Abstract
Pancreatic islet α-cell development and glucagon production are mainly regulated by Pax6 in the homeobox gene families. However, the molecular mechanism fine-tuning the regulation of these events in α-cell still remains unclear. In ocular cells, Pax6 transcription is regulated by CTCF through its binding to specific sites in Pax6 promoter. In this study, CTCF-mediated regulations of islet α-cell development and glucagon production were investigated in both CTCF transgenic mice and α-TC-1-6 cells. Over-expression of CTCF in transgenic mice affected development of pancreatic islets by significantly suppressing α-cell population in both embryonic and adult pancreases. The effect of CTCF on Pax6 gene expression and subsequently, on pro-glucagon production was however, examined in pancreatic islet α-cells. Over-expression and knock-down of CTCF directly affected Pax6 expression. More importantly, the CTCF binding sites upstream from Pax6 p0 promoter were required for regulating p0 promoter activity in islet α-cells. Stimulation of α-cells with insulin resulted in a significant increase in CTCF expression and a decrease in Pax6 expression, and consequently suppressed pro-glucagon expression. In contrast, these insulin-induced effects were blocked by knockdown of CTCF mRNA with specific siRNA in α-cells. Altogether, our results demonstrated for the first time that CTCF functions as a switch-like molecule between the insulin signaling and the regulations of Pax6 and glucagon expression in pancreatic islet α-cells.
Keywords: Pax6, gene regulation, insulin, islet cells, gene transcription, α-cell
Introduction
Glucagon and insulin, two of the most important pancreatic hormones secreted by α- and β-cells located in the islet of Langerhans, are essential for maintenance of glucose homeostasis. They provide opposing effects on controlling the level of blood glucose, one of the most complex endocrine systems in human body [1, 2]. Many immerging evidences support the idea that insulin inhibits glucagon gene transcription and secretion in α-cells [3-10]. Lots of signaling pathways downstream from the insulin receptor were proven to involve gene regulation directed by insulin [11-19]. Among these, PI3 Kinase/PKB pathway is a well-known example that is required for the regulation of glucagon expression [20]. Despite so much data has been published, the molecular mechanism involving glucagon expression in islet α-cells is still, however, not fully understood.
Development and function of pancreatic islet α and β cells are regulated by several transcription factors [21-23]. One of these transcription factors is the homeobox gene product Pax6 (p46), which is initially found in pancreatic progenitor cells. Following pancreatic development, Pax6 is found in all pancreatic endocrine cells and concomitant with the onset of hormone production [24]. Small eye (SEYNeu) mutant mice exhibit markedly reduced numbers of islet α-cells, providing evidence that Pax6 is essential for the formation of the pancreatic α-cell lineage [21-23]. Therefore, Pax6 is suggested to play an important role for directional differentiation of pancreatic progenitor cells to islet α-cells [25]. Furthermore, in a developed pancreas, SEYNeu mutant mice result a decreased production of pancreatic glucagon, suggesting that Pax6 also regulates α-cell endocrine function. The promoter region of the pro-glucagon gene contains 4 cis-acting elements (G1 to G4) that interact with specific transcription factors [26, 27]. G2, G3 and G4 confer as binding sites for islet-specific factors, while G1 element restricts to pro-glucagon specific gene expression in α-cells [28, 29]. As a major transcription factor for insulin-mediated inhibition of glucagon gene, Pax6 binds to the G1 and G3 elements and trans-activates the pro-glucagon gene expression [30] [31].
Two tissue-specific promoters, p0 and p1, are utilized to regulate the Pax6 transcription. However, p0 is the sole promoter responsible for the Pax6 expression in the eyes and pancreas. Pax6 p0 promoter activity is negatively controlled by a CCCTC binding factor, CTCF. This protein is an evolutionarily conserved and ubiquitously expressed zinc finger protein [32, 33]. It plays roles in many epigenetic regulations of gene expression [34-36]. CTCF functions as an insulator such as controling IGF-2 and H19 imprinting through a DNA methylation-sensitive mechanism [37-39]. It binds to many promoters in the human genome to regulate gene transcriptions, and consequently controls activities of many vital regulators for differentiation, cellular senescence, cell cycle control and progression [32, 45, 46]. More recent studies have revealed that CTCF is involved in cancer cell proliferation, tumor suppression, and apoptosis [40-43]. Human CTCF gene possesses a 4.1kb mRNA and a long open reading frame (~2.2 kb) encoding 728 amino acids with a predicted molecular weight (MW) of 82 kDa [44]. Previously, our lab has demonstrated that CTCF negatively regulates Pax6 promoter activity by interacting with a repressor located between ectoderm enhancer (EE) and the Pax6 p0 promoter [47, 48]. CTCF is regulated by EGF and insulin through activation of ERK and AKT signaling cascades [49]. Furthermore, over-expression of CTCF in transgenic mice results in a small eye phenotype that is similar to SEYNeu mice of which Pax6 was mutated [47].
In the present study, we report that pancreatic islet α-cell development and glucagon production were affected by overexpression of CTCF in transgenic mice. Glucagon production and its responses to insulin were also regulated by alterations of CTCF expression in pancreatic islet α cells. In these cells, CTCF was up-regulated by insulin to inhibit Pax6 expression through interaction with the Pax6 P0 promoter. In conclusion, insulin-induced increase and decrease in CTCF expression in α cells resulted in down- and up-regulation of Pax6 and glucagon production, respectively.
Materials and Methods
CTCF transgenic mice preparation
Full-length cDNA encoding human CTCF was subcloned into pcDNA4-to-A expression vector containing a CMV promoter (pcDNA4-CTCF, Invitrogen, CA). Fertilized eggs from B6 donor mice were injected with a linearized Mul1 and Not1 fragment using a micro dispenser. There were total of 4 CTCF transgenic mouse lines generated for the study. Some littermates injected with pcDNA4-to-A vector only were used as controls. Injected eggs were cultured in vitro for 2 days before they were transplanted into the ovarian duct of maternal recipient mice. Embryos were collected from the uterus at day 14. The genotypes of CTCF transgenic mice were confirmed by genomic screenings with a PCR-based procedure. The wildtype siblings from the same litter were used as controls. The experimental procedures performed in this study were in compliance with the NIH Guide for the Care and Use of Laboratory Animals and were approved by the Animal Use Committee at the Los Angeles Biomedical Research Institute's at Harbor-UCLA. The animal facility is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC).
Preparation of pancreatic tissue sections
Pancreatic tissues collected from both mouse embryos and adult mice were fixed with 4% paraformadehyde in PBS and embedded in paraffin blocks as described in Current Protocols in Molecular Biology. Briefly, pancreatic samples were dehydrated by a serial incubation in 50%, 70%, 85%, 95% and 100% ethanol (30 min for each concentration). After dehydration, tissues were incubated in xylene for 1 h. All samples were transferred into xylene/paraffin (50/50) blocks overnight before tissue samples were individually embedded in wax-blocks for processing to thick sections (6.0 μm).
Immunohistochemistry
Tissue sections were membrane-permeated by treatment with 10 mM sodium citrate solution (pH 6.0) for 5 min at 95 °C. Tissue sections were transferred into 0.03% H2O2 solution for 20 min to deactivate endogenous peroxidase followed by 1 h incubation in 10% normal horse serum (NHS) in PBS at 22 °C. For immunostaining experiments, tissue samples were incubated overnight at 4 °C with primary antibodies including IgGs of anti-insulin (1:500, Santa Cruz, CA), anti-glucagon (1:500, Santa Cruz), anti-CTCF (1:200, UpState) and anti-Pax6 (1:200, Covence). After rinsing with PBS, biotin-conjugated secondary antibody (1:100, Santa Cruz) was applied for 1 h at 22 °C. Positively stained cells in tissue sections were visualized by using an ABC kit (Santa Cruz) and an Olympus Fluoview microscope. Cell numbers in pancreatic islet sections were quantified per mm2 by measuring two longer axes and two shorter axes of each islet. Areas of islets were calculated using an equation of: area (mm2) = (islet radius)2 × π. The islet radius was determined by: long axes 1 + long axes 2 + short axes 1 + short axes 2)/(4 × 2.2). Glucagon-positive α cells in each islet were counted and normalized by islet areas (mm2). At least ten islets per section and three sections from each pancreas were counted.
Cell culture and transfection
Mouse pancreatic α-TC-1-6 cell line was cultured in RPMI-1640 medium supplemented with 10% FBS in a humidified incubator with 5% CO2 at 37 °C. Cells were synchronized by serum-starvation for at least 24 hr prior to various treatments as indicated. Gene-specific expression vectors and promoter-containing lacZ vectors were introduced into α-cells by transfection using a Lipofectin kit (Invitrogen, CA) as instructed by manufacture. For CTCF knocking down experiments, 25 nM CTCF-specific siRNA and 12 μL transfection reagent (HiPerFect; H301705; Qiagen) in 100 μL culture medium without serum. Transfection complexes in the mixture were formed after 10 minutes of incubation at room temperature. The mixture was evenly and slowly dropped into cultured cells. Transfected cells were cultured under normal growth conditions for 48 to 84 hours before the experiments. Control cells were transfected with nonsilencing siRNA using the method described above.
Western blot analysis
Cells (5×105) were treated as indicated and harvested in 0.5 ml lysing buffer which contains 137mM NaCl, 1.5mM MgCl2, 2mM EDTA, 10mM Na-pyrophosphate, 25mM β-glycerophosphate, 10% glycerol, 1% Triton X-100, 1mM Naorthovanadate, 1mM phenylmethylsulfonyl fluoride, and 20mM Tris, pH 7.5. Cell lysates were pre-cleared by centrifugation at 13,000xg, 4C for 20 min and boiled in sample buffer. Equal amounts of the protein samples were fractionated by electrophoresis with 7% and 15% SDS-PAGE gels. The fractionated proteins were transferred onto PDF membranes, probed with primary antibodies against pro-glucagon, CTCF and Pax6 (Santa Cruz Biotechnology, Santa Cruz, CA). HRP-conjugated secondary antibodies were applied at 1:1000 dilution and targeted protein bands were visualized using Western blotting Luminol Reagent (Santa Cruz Biotechnology, CA).
RNA purification and RT-qPCR
Cells were treated with insulin for periods of time as indicated and harvested. Total RNA was purified by Trizol reagent (Bio-Rad) and treated with RNase-free DNaseI to remove contaminated DNAs before being reverse-transcribed into cDNA with MMLv RT kit (Promega). qPCR was performed by combining cDNA with mouse primer sets for CTCF, Pax6 and Pro-glucagon and Taq polymerase mix from Applied Biosystem, Inc. The experiments were performed multiple times and each set of data was represented as a mean of all the experiments +/− SE.
β-galactosidase assay for Pax6 promoter activity
Pax6 P0 promoter and its deletion mutant constructs were transfected into α-cells with either the control or CTCF over-expression plasmid. Cells were harvested 48 h after transfection by rinsing two times with ice-cold PBS and suspended in lysis buffer containing 100 mM potassium phosphate buffer with 1 mM dithiothreitol, pH 7.8. Cell lysates were pre-cleared by centrifugation at 13,000×g for 5 min and the supernatants were incubated with a chemiluminescent substrate, galacton-star (invitrogen, CA) to determine β-galactosidase activity by a luminometer. pBRL-TK was co-transfected into the cells and served as an internal control to normalize the Pax6 P0 reporter lacZ activity. The leuciferase activity for internal controls was measured by using a Dual-Luciferase reporter assay system (Promega, CA). The experiments were performed at least 3 times and each set of data was plotted as a mean of all experiments +/− SE. Significant differences between the control groups and experimental groups were determined by One-way ANOVA and Student's t test at p<0.05.
Results
Effect of CTCF overexpression on islet cell development and α-cell population in adult mice
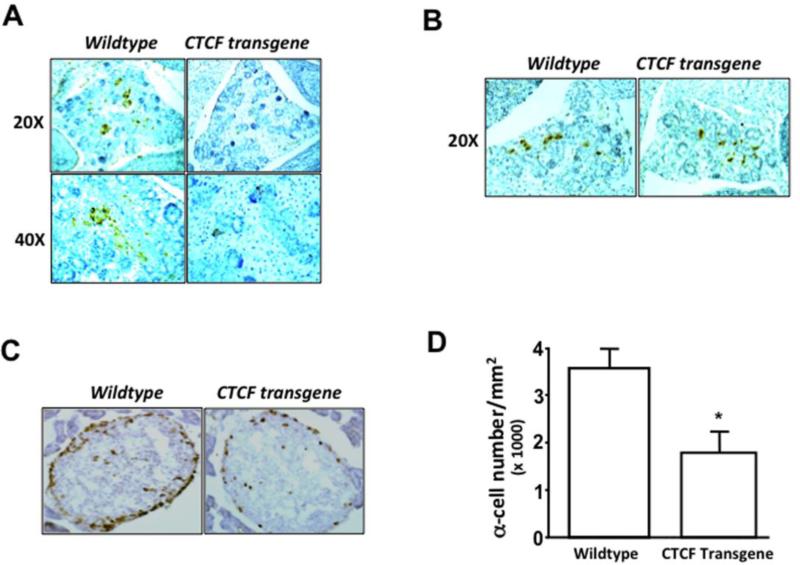
Fourteen-day (E14) mouse embryonic samples were obtained from the wildtype and the sibling CTCF transgenic animals in the same litter. The transgene encoding CTCF in mouse tissues were verified by PCR detection. There was no morphological defects observed in pancreatic islets during embryonic developments observed from Homatoxylin-Errosin (H-E)-stained tissue sections (data not shown). However, immunostaining using the specific antibody against proglucagon showed glucagon-positive α cells in pancreatic islets only in wildtype mice instead of CTCF transgenic mice (Fig. 1A). In addition, pancreatic sections were also stained with specific antibody against insulin. It showed a normal expression level of insulin-positive β cells in pancreatic islets of both wildtype and CTCF transgenic mice (Fig. 1B). These data suggest that over-expression of CTCF transgene in mice results in suppression of glucagon-positive α cells without affecting insulin-positive β cells in developmental pancreatic islets. Furthermore, pancreatic tissue sections of 5-6 week-old mice were examined by immunostaining with both anti-proglucagon and anti-insulin antibodies. Positively stained islet cells were counted in each section under a microscope. We found that there was no difference in pancreatic islet β cell population between wild-type and CTCF transgenic mice (data not shown). However, numbers of glucagon-staining positive α cells surrounded at edges of islets were markedly decreased in CTCF transgenic mice compared with its wildtype counterpart (Fig. 1C). The population of α cells was calculated from 10 islets in each slide section and at least 3 sections from each animal. After comparing 20 wildtype and 15 CTCF transgenic mice in isolated pancreatic tissues, we found that the population of islet α cells was significantly decreased from 35.6±4 ×1000/mm2 in the pancreas of wildtype mouse to 20.7±5 ×1000/mm2 (approximately 42%) in the pancreas of CTCF transgenic mouse (Fig. 1D). These results suggest that CTCF plays an important role in the development of pancreatic islet α cells.
Figure 1. Effect of over-expressing CTCF on mouse islet α cell development.
(A) Immunostaining of pro-glucagon-containing cells in pancreatic islets. (B) Immunostaining of insulin-containing cells in pancreatic islets. Tissue sections were prepared from embryonic pancreases of sibling wild-type and CTCF transgenic mice at day 14 (E14). Pancreatic tissues were stained with anti-glucagon and anti-insulin antibodies. (C) Identification of pro-glucagon-positive α cells in pancreatic islets of adult mice by immunostaining with anti-glucagon antibodies. (D) Comparison of α cell populations in pancreatic islets of sibling wildtype and CTCF transgenic mice. Cell numbers were quantified by measuring pro-glucagon-positive cells per mm2. Pancreatic tissue sections were prepared from 4-5 months old wildtype and CTCF transgenic mice. Symbol “*” represents a significant difference (p<0.05, n=15).
Effect of altered CTCF levels on Pax6 expression in islet α-cells
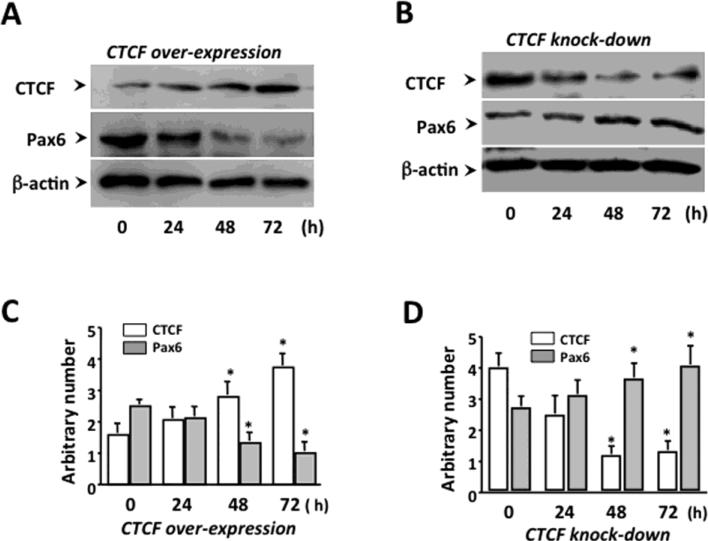
Pax6 plays key roles in the control of pancreatic islet α cell development and glucagon production and is regulated by CTCF through transcription control. To explore the roles of CTCF in the regulation of islet α-cell function mediated through Pax6, mouse pancreatic islet α-cell line was used to detect cellular Pax6 expression in CTCF-altered states. Western blot analysis showed that transfection of CTCF over-expression construct increased CTCF protein level in α-cells and significantly down-regulated the endogenous Pax6 protein level at 48 and 72 hours after over-expressing CTCF (Fig. 2A&C). In contrast, knockdown of CTCF with CTCF-specific siRNA resulted in a decreased CTCF protein level, and significantly up-regulated Pax6 expression within 48 to 72 hrs after knocking down the CTCF (Fig. 2B&D).
Figure 2. Effects of altered CTCF levels on Pax6 expression in islet α cells.
(A) Effect of CTCF over-expression on the suppression of Pax6 expression. (B) Effect of knocking down CTCF on Pax6 expression. (C) Statistical analysis of suppressing Pax6 expression by over-expression of CTCF. (D) Statistical analysis of knocking down CTCF affecting Pax6 expression. Pancreatic α-cells were transfected with cDNA encoding full-length CTCF and siRNA specific to CTCF to over-express CTCF and to knock down CTCF, respectively. Cells were lysed and CTCF and Pax6 expression levels were analyzed by Western blots. All of the bar-graphs on the right depict the densitometry analysis of the corresponding bands on the figures. Band intensity was normalized against its internal control β-actin band. Symbol “*” represents a significant difference (p<0.05, n=4).
Effect of CTCF on Pax6 promoter activity in islet α-cells
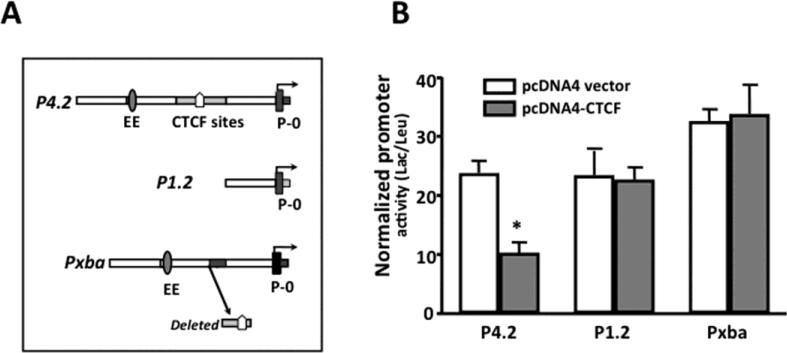
The promoter activity of Pax6 was studied in α-cells by transfecting the cells with a Pax6 P4.2 reporter construct (wild type) and two mutants of the reporter constructs, P1.2 and Pxba. P1.2 contains upstream EE and CTCF-binding motif deletion and Pxba has a defined CTCF-binding motif deletion (Fig. 3A). In addition, α-cells were co-transfected with pcDNA4-hCTCF construct expressing full-length CTCF. The pcDNA4 vector alone was included in the other group of α-cells as a control experiment. Over-expression of CTCF resulted in significant decrease in Pax6P4.2 reporter activity, but it did not affect reporter activities of P1.2 and Pxba deletion mutants that lack CTCF binding domain located at 1.1 kb upstream from Pax6 p0 promoter (Fig. 3B). Therefore, our data indicate that CTCF plays a functional role in control of Pax6 expression at transcriptional level in pancreatic islet α-cells.
Figure 3. Effect of over-expressed CTCF on Pax6 promoter activity in islet α-cells.
(A) Wild-type (P4.2) and its deletion mutants (P1.2 and Pxba) of Pax6 P0 promoter reporter constructs are diagramed schematically on the left. (B) Over-expressed CTCF decreased the Pax6 P0 promoter activity only in the presence of an intact up-stream CTCF binding site. Pax6 P0 promoter reporter (P4.2) and its deletion mutants (P1.2 and Pxba) that lack the CTCF binding region were co-transfected with pcDNA4-CTCF into α-cells by lipofections. The pcDNA4 vector was transfected into α-cells to replace pcDNA4-CTCF as a negative control. The pRL-TK vector was included in all the transfections as internal controls. β-galactosidase and leuciferase activities were determined 48 h after transfection. β-galactosidase activities were normalized to those of the RL luciferase as a transfection efficiency control and are shown on the right. Data represent the mean of at least 2 independent experiments. Symbol “*” indicates significant difference between CTCF over-expression and the vector-transfected control cells (p<0.05, n= 4).
Effect of insulin-induced CTCF up-regulation on Pax6 expression
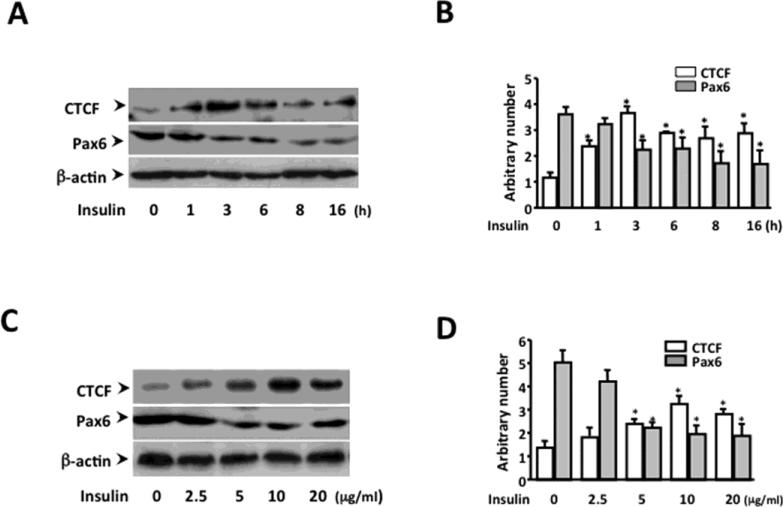
Under normal physiological conditions, glucose stimulates insulin secretion from β-cells; and insulin suppresses the release of glucagon from α-cells in pancreatic islets. It has been shown that pancreatic islets require insulin prior to responding to glucose deprivation for releasing glucagon [51-53]. To study the time course of the direct effect of insulin, pancreatic islet α-cells were treated with 20 μg/ml of insulin for periods of time as indicated (Fig. 4A). After insulin treatment for 1 hour, the CTCF expression level was increased 2-time higher, and reached the peak level (4-fold higher) in 3 hours. On the other hand, the expression of Pax6 was decreased in 3 hours after the insulin treatment (Fig. 4B). In insulin dose-response studies, application of 2.5 to 5.0 μg/ml insulin onto α-cells stimulated a subtle increase in CTCF expression, and a peak expression level of CTCF was obtained by 10 μg/ml. In addition, application of insulin induced a significant suppression of Pax6 expression at a dosage of 5 and 10 μg/ml insulin (Fig. 4C and 4D). The data suggested that there was a closely reciprocal relationship between CTCF and Pax6 expression levels in insulin-induced α-cells following dose- and time- dependent patterns. To eliminate the possible effects of glucose variations in growth media on CTCF and Pax6expression, different concentrations of glucose were applied to α-cells. We found that neither the physiological concentration of glucose in the growth media nor non-physiologically higher glucose concentrations (up to 18g/L) would affect CTCF or Pax6 expression (data not shown). This result indicated that glucose concentrations used in this experiment doesn't have any effect on CTCF and Pax6 expression in pancreatic islet α-cells. The finding reveals that insulin up-regulates CTCF expression and in parallel to suppress Pax6 expression, which is consistent with the results from manipulating CTCF level shown in the previous figures in these cells.
Figure 4. Effect of insulin on CTCF and Pax6 expression in pancreatic islet α cells.
(A) Time course experiment of insulin-induced changes in the expressions of CTCF and Pax6. (B) Statistic analysis of insulin-induced CTCF and Pax6 expression following a time course. (C) Dose effect of insulin on CTCF and Pax6 expression. (D) Statistic analysis of various insulin dosage-induced changes of CTCF and Pax6 expression. Islet α-cells were starved and treated with insulin at different time points or grown in media with different concentrations of insulin as indicated. Cells were lysed and proteins were solubilized in sample buffer before fractionated onto a SDS-PAGE. Western blots were performed. Bar graphs on the right depict the densitometry analysis of the corresponding bands on the figures. Band intensity was normalized against its internal control α-actin band. Symbol “*” indicates significant difference between insulin-induced and mock-induced α-cells (p<0.05, n= 3)
Effect of insulin on CTCF, Pax6 and proglucagon mRNA expressions
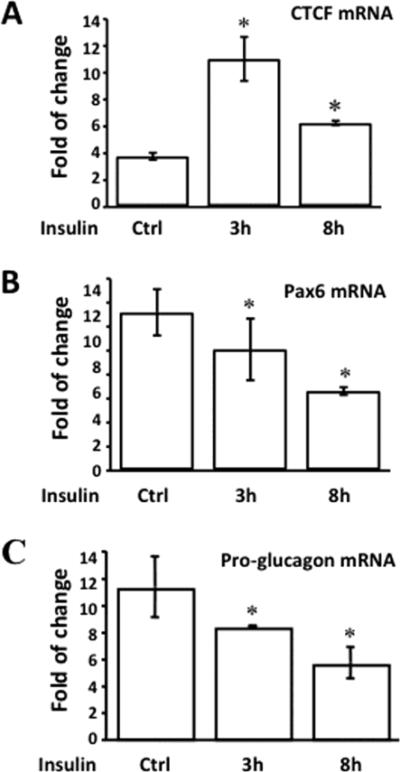
Steady-state RNA levels in islet α-cells after the treatment of insulin were determined by quantitative real-time PCR (RT-qPCR) (Fig. 5). The CTCF mRNA level was increased approximately 4 folds after the treatment of the cells with insulin for 3 hours (Fig. 5A). On the other hand, both Pax6 and pro-glucagon mRNA levels were decreased along with insulin treatments (Fig. 5B & 5C). The results suggested that up-regulation of CTCF and down-regulation of Pax6 and glucagon in α-cells by insulin were proven to be evident at the RNA level.
Figure 5. Effect of insulin on steady-state mRNA levels of CTCF, Pax6 and pro-glucagon in pancreatic islet α cells.
(A) Steady-state mRNA level of CTCF. (B) Steady-state mRNA level of Pax6. (C) Steady-state mRNA level of pro-glucagon. Islet α-cells were starved and treated with insulin (5μg/ml) for different periods of time as indicated. Cells were lysed and RNAs were purified with Trizol. Reverse-transcription was performed and cDNAs were used for qPCR with primer sets for CTCF, Pax6 and pro-glucagon. Symbol “*” represents significant differences in the absence and presence of insulin stimulation (p<0.05, n=6).
Effect of knocking down CTCF or Pax6 on insulin-suppressed glucagon production
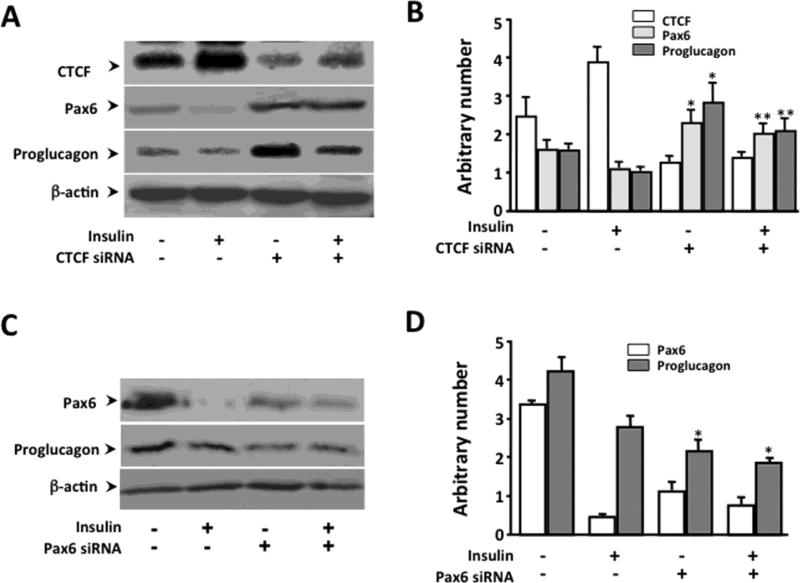
Next, we examined whether the inhibitory effect of insulin on production of pro-glucagon is mediated by a mechanism involving CTCF-regulated Pax6 activity. It has been known from previous studies that activation of PI3 kinase/PKB pathway has been shown to down-regulate Pax6 activity and eventually, suppress the promoter activity of glucagon [20]. However, little is known about the connection between this signaling pathway and Pax6 that regulates glucagon production in α-cells. We have demonstrated that insulin induced an increase in CTCF level, and later resulted in suppression of Pax6 expression. Pax6 promoter activity is sensitive to the level of CTCF only if the CTCF binding sites are present. To further verify the effect of insulin-activated CTCF on suppressing Pax6 and glucagon production, CTCF was knocked down in α-cells by transfecting cells with CTCF-specific siRNA. Diminished CTCF effectively abolished the effect of insulin on suppression of Pax6 expression and insulin-induced suppression of proglucagon production (Fig. 6A&B). The effect of Pax6 on glucagon production was also verified by knocking down Pax6 mRNA by transfecting α-cells with CTCF-specific siRNA. Suprression of Pax6 expression by knockdown of Pax6 mRNA effectively reduced pro-glucagon production (Fig. 6C&D). Thees results demonstrate that CTCF plays a crucial role in mediating insulin-induced down-regulation of Pax6 expression, and subsequently suppresses glucagon synthesis.
Figure 6. Effect of knocking down CTCF or Pax6 on insulin-induced suppression of pro-glucagon production.
(A) Effect of knocking down CTCF on insulin-induced suppression of Pax6 expression and pro-glucagon production. (B) Statistical analysis of knocking down CTCF affecting insulin-induced suppression of Pax6 expression and pro-glucagon production. (C) Effect of knocking down Pax6 on pro-glucagon expression. (D) Statistical analysis of knocking down Pax6 affecting pro-glucagon expression. CTCF or Pax6 specific siRNA and control siRNA were transfected into α-cells by lipofection, and the cells were treated with insulin and collected 48 h after transfection. CTCF, Pax6 and pro-glucagon expressions in α-cells were detected by Western analysis. Symbols “*” and “**” represent significant differences in the absence and presence of insulin in CTCF or Pax6 siRNA-knocking down cells (p<0.05, n=3).
Discussion
Glucagon and insulin are produced in pancreatic islet by α and β cells, respectively. They are essential for maintenances of the glucose homeostasis. Previous studies showed that glucagon production is regulated by Pax6 at transcriptional level [25, 54, 55]. However, the molecular mechanism of Pax6 regulation in pancreatic islet α-cells is still unknown. There is a consistently reciprocal relationship in the expression pattern between CTCF and Pax6 found in many cells. In the present study, we report that CTCF functioned as a switch-like molecule upon insulin stimulation to negatively regulate Pax6 expression, and consequently gave rise to a less proglucagon production by pancreatic islet α-cells. The regulatory relationship in α-cells was predominately at transcriptional level as CTCF manipulate Pax6 promoter through its up-stream binding motifs (Fig. 3). This is consistent with the previous finding that CTCF binds to a pivotal cis-acting regulatory DNA element on Pax6 promoter and down-regulates Pax6 by blocking the communication between the ectoderm enhancer (EE) and Pax6 p0 promoter [37, 47, 56].
