Abstract
Purpose
To construct predictive models using comprehensive tumor features for the evaluation of tumor response to neoadjuvant chemoradiotherapy (CRT) in patients with esophageal cancer.
Methods and Materials
This study included 20 patients who underwent trimodality therapy (CRT + surgery) and had 18F-FDG PET/CT scans both before and after CRT. Four groups of tumor features were examined: (1) conventional PET/CT response measures (SUVmax, tumor diameter, etc.); (2) clinical parameters (TNM stage, histology, etc.) and demographics; (3) spatial-temporal PET features, which characterize tumor SUV intensity distribution, spatial patterns, geometry, and associated changes resulting from CRT; and (4) all features combined. An optimal feature set was identified with recursive feature selection and cross-validations. Support vector machine (SVM) and logistic regression (LR) models were constructed for prediction of pathologic tumor response to CRT, using cross-validations to avoid model over-fitting. Prediction accuracy was assessed via area under the receiver operating characteristic curve (AUC), and precision was evaluated via confidence intervals (CIs) of AUC.
Results
When applied to the 4 groups of tumor features, the LR model achieved AUCs (95% CI) of 0.57 (0.10), 0.73 (0.07), 0.90 (0.06), and 0.90 (0.06). The SVM model achieved AUCs (95% CI) of 0.56 (0.07), 0.60 (0.06), 0.94 (0.02), and 1.00 (no misclassifications). Using spatial–temporal PET features combined with conventional PET/CT measures and clinical parameters, the SVM model achieved very high accuracy (AUC 1.00) and precision (no misclassifications), significantly better than using conventional PET/CT measures or clinical parameters and demographics alone. For groups with a large number of tumor features (groups 3 and 4), the SVM model achieved significantly higher accuracy than the LR model,
Conclusions
The SVM model using all features including spatