the correlation between the measured neutralization sensitivity of viruses containing these Envs and the neutralization sensitivity predicted either by Ab-Env binding measurements alone or by the combined contributions of Ab-Env binding, ER and PF (Equation 2). As shown in Figure 5A, the correlations between the predicted and measured values of neutralization were significantly stronger when all three parameters were used in the prediction. Furthermore, Equation 2 allowed an accurate prediction of the absolute increase in sensitivity caused by given changes in ER, PF and binding. These results suggest that Equation 2 accurately incorporates the factors that affect inhibition of HIV-1 by Abs and describes the relationships among them.
Figure 5. Explanatory Capability of Different Models of HIV-1 Neutralization.
(A) Correlations between the measured relative neutralization sensitivity of HXBc2 and HXBc2(AQR) viruses and the relative neutralization sensitivity predicted by a model based on Equation 2 (top panel) or a model based only on Ab-Env binding (bottom panel). (B) Correlations between predicted and measured levels of inhibition. Nine closely-matched variants of the AD8 Env that differ in ER were tested for their sensitivity to each of the indicated Abs. For each Ab, we show the correlation between the measured neutralization sensitivity and the neutralization sensitivity predicted by models based on Ab-Env binding alone, ER alone, or the multiple parameters in Equation 2. Spearman rank-order correlation coefficients are reported and are colored according to the strength of the correlation, as detailed in the key. (C) Correlations between the measured neutralization sensitivity of the nine AD8 variants to the low-PF Abs (upper row) and the high-PF Abs (lower row) and the neutralization sensitivity predicted by a model based on Ab-Env binding alone (left panels) or based on Equation 2 (right panels). To allow a quantitative comparison of the neutralization obtained with different Abs, all data for each Env variant are expressed as the fold change in the measured or calculated value relative to the value obtained for the AD8 Env, which is assigned a value of 1. Spearman rank-order correlation coefficient, rS; P value, two-tailed T-test. The PF values of the Abs used for the calculations in this figure were measured on the AD8 Env.
To test the accuracy of our model further, we examined a panel of nine closely matched variants of the AD8 Env (Haim et al., 2011). These variants differ in the level of ER and, in a few cases, in the integrity of specific epitopes. This allowed us to examine the model accuracy when both binding and ER are changing. Neutralization of all the virus variants by different monoclonal Abs and CD4-Ig was tested. We measured the binding of the Abs to the Env variants expressed on HOS cell surfaces. Finally, we assessed the ER of each variant by measuring sCD4 reactivity (Haim et al., 2011). For each Ab tested, we examined the correlation for the entire panel of Envs between the measured neutralization sensitivity and three different potential predictors of neutralization: i) Ab-Env binding, ii) Env reactivity, and iii) the expression in Equation 2. The Spearman rank order coefficients for the different correlations were compared as measures of the predictive value of each expression for virus neutralization by each Ab (Figure 5B). For the Abs examined, Equation 2 explained the neutralization data as well or better than Ab-Env binding or ER alone, with only a few exceptions. For example, the binding of the b12 and VRC01 Abs, which target the CD4 binding site, is decreased by the N197S change in gp120. Consequently, Envs in the panel that contain serine 197 exhibited significantly reduced binding of these Abs, which was directly reflected by poor neutralization. Thus, for b12 and VRC01, there was a very good correlation between Ab-Env binding and inhibition. Furthermore, the relatively low PF of these Abs minimized the contribution of Env reactivity to virus neutralization. Nonetheless, even in these cases, Equation 2 performed only marginally worse than binding alone as a predictor of virus inhibition.
The relationship between the neutralization data from this panel of nine AD8 Env variants and either Ab-Env binding or Equation 2 was examined separately for the low-PF and high-PF Abs. For both low-PF and high-PF Abs, an excellent correlation was found between the observed virus neutralization and that predicted by Equation 2 (Figure 5C, right panels). By contrast, Ab-Env binding alone accurately predicted neutralization of the viruses by low-PF Abs, but not by high-PF Abs (Figure 5C, left panels). In summary, incorporation of Ab-Env binding, ER and PF into a model explains the inhibition of viruses with a range of neutralization sensitivities by Abs with different Env-binding characteristics.
Reversibility of Inhibition by High-PF and Low-PF Abs
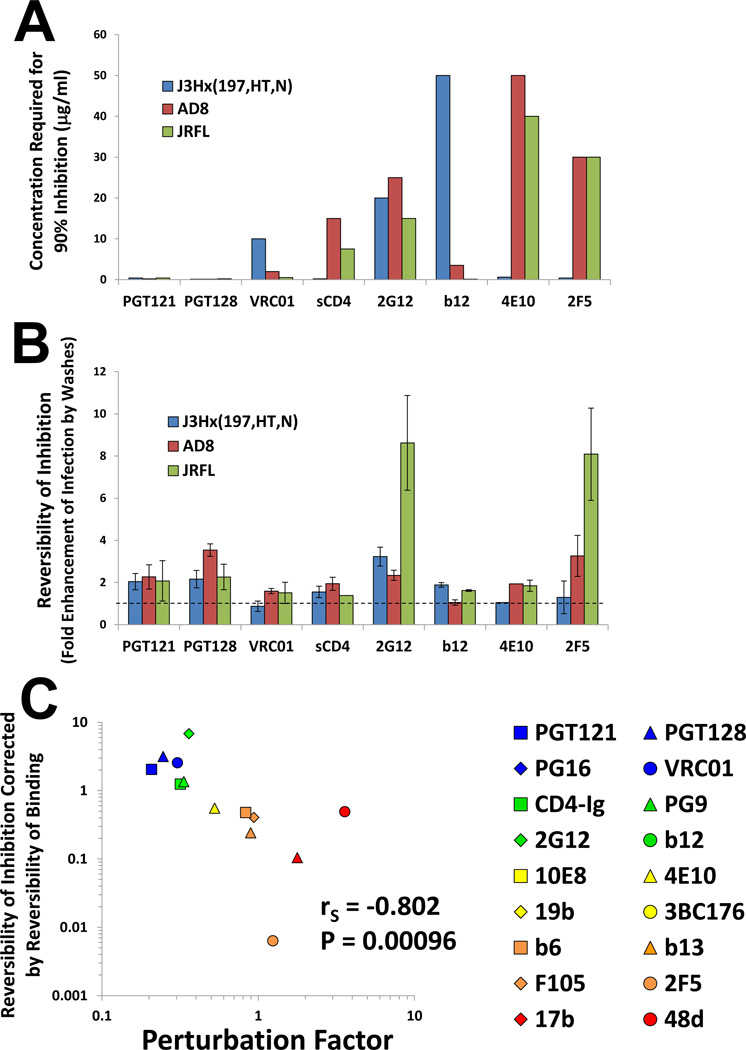
Some high-PF Abs bound relatively weakly to the high-ER Envs but inhibited the infectivity of the corresponding virus very efficiently (Figures 3A and 4A). We hypothesized that inhibition of high-ER viruses by the high-PF Abs may involve irreversible inactivation; by contrast, inhibition by the low-PF Abs, even of high-ER viruses, is effected by a reversible occupancy of the Env spike. To test these hypotheses, we examined the reversibility of inhibition for the different Abs and Envs. To this end, we directly attached viruses to protein-binding plates and could thus wash the virus after exposure to Ab. Infection of cells was measured either in the presence of the Ab or after extensively washing the virus following Ab exposure. We compared the degree of inhibition of the virus continuously exposed to the Ab with the inhibition of virus that was washed before cells were added. We interpreted increases in the infectivity of the washed samples to be indicative of reversible inhibition.
Binding the virus to the plate affected neither its infectivity nor the concentration of Ab required to achieve 90% inhibition (Figure 6A and data not shown). Figure 6B and Figure S3 show that some Abs, such as the previously reported 2G12 Ab (Platt et al., 2012), demonstrated high degrees of reversibility. The absence of an effect of washes on inhibition could be attributed to either low dissociation of Ab or to irreversible inactivation. We therefore sought to normalize the reversibility of inhibition by measuring the reversibility of virus binding, which can be quantitated by the amount of Ab bound before washes relative to Ab bound after washes. However, control experiments showed that the level of Ab bound before the washes cannot be accurately determined due to high background relative to the signal.
Figure 6. Reversibility of Ab-Env Binding and HIV-1 Inhibition.
(A,B) Viruses bound to protein-binding plates were incubated with Ab at axconcentration that neutralizes 90% of free virus (shown in (A)). Cf2Th CD4+CCR5+ target cells were then added to the viruses either in the presence of Ab or after washing the virus extensively. The virus-cell mixture was then incubated at 37°C for 1 hour to allow entry, after which cells were trypsinized to halt entry and further cultured for two days. Infectivity was measured by luciferase activity. For the absolute infection values, see Figure S3. (B) The reversibility of inhibition was calculated by dividing the infectivity of washed virus by the infectivity of unwashed virus. Data are represented as mean +/− SEM.. (C) Correlation between the reversibility of inhibition of the J3Hx(197,HT,N) virus (corrected for the reversibility of Ab-Env binding) and the PF measured on the AD8 Env. Spearman rank-order correlation coefficient, rS; P value, two-tailed T test. Figure 6 is related to Figure S3.