Furthermore, the expression of CTCF in α-cells is proven to be regulated by insulin, which is similar to the previous findings that CTCF expression is mediated by epidermal growth factor- and insulin-induced Erk and PI3K/PKB activation in corneal epithelial and hematopoietic myeloid cells [50, 57]. A publication showed that the activation of PI3-kinase/PKB pathway is essential to the glucagon synthesis in islet α-cells [20]. Activation of PKB by insulin in α-cells is also observed in our lab (data not shown). Thus, it needs further studies to understand if there is a relationship between upstream PI3K/PKB pathway and downstream regulation of CTCF, Pax6, and glucagon expressions by insulin in islet α-cells. Diao, et al. reported that low concentration of glucose up-regulates glucagon secretion in pancreatic a cells [9]. In our study in the physiological condition, the glucose concentration in the growth medium is 10 times higher than the glucose concentration that they have used to observe both the insulin and glucagon effects. We have also performed control experiments in very high glucose concentrations. Our result showed that changes of the glucagon level in α-cells were independent to the tested glucose concentrations presented in the growth media.
In the present study, a mouse model with over-expressed CTCF affected pancreatic islet development by suppressing pro-glucagon-positive α cells (Fig 1). This result further emphasizes that there must be a physiological role for CTCF to play in pancreatic α-cell function and glucagon production. An insulin-induced increase in CTCF or over-expressing CTCF in α-cells effectively suppresses Pax6 expression, resulting in decreased productions of glucagon. These results suggest that there is an insulin-induced feedback mechanism for regulation of glucagon production. In diabetic condition, some patients suffer from hyperglycemia resulting from hyperglucagonemia. This elevated glucagon level may be possible due to the abnormal level of insulin, which is required for inhibition of the glucagon synthesis and secretion. Therefore, to understand clearly the mechanism of glucagon regulation in α-cells may bring insights to help control of diabetes. Thus, we believe that it is very likely insulin-induced suppression of glucagon production mediated through CTCF may have important physiological and pathological implications that can be served for therapeutic purpose.
Acknowledgments
We appreciate Dr. R. Paul Robertson (Pacific Northwest Research Institute) for providing us pancreatic islet cells.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lefebvre PJ. Glucagon and its family revisited. Diabetes Care. 1995;18:715–730. doi: 10.2337/diacare.18.5.715. [DOI] [PubMed] [Google Scholar]
- 2.Unger RH, Orci L. Glucagon and the A cell: physiology and pathophysiology (first two parts) N Engl J Med. 1981;304:1518–1524. doi: 10.1056/NEJM198106183042504. [DOI] [PubMed] [Google Scholar]
- 3.Baulmann D, Ohlmann A, Flugel-Koch C, Goswami S, Cvekl A, Tamm E. Pax6 heterozygous eyes show defects in chamber angle differentiation that are associated with a wide spectrum of other anterior eye segment abnormalities. Mech Dev. 2002;118:3. doi: 10.1016/s0925-4773(02)00260-5. [DOI] [PubMed] [Google Scholar]
- 4.Bonner-Weir S, Orci L. New perspectives on the microvasculature of the islets of Langerhans in the rat. Diabetes. 1982;31:883–889. doi: 10.2337/diab.31.10.883. [DOI] [PubMed] [Google Scholar]
- 5.Samols E, Stagner JI. Intra-islet regulation. Am J Med. 1988;85:31–35. doi: 10.1016/0002-9343(88)90395-6. [DOI] [PubMed] [Google Scholar]
- 6.Stagner JI, Samols E. Retrograde perfusion as a model for testing the relative effects of glucose versus insulin on the A cell. J Clin Invest. 1986;77:1034–1037. doi: 10.1172/JCI112356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawamori D, Kurpad AJ, Hu J, Liew CW, Shih JL, Ford EL, Herrera PL, Polonsky KS, McGuinness OP, Kulkarni RN. Insulin signaling in alpha cells modulates glucagon secretion in vivo. Cell Metab. 2009;9:350–361. doi: 10.1016/j.cmet.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leung YM, Ahmed I, Sheu L, Gao X, Hara M, Tsushima RG, Diamant NE, Gaisano HY. Insulin regulates islet alpha-cell function by reducing KATP channel sensitivity to adenosine 5′-triphosphate inhibition. Endocrinology. 2006;147:2155–2162. doi: 10.1210/en.2005-1249. [DOI] [PubMed] [Google Scholar]
- 9.Diao J, Asghar Z, Chan CB, Wheeler MB. Glucose-regulated glucagon secretion requires insulin receptor expression in pancreatic alpha-cells. J Biol Chem. 2005;280:33487–33496. doi: 10.1074/jbc.M506276200. [DOI] [PubMed] [Google Scholar]
- 10.Schrader H, Menge BA, Breuer TG, Ritter PR, Uhl W, Schmidt WE, Holst JJ, Meier JJ. Impaired glucose-induced glucagon suppression after partial pancreatectomy. J Clin Endocrinol Metab. 2009;94:2857–2863. doi: 10.1210/jc.2009-0826. [DOI] [PubMed] [Google Scholar]
- 11.Banfi C, Eriksson P, Giandomenico G, Mussoni L, Sironi L, Hamsten A, Tremoli E. Transcriptional regulation of plasminogen activator inhibitor type 1 gene by insulin: insights into the signaling pathway. Diabetes. 2001;50:1522–1530. doi: 10.2337/diabetes.50.7.1522. [DOI] [PubMed] [Google Scholar]
- 12.Guo S, Rena G, Cichy S, He X, Cohen P, Unterman T. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J Biol Chem. 1999;274:17184–17192. doi: 10.1074/jbc.274.24.17184. [DOI] [PubMed] [Google Scholar]
- 13.Kotani K, Ogawa W, Hino Y, Kitamura T, Ueno H, Sano W, Sutherland C, Granner DK, Kasuga M. Dominant negative forms of Akt (protein kinase B) and atypical protein kinase Clambda do not prevent insulin inhibition of phosphoenolpyruvate carboxykinase gene transcription. J Biol Chem. 1999;274:21305–21312. doi: 10.1074/jbc.274.30.21305. [DOI] [PubMed] [Google Scholar]