As an alternative, we examined the binding of each Ab at a concentration that induces a fixed level (90%) of virus neutralization. We assume that, for a given level of inhibition, the initial level of binding is identical, subsequent to which the Ab can remain bound (for reversible inhibition) or detach (for irreversible inhibition). As the level of binding before the washes is adjusted to be constant in all samples, we can quantitate the relative reversibility of binding by measuring the amount of Ab bound after the washes (see detailed description in the Extended Experimental Procedures Section).
The impact of PF on the reversibility of inhibition was examined using the highER Env J3Hx(197,HT,N), which is susceptible to neutralization by high-PF and low-PF Abs. A very strong inverse correlation was observed between the reversibility of inhibition (corrected for the reversibility of binding) and the PF of each Ab (Figure 6C). That “reversibility of neutralization” is a continuous rather than binary variable suggests that some Abs inhibit virus-infection by both reversible and non-reversible mechanisms. Thus, high-PF Abs can inhibit low-ER Envs but require high concentrations to do so, presumably to achieve the necessary level of Env occupancy (see inhibition of AD8 and JR-FL by 2F5 in Figures 6A and 6B). Inhibition of these low-ER viruses by this high-PF Ab was reversible, whereas its inhibition of the high-ER J3Hx(197,HT,N) virus was non-reversible. Thus, some Abs can inhibit by both occupancy- and perturbation-based mechanisms.
DISCUSSION
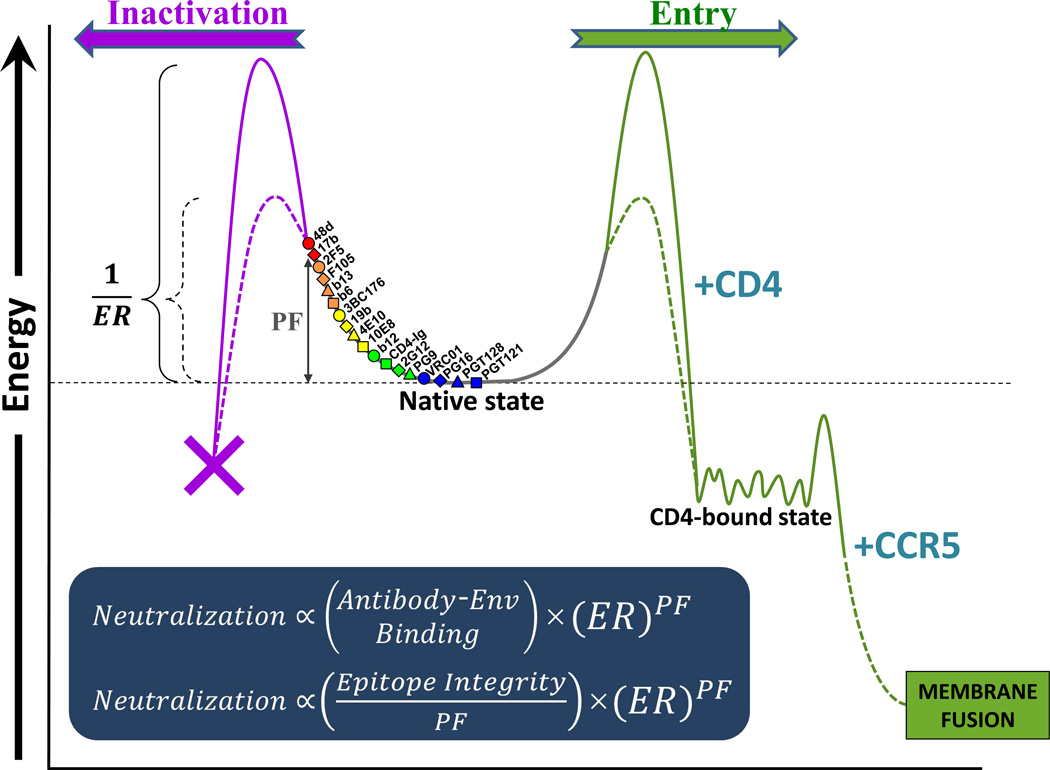
The HIV-1 Env trimer is a high-potential energy molecular machine. Receptor binding releases the energy stored in Env to drive virus-cell membrane fusion and virus entry (Blumenthal et al., 2012). Because premature loss of the potential energy of unliganded Env would result in functional inactivation, this metastable state of Env must be maintained until receptor engagement at the target cell surface. Activation energy barriers, formed by the interactions that maintain the structural integrity of the unliganded state of the trimer, prevent the metastable Env from transitioning to lowerenergy, functional or non-functional states (Figure 7). The magnitude of these activation energy barriers determines Env reactivity (ER), which influences both HIV-1 activation (by CD4) and inactivation (by Abs, other inhibitory ligands, or exposure to cold) (Haim et al., 2011).
Figure 7. Model of HIV-1 Neutralization by Abs.
The efficiency of HIV-1 neutralization is determined by the capacity of the Ab to engage Env and to modulate its conformation. The metastable HIV-1 Env is maintained in a high-potential-energy state by activation energy barriers. The magnitude of these activation barriers determines the degree of Env reactivity (ER), which is defined as the propensity of Env to transition to lower-energy states upon perturbation. The sampling frequency of the Env conformations capable of binding each Ab reflects the degree of structural change (perturbation) required to achieve these conformations from the basal state of unliganded Env. Abs with low perturbation factors (PF) (colored blue and green) recognize lower-energy (highly sampled) conformations and inhibit infection by reversibly preventing transitions down the productive (entry) pathway. Such low-PF Abs are less affected by the level of ER because little conformational perturbation of Env is required for binding. Inhibition by high-PF Abs (colored orange and red) is primarily determined by the level of ER because inhibition is mainly effected by promoting irreversible transitions down non-productive pathways.
HIV-1 neutralization represents the product of Ab binding to the Env trimer and the consequences of that binding. Each of these factors is potentially influenced by viral and Ab variables. Ab binding to the Env trimer depends upon the affinity of the Ab for its unconstrained epitope and upon the degree of conformational change in the Env trimer required for Ab binding. The latter property, herein defined as the perturbation factor (PF), can be assessed by measuring Ab binding to glutaraldehyde (GA)-crosslinked versus untreated cell-surface HIV-1 Env trimers. The crosslinking analysis provides an indication of the relative occupancy of different conformational states by the unliganded HIV-1 Env trimer (Yuan et al., 2006). The relative free energies of these conformational states can be calculated based on the Boltzmann distribution (Guggenheim, 1955). The conformations favored by the low-PF Abs are lower in energy and are sampled more frequently by the unliganded Env trimer; conversely, the conformations favored by the high-PF Abs have high free energies and are sampled less frequently (Figure 7). In this view, the major Env conformations (unliganded, CD4-bound, coreceptor-bound, fusion-active) consist of a collection of conformers, each with a characteristic energy, sampling frequency and ability to be recognized by particular ligands. Some of the Env conformers in the unliganded state presumably share structural features with conformers in the CD4-bound state; for example, Abs against CD4-induced epitopes can still recognize Env fixed in the unliganded state, albeit at a low efficiency. The many different states of Env contribute to the apparently continuous nature of the PF variable.
Empirically, we found that, for primary HIV-1 isolates with low ER, the reciprocal of the PF predicts neutralization as well or better than equilibrium Ab-Env binding, consistent with the importance of Ab on-rate to Env trimer binding and HIV-1 -neutralizing potency (Sattentau and Moore, 1995; Steckbeck et al., 2005). Under these circumstances, Ab-Env binding in Equation 2 can be replaced as follows:
| (3) |
Equation 3 suggests that the PF of the Ab affects neutralization in opposite ways, depending upon the ER of the virus. For low-ER viruses, Ab-Env binding is the dominant determinant of neutralization; this situation favors Abs with low PF values that can achieve high on-rates. For viruses with high ER, the degree of structural perturbation of Env required for Ab binding to the Env trimer represents the dominant determinant of neutralization; this situation favors Abs with high PF values, which can trap the highly reactive Envs in conformations that are prone to irreversible inactivation.