- 14.Leibiger B, Leibiger IB, Moede T, Kemper S, Kulkarni RN, Kahn CR, de Vargas LM, Berggren PO. Selective insulin signaling through A and B insulin receptors regulates transcription of insulin and glucokinase genes in pancreatic beta cells. Mol Cell. 2001;7:559–570. doi: 10.1016/s1097-2765(01)00203-9. [DOI] [PubMed] [Google Scholar]
- 15.Leibiger IB, Leibiger B, Moede T, Berggren PO. Exocytosis of insulin promotes insulin gene transcription via the insulin receptor/PI-3 kinase/p70 s6 kinase and CaM kinase pathways. Mol Cell. 1998;1:933–938. doi: 10.1016/s1097-2765(00)80093-3. [DOI] [PubMed] [Google Scholar]
- 16.Liao J, Barthel A, Nakatani K, Roth RA. Activation of protein kinase B/Akt is sufficient to repress the glucocorticoid and cAMP induction of phosphoenolpyruvate carboxykinase gene. J Biol Chem. 1998;273:27320–27324. doi: 10.1074/jbc.273.42.27320. [DOI] [PubMed] [Google Scholar]
- 17.Lochhead PA, Coghlan M, Rice SQ, Sutherland C. Inhibition of GSK-3 selectively reduces glucose-6-phosphatase and phosphatase and phosphoenolypyruvate carboxykinase gene expression. Diabetes. 2001;50:937–946. doi: 10.2337/diabetes.50.5.937. [DOI] [PubMed] [Google Scholar]
- 18.Scassa ME, Guberman AS, Varone CL, Canepa ET. Phosphatidylinositol 3-kinase and Ras/mitogen-activated protein kinase signaling pathways are required for the regulation of 5-aminolevulinate synthase gene expression by insulin. Exp Cell Res. 2001;271:201–213. doi: 10.1006/excr.2001.5386. [DOI] [PubMed] [Google Scholar]
- 19.Schmoll D, Walker KS, Alessi DR, Grempler R, Burchell A, Guo S, Walther R, Unterman TG. Regulation of glucose-6-phosphatase gene expression by protein kinase Balpha and the forkhead transcription factor FKHR. Evidence for insulin response unit-dependent and -independent effects of insulin on promoter activity. J Biol Chem. 2000;275:36324–36333. doi: 10.1074/jbc.M003616200. [DOI] [PubMed] [Google Scholar]
- 20.Schinner S, Barthel A, Dellas C, Grzeskowiak R, Sharma SK, Oetjen E, Blume R, Knepel W. Protein kinase B activity is sufficient to mimic the effect of insulin on glucagon gene transcription. J Biol Chem. 2005;280:7369–7376. doi: 10.1074/jbc.M408560200. [DOI] [PubMed] [Google Scholar]
- 21.Sander M, Neubuser A, Kalamaras J, Ee HC, Martin GR, German MS. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes Dev. 1997;11:1662–1673. doi: 10.1101/gad.11.13.1662. [DOI] [PubMed] [Google Scholar]
- 22.Sander M, German MS. The beta cell transcription factors and development of the pancreas. J Mol Med. 1997;75:327–340. doi: 10.1007/s001090050118. [DOI] [PubMed] [Google Scholar]
- 23.Gosmain Y, Marthinet E, Cheyssac C, Guerardel A, Mamin A, Katz LS, Karim B, Philippe J. PAX6 controls the expression of critical genes involved in pancreatic {alpha}-cell differentiation and function. J Biol Chem. 2010 doi: 10.1074/jbc.M110.147215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamasaki A, Yamada Y, Kurose T, Ban N, Nagashima K, Takahashi A, Fujimoto S, Shimono D, Fujiwara M, Toyokuni S, Seino Y, Inagaki N. Adult pancreatic islets require differential pax6 gene dosage. Biochem Biophys Res Commun. 2007;353:40–46. doi: 10.1016/j.bbrc.2006.11.105. [DOI] [PubMed] [Google Scholar]
- 25.St-Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A, Gruss P. Pax6 is required for differentiation of glucagon-producing alpha-cells in mouse pancreas. Nature. 1997;387:406–409. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- 26.Philippe J, Drucker DJ, Knepel W, Jepeal L, Misulovin Z, Habener JF. Alpha-cell-specific expression of the glucagon gene is conferred to the glucagon promoter element by the interactions of DNA-binding proteins. Mol Cell Biol. 1988;8:4877–4888. doi: 10.1128/mcb.8.11.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Philippe J. Insulin regulation of the glucagon gene is mediated by an insulin-responsive DNA element. Proc Natl Acad Sci U S A. 1991;88:7224–7227. doi: 10.1073/pnas.88.16.7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morel C, Cordier-Bussat M, Philippe J. The upstream promoter element of the glucagon gene, G1, confers pancreatic alpha cell-specific expression. J Biol Chem. 1995;270:3046–3055. doi: 10.1074/jbc.270.7.3046. [DOI] [PubMed] [Google Scholar]
- 29.Cordier-Bussat M, Morel C, Philippe J. Homologous DNA sequences and cellular factors are implicated in the control of glucagon and insulin gene expression. Mol Cell Biol. 1995;15:3904–3916. doi: 10.1128/mcb.15.7.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ritz-Laser B, Estreicher A, Klages N, Saule S, Philippe J. Pax-6 and Cdx-2/3 interact to activate glucagon gene expression on the G1 control element. J Biol Chem. 1999;274:4124–4132. doi: 10.1074/jbc.274.7.4124. [DOI] [PubMed] [Google Scholar]
- 31.Grzeskowiak R, Amin J, Oetjen E, Knepel W. Insulin responsiveness of the glucagon gene conferred by interactions between proximal promoter and more distal enhancer-like elements involving the paired-domain transcription factor Pax6. J Biol Chem. 2000;275:30037–30045. doi: 10.1074/jbc.M000984200. [DOI] [PubMed] [Google Scholar]
- 32.Filippova GN, Fagerlie S, Klenova EM, Myers C, Dehner Y, Goodwin G, Neiman PE, Collins SJ, Lobanenkov VV. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol Cell Biol. 1996;16:2802–2813. doi: 10.1128/mcb.16.6.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filippova GN, Lindblom A, Meincke LJ, Klenova EM, Neiman PE, Collins SJ, Doggett NA, Lobanenkov VV. A widely expressed transcription factor with multiple DNA sequence specificity, CTCF, is localized at chromosome segment 16q22.1 within one of the smallest regions of overlap for common deletions in breast and prostate cancers. Genes Chromosomes Cancer. 1998;22:26–36. [PubMed] [Google Scholar]
- 34.Baniahmad A, Steiner C, Kohne AC, Renkawitz R. Modular structure of a chicken lysozyme silencer: involvement of an unusual thyroid hormone receptor binding site. Cell. 1990;61:505–514. doi: 10.1016/0092-8674(90)90532-j. [DOI] [PubMed] [Google Scholar]