For the neutralization of the majority of naturally occurring HIV-1 strains with low ER values, the negative contribution of PF to Env trimer binding outweighs the potentially positive contribution of PF to the consequences of Ab binding. In this situation, the epitopes recognized by weakly neutralizing (high-PF) Abs are unavailable and not easily induced on the Env trimer (Figure 7). By contrast, potently neutralizing (low-PF) Abs do not require the unliganded Env trimer to change significantly in structure for efficient binding to occur. For example, potently neutralizing Abs directed against the CD4-binding site (CD4BS) of gp120 recognize the unliganded state of the HIV-1 Env trimer more effectively than less potently neutralizing Abs targeting this gp120 region (Chen et al., 2009). Such differences in binding to the intact trimer spike among the b12, b13 and F105 Abs were indeed reflected in their PF values. Even though some potent CD4BS Abs like VRC01 can induce the CD4-bound state in monomeric gp120, this induced conformational change is not necessary for efficient Env trimer binding (Zhou et al., 2010). Thus, the inhibitory capacity of an Ab for most primary HIV-1 isolates is predominantly determined by the degree of perturbation of Env conformation that is required for its binding.
In addition to Ab binding efficiency to Env, HIV-1 neutralization is also influenced by the consequences of Ab binding (i.e., the degree of perturbation of Env structure associated with Ab binding). The contribution of this factor to HIV-1 neutralization is strongly influenced by properties of both virus and Ab. HIV-1 strains differ in their requirements for the level of CD4 on target cells, and CD4 dependence is inversely related to general neutralization sensitivity (Kolchinsky et al., 2001; Zhang et al., 2002). This relationship between CD4 independence and neutralization sensitivity is explained by differences in ER (Haim et al., 2011). ER is defined as the propensity of Env to undergo conformational changes when perturbed by ligand binding or exposure to cold. Although the PF formally describes the degree of Env conformational changes that is required for Ab engagement and not necessarily the degree of change that is induced by the Ab, the binding of high-PF Abs is expected to trap Env in less favored conformational states and thus, to modulate Env conformation. It is satisfying to observe that the PF, an Ab property that governs the impact of ER on neutralization, reflects the ability of the Ab to “perturb” Env conformational states. In this light, CD4 and CCR5 can be viewed as perturbing factors that disrupt metastable Env states to drive the entry process. How natural HIV-1 variants regulate their reactivity to the receptors versus Abs is of great interest for future studies.
That Abs can promote inhibitory consequences beyond Env trimer binding is supported by the apparent irreversibility of HIV-1 neutralization in some instances. Irreversible inhibition was associated with neutralization of high-ER viruses by high-PF Abs. This observation suggests that the induction of Env conformational change by Ab binding is important for irreversible virus inactivation. Ab-induced movement of Env from metastable states to lower-energy states could explain the irreversibility of the inhibitory effect (Figure 7). Such a mechanism is reminiscent of the premature transition of Env to the labile CD4-bound state that underlies the irreversible inhibition of HIV-1 entry seen for some small-molecule CD4-mimetic compounds or soluble forms of CD4 (Haim et al., 2009).
The relationship between HIV-1 neutralization and binding, PF and ER implied by Equation 2 does not take into account the specific Env binding site of the Ab. Hypothetically, Ab binding to functional elements of Env might result in a gain in neutralization potency. For example, an Ab that competes with CD4 might be more effective in neutralizing HIV-1 than an Ab that binds a non-functional element on the Env trimer. However, consideration of the above three factors in Equation 2 accurately explains the sensitivity of a wide range of HIV-1 variants to inhibition by Abs of different potencies. The steric effects of trimer-bound Ab on the fusion of the viral and target cell membranes may be much more significant than the specific impact of Ab binding to its epitope (Yang et al., 2006). Should future work demonstrate that binding to a specific Env element results in more effective inhibition of HIV-1 entry, Equation 2 can be modified to include such a factor. Similarly, incorporation of the stoichiometry of Ab engagement of the Env trimer, which may differ among Abs (Loving et al., 2013; Yang et al., 2005), may improve the precision of the above expression.
Our findings describe quantitatively two separate pathways that allow inactivation of Env function, through Ab engagement of the functional Env spike and through perturbation of Env structure. This basic mechanistic understanding should help the design of immunogens for an AIDS vaccine and inhibitors of HIV-1 entry into cells.
EXPERIMENTAL PROCEDURES
Full methods are available in Extended Experimental Procedures.
Abs and Soluble Forms of CD4
The broadly neutralizing Abs IgG1 b12 and VRC01 recognize the CD4-binding site of gp120 (Chen et al., 2009; Zhou et al., 2010)). The b6, b13 and F105 Abs also target the CD4-binding site but generally do not neutralize primary HIV-1 isolates efficiently (Chen et al., 2009). The 17b and 48d Abs recognize gp120 epitopes that are induced by CD4 binding. Ab 3BC176 targets a still incompletely defined gp120 epitope that is exposed in part by CD4 binding. Abs 2F5, 4E10 and 10E8 recognize the membrane-proximal external region (MPER) of gp41. The 2G12, PG9, PG16, PGT121 and PGT128 Abs recognize glycan-dependent epitopes on gp120.
The CD4-Ig fusion protein is composed of the Fc region of human IgG1 linked to two copies of the two N-terminal domains of the CD4 molecule. The soluble form of CD4 (sCD4) is composed of domains D1-D4 of this molecule and was purified as previously described (Haim et al., 2011).
Env Constructs
The Envs of the AD8, JR-FL and HXBc2 isolates of HIV-1 (accession numbers AF004394, U63632 and K03455, respectively) were expressed from the pSVIIIenv vector. The J3Hx(197) variant is a chimera between the Envs of the AD8 and HXBc2 strains and contains an Asn to Ser change at position 197 (Haim et al., 2011; Kolchinsky et al., 2001). The J3Hx(197,HT,N) construct is identical to J3Hx(197) except for additional changes: Env residues 625 and 626 are changed from Asn-Met to His-Thr and Env residue 674 from Asp to Asn.
The Hx(AQR) construct contains a deletion of the amino acid residues Gln-Arg 310–311 at the tip of the V3 loop of the HXBc2 Env. The Envs of three transmitted/founder HIV-1 subtype C viruses were studied: isolate ZM249M from Zambia, isolate 704809221 from South Africa and isolate 703010217 from Malawi (accession numbers EU166862, FJ444116 and FJ443589, respectively).
Complete descriptions of the antibodies and Env variants can be found in the Extended Experimental Procedures.
Cell-based ELISA
HOS cells were seeded in 96-well plates and were transfected the next day with 0.06 µg of an Env-expressing plasmid and 0.008 µg of a Tat-expressing plasmid per well using Effectene reagent, as previously described (Haim et al., 2011). Three days later, cells were incubated with the indicated primary Abfor 30 min at 37°C. Cells were then washed and incubated with a secondary horseradish peroxidase (HRP)-conjugated Ab for 45 min at room temperature. Cells were washed and HRP enzyme activity was determined after addition of Western Lightning reagents (Perkin Elmer) with a Mithras LB 940 luminometer (Berthold Technologies).
GA Fixation of Cell-surface Env
Three days after transfection, HOS cells were washed twice with fixation buffer (140 mM NaCl, 1.8 mM CaCl2, 1 mM MgCl2 and 10 mM Hepes, pH 7.4) and then incubated with fixation buffer containing 5 mM GA for 15 min at room temperature. As controls, some samples were incubated with fixation buffer with no GA added. GA activity was halted by addition of 25 mM glycine in fixation buffer, which was added to all samples. Subsequently, fixed and non-fixed samples were examined for binding of Abs using the cell-based ELISA method described above. Complete details can be found in Extended Experimental Procedures.
Measurement of the On-rate of Ab Binding to Cell-surface Env
HOS cells cultured in 96-well plates were transfected with the AD8 Env-expressing plasm id. Three days later, cells were incubated with different Abs at 5 µg/ml for different time periods, from 2 to 45 minutes. After incubation of the last sample, all samples were washed and Ab binding was measured using an HRP-conjugated secondary Ab, as described above.
Neutralization of Recombinant HIV-1
Single-round, recombinant HIV-1 viruses that express the luciferase gene were generated by transfection of 293T cells, as described (Haim et al., 2011). For neutralization tests, viruses were exposed to Abs for 1 hr at 37°C prior to the addition of target cells, either CD4+CCR5+ or CD4+CXCR4+ Cf2Th cells. After two days at 37°C, luciferase activity in the target cells was measured. Details of the neutralization assays can be found in Extended Experimental Procedures.
Reversibility of Ab Binding and Inhibition
Recombinant HIV-1 expressing luciferase was purified by ultracentrifugation through a 30% sucrose cushion and then spinoculated onto 96-well protein-binding plates (PerkinElmer). Plate-bound viruses were then incubated with Ab (plus HRP-conjugated Protein G for binding assays) at a 90% inhibitory concentration for 2 hours at 37°C. The virus-Ab complexes were washed extensively before measuring bound Ab (HRP) or adding target cells and measuring infection (luciferase), as described above. Details can be found in Extended Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Antibodies (Abs) differ in the degree of HIV-1 Env perturbation required for binding
Minimally perturbing Abs bind more efficiently and inhibit more effectively
Highly perturbing Abs mainly inhibit perturbation-sensitive “reactive” Envs
Integrating Ab binding & perturbation capacity improves predictions of HIV-1 inhibition
ACKNOWLEDGMENTS
We thank Ms. Yvette McLaughlin and Ms. Elizabeth Carpelan for manuscript preparation. H.H. was supported by an NRSA Postdoctoral Training Program in AIDS Research (NIH T32 AI007386). The National Institutes of Health (AI24755, AI67854 and CFAR Award AI060354), the International AIDS Vaccine Initiative and the late William F. McCarty-Cooper supported this research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures and 3 figures and can be found with this article online.