- 35.Bell KD, Campbell RJ, Bourne WM. Pathology of late endothelial failure: late endothelial failure of penetrating keratoplasty: study with light and electron microscopy. Cornea. 2000;19:40–46. doi: 10.1097/00003226-200001000-00009. [DOI] [PubMed] [Google Scholar]
- 36.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 37.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 38.Nakagawa H, Chadwick RB, Peltomaki P, Plass C, Nakamura Y, de La Chapelle A. Loss of imprinting of the insulin-like growth factor II gene occurs by biallelic methylation in a core region of H19-associated CTCF-binding sites in colorectal cancer. Proc Natl Acad Sci U S A. 2001;98:591–596. doi: 10.1073/pnas.011528698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Percec I, Bartolomei MS. Genetics. Do X chromosomes set boundaries? Science. 2002;295:287–288. doi: 10.1126/science.1068663. [DOI] [PubMed] [Google Scholar]
- 40.Qi CF, Martensson A, Mattioli M, Dalla-Favera R, Lobanenkov VV, Morse HC., 3rd CTCF functions as a critical regulator of cell-cycle arrest and death after ligation of the B cell receptor on immature B cells. Proc Natl Acad Sci U S A. 2003;100:633–638. doi: 10.1073/pnas.0237127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rasko JE, Klenova EM, Leon J, Filippova GN, Loukinov DI, Vatolin S, Robinson AF, Hu YJ, Ulmer J, Ward MD, Pugacheva EM, Neiman PE, Morse HC, 3rd, Collins SJ, Lobanenkov VV. Cell growth inhibition by the multifunctional multivalent zinc-finger factor CTCF. Cancer Res. 2001;61:6002–6007. [PubMed] [Google Scholar]
- 42.Docquier F, Farrar D, D'Arcy V, Chernukhin I, Robinson AF, Loukinov D, Vatolin S, Pack S, Mackay A, Harris RA, Dorricott H, O'Hare MJ, Lobanenkov V, Klenova E. Heightened expression of CTCF in breast cancer cells is associated with resistance to apoptosis. Cancer Res. 2005;65:5112–5122. doi: 10.1158/0008-5472.CAN-03-3498. [DOI] [PubMed] [Google Scholar]
- 43.Fiorentino FP, Giordano A. The tumor suppressor role of CTCF. J Cell Physiol. 2011 doi: 10.1002/jcp.22780. [DOI] [PubMed] [Google Scholar]
- 44.Klenova EM, Nicolas RH, U S, Carne AF, Lee RE, Lobanenkov VV, Goodwin GH. Molecular weight abnormalities of the CTCF transcription factor: CTCF migrates aberrantly in SDS-PAGE and the size of the expressed protein is affected by the UTRs and sequences within the coding region of the CTCF gene. Nucleic Acids Res. 1997;25:466–474. doi: 10.1093/nar/25.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Awad TA, Bigler J, Ulmer JE, Hu YJ, Moore JM, Lutz M, Neiman PE, Collins SJ, Renkawitz R, Lobanenkov VV, Filippova GN. Negative transcriptional regulation mediated by thyroid hormone response element 144 requires binding of the multivalent factor CTCF to a novel target DNA sequence. J Biol Chem. 1999;274:27092–27098. doi: 10.1074/jbc.274.38.27092. [DOI] [PubMed] [Google Scholar]
- 46.Burcin M, Arnold R, Lutz M, Kaiser B, Runge D, Lottspeich F, Filippova GN, Lobanenkov VV, Renkawitz R. Negative protein 1, which is required for function of the chicken lysozyme gene silencer in conjunction with hormone receptors, is identical to the multivalent zinc finger repressor CTCF. Mol Cell Biol. 1997;17:1281–1288. doi: 10.1128/mcb.17.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li T, Lu Z, Lu L. Regulation of eye development by transcription control of CCCTC binding factor (CTCF) J Biol Chem. 2004;279:27575–27583. doi: 10.1074/jbc.M313942200. [DOI] [PubMed] [Google Scholar]
- 48.Li T, Lu Z, Lu L. Pax6 regulation in retinal cells by CCCTC binding factor. Invest Ophthalmol Vis Sci. 2006;47:5218–5226. doi: 10.1167/iovs.06-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li T, Lu L. Functional role of CCCTC binding factor (CTCF) in stress-induced apoptosis. Exp Cell Res. 2007;313:3057–3065. doi: 10.1016/j.yexcr.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li T, Lu L. Epidermal growth factor-induced proliferation requires down-regulation of Pax6 in corneal epithelial cells. J Biol Chem. 2005;280:12988–12995. doi: 10.1074/jbc.M412458200. [DOI] [PubMed] [Google Scholar]
- 51.Hope KM, Tran PO, Zhou H, Oseid E, Leroy E, Robertson RP. Regulation of alpha-cell function by the beta-cell in isolated human and rat islets deprived of glucose: the “switch-off” hypothesis. Diabetes. 2004;53:1488–1495. doi: 10.2337/diabetes.53.6.1488. [DOI] [PubMed] [Google Scholar]
- 52.Zhou H, Tran PO, Yang S, Zhang T, LeRoy E, Oseid E, Robertson RP. Regulation of alpha-cell function by the beta-cell during hypoglycemia in Wistar rats: the “switch-off” hypothesis. Diabetes. 2004;53:1482–1487. doi: 10.2337/diabetes.53.6.1482. [DOI] [PubMed] [Google Scholar]
- 53.Raju B, Cryer PE. Loss of the decrement in intraislet insulin plausibly explains loss of the glucagon response to hypoglycemia in insulin-deficient diabetes: documentation of the intraislet insulin hypothesis in humans. Diabetes. 2005;54:757–764. doi: 10.2337/diabetes.54.3.757. [DOI] [PubMed] [Google Scholar]
- 54.Hill ME, Asa SL, Drucker DJ. Essential requirement for Pax6 in control of enteroendocrine proglucagon gene transcription. Mol Endocrinol. 1999;13:1474–1486. doi: 10.1210/mend.13.9.0340. [DOI] [PubMed] [Google Scholar]
- 55.Kim EA, Noh YT, Ryu MJ, Kim HT, Lee SE, Kim CH, Lee C, Kim YH, Choi CY. Phosphorylation and transactivation of Pax6 by homeodomain-interacting protein kinase 2. J Biol Chem. 2006;281:7489–7497. doi: 10.1074/jbc.M507227200. [DOI] [PubMed] [Google Scholar]
- 56.Bell AC, West AG, Felsenfeld G. Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science. 2001;291:447–450. doi: 10.1126/science.291.5503.447. [DOI] [PubMed] [Google Scholar]
- 57.Gao J, Li T, Lu L. Functional Role of CCCTC Binding Factor in Insulin-stimulated Cell Proliferation. Cell Proliferation. 2007 doi: 10.1111/j.1365-2184.2007.00472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]