REFERENCES
- Blumenthal R, Durell S, Viard M. HIV entry and envelope glycoproteinmediated fusion. J. B. Chem. 2012;287:40841–40849. doi: 10.1074/jbc.R112.406272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Kwon YD, Zhou T, Wu X, O'Dell S, Cavacini L, Hessell AJ, Pancera M, Tang M, Xu L, et al. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science. 2009;326:1123–1127. doi: 10.1126/science.1175868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guggenheim EA. Boltzmann's distribution law. Amsterdam, New York: North-Holland Pub. Co.; Interscience Publishers; 1955. [Google Scholar]
- Haim H, Salas I, Sodroski J. Proteolytic processing of the human immunodeficiency virus envelope glycoprotein precursor decreases conformational flexibility. J. Virol. 2013;87:1884–1889. doi: 10.1128/JVI.02765-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haim H, Si Z, Madani N, Wang L, Courter JR, Princiotto A, Kassa A, DeGrace M, McGee-Estrada K, Mefford M, et al. Soluble CD4 and CD4-mimetic compounds inhibit HIV-1 infection by induction of a short-lived activated state. PLoS Pathog. 2009;5:e1000360. doi: 10.1371/journal.ppat.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haim H, Strack B, Kassa A, Madani N, Wang L, Courter JR, Princiotto A, McGee K, Pacheco B, Seaman MS, et al. Contribution of intrinsic reactivity of the HIV-1 envelope glycoproteins to CD4-independent infection and global inhibitor sensitivity. PLoS Pathog. 2011;7:e1002101. doi: 10.1371/journal.ppat.1002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammes GG, Chang YC, Oas TG. Conformational selection or induced fit: a flux description of reaction mechanism. Proc. Natl. Acad. Sci. USA. 2009;106:13737–13741. doi: 10.1073/pnas.0907195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasse PJ, Sattentau QJ. Occupancy and mechanism in antibody-mediated neutralization of animal viruses. J. Gen. Virol. 2002;83:2091–2108. doi: 10.1099/0022-1317-83-9-2091. [DOI] [PubMed] [Google Scholar]
- Kolchinsky P, Kiprilov E, Sodroski J. Increased neutralization sensitivity of CD4-independent human immunodeficiency virus variants. J. Virol. 2001;75:2041–2050. doi: 10.1128/JVI.75.5.2041-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T, et al. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc. Natl. Acad. Sci. USA. 2012;109:5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loving R, Sjoberg M, Wu SR, Binley JM, Garoff H. Inhibition of the HIV-1 spike by single-PG9/16-antibody binding suggests a coordinated-activation model for its three protomeric units. J. Virol. 2013;87:7000–7007. doi: 10.1128/JVI.00530-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR. The cat and mouse of HIV-1 antibody escape. PLoS Pathog. 2009;5:e1000592. doi: 10.1371/journal.ppat.1000592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Snyder SW, Weislow OS, Belay SM, Belshe RB, Schwartz DH, Clements ML, Dolin R, Graham BS, Gorse GJ, et al. Immunization with envelope subunit vaccine products elicits neutralizing antibodies against laboratoryadapted but not primary isolates of human immunodeficiency virus type 1. The National Institute of Allergy and Infectious Diseases AIDS Vaccine Evaluation Group. The J. Infect. Dis. 1996;173:340–348. doi: 10.1093/infdis/173.2.340. [DOI] [PubMed] [Google Scholar]
- Montefiori DC, Mascola JR. Neutralizing antibodies against HIV-1: can we elicit them with vaccines and how much do we need? Curr. Opin. HIV and AIDS. 2009;4:347–351. doi: 10.1097/COH.0b013e32832f4a4d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JP, Cao Y, Qing L, Sattentau QJ, Pyati J, Koduri R, Robinson J, Barbas CF, Burton DR, 3rd, Ho DD. Primary isolates of human immunodeficiency virus type 1 are relatively resistant to neutralization by monoclonal antibodies to gp120, and their neutralization is not predicted by studies with monomeric gp120. J. Virol. 1995;69:101–109. doi: 10.1128/jvi.69.1.101-109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pancera M, Wyatt R. Selective recognition of oligomeric HIV-1 primary isolate envelope glycoproteins by potently neutralizing ligands requires efficient precursor cleavage. Virology. 2005;332:145–156. doi: 10.1016/j.virol.2004.10.042. [DOI] [PubMed] [Google Scholar]
- Parren PW, Mondor I, Naniche D, Ditzel HJ, Klasse PJ, Burton DR, Sattentau QJ. Neutralization of human immunodeficiency virus type 1 by antibody to gp120 is determined primarily by occupancy of sites on the virion irrespective of epitope specificity. J. Virol. 1998;72:3512–3519. doi: 10.1128/jvi.72.5.3512-3519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt EJ, Gomes MM, Kabat D. Kinetic mechanism for HIV-1 neutralization by antibody 2G12 entails reversible glycan binding that slows cell entry. Proc. Natl. Acad. Sci. USA. 2012;109:7829–7834. doi: 10.1073/pnas.1109728109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugach P, Kuhmann SE, Taylor J, Marozsan AJ, Snyder A, Ketas T, Wolinsky SM, Korber BT, Moore JP. The prolonged culture of human immunodeficiency virus type 1 in primary lymphocytes increases its sensitivity to neutralization by soluble CD4. Virology. 2004;321:8–22. doi: 10.1016/j.virol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Sattentau QJ, Moore JP. Human immunodeficiency virus type 1 neutralization is determined by epitope exposure on the gp120 oligomer. J. Exp. Med. 1995;182:185–196. doi: 10.1084/jem.182.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckbeck JD, Orlov I, Chow A, Grieser H, Miller K, Bruno J, Robinson JE, Montelaro RC, Cole KS. Kinetic rates of antibody binding correlate with neutralization sensitivity of variant simian immunodeficiency virus strains. J. Virol. 2005;79:12311–12320. doi: 10.1128/JVI.79.19.12311-12320.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong T, Crooks ET, Osawa K, Binley JM. HIV-1 virus-like particles bearing pure env trimers expose neutralizing epitopes but occlude nonneutralizing epitopes. J. Virol. 2012;86:3574–3587. doi: 10.1128/JVI.06938-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, et al. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Wrin T, Loh TP, Vennari JC, Schuitemaker H, Nunberg JH. Adaptation to persistent growth in the H9 cell line renders a primary isolate of human immunodeficiency virus type 1 sensitive to neutralization by vaccine sera. J. Virol. 1995;69:39–48. doi: 10.1128/jvi.69.1.39-48.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Kwong PD, Desjardins E, Sweet RW, Robinson J, Hendrickson WA, Sodroski JG. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- Yang X, Kurteva S, Lee S, Sodroski J. Stoichiometry of antibody neutralization of human immunodeficiency virus type 1. J. Virol. 2005;79:3500–3508. doi: 10.1128/JVI.79.6.3500-3508.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lipchina I, Cocklin S, Chaiken I, Sodroski J. Antibody binding is a dominant determinant of the efficiency of human immunodeficiency virus type 1 neutralization. J. Virol. 2006;80:11404–11408. doi: 10.1128/JVI.01102-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Bazick J, Sodroski J. Characterization of the multiple conformational States of free monomeric and trimeric human immunodeficiency virus envelope glycoproteins after fixation by cross-linker. J. Virol. 2006;80:6725–6737. doi: 10.1128/JVI.00118-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang PF, Bouma P, Park EJ, Margolick JB, Robinson JE, Zolla-Pazner S, Flora MN, Quinnan GV., Jr A variable region 3 (V3) mutation determines a global neutralization phenotype and CD4-independent infectivity of a human immunodeficiency virus type 1 envelope associated with a broadly crossreactive, primary virus-neutralizing antibody response. J. Virol. 2002;76:644–655. doi: 10.1128/JVI.76.2.644-655.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Georgiev I, Wu X, Yang ZY, Dai K, Finzi A, Kwon YD, Scheid JF, Shi W, Xu L, et al. Structural basis for broad and potent neutralization of HIV-1 by antibody VRC01. Science. 2010;329:811–817. doi: 10.1126/science.1192819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwick MB, Burton DR. HIV-1 neutralization: mechanisms and relevance to vaccine design. Curr. HIV Res. 2007;5:608–624. doi: 10.2174/157016207782418443. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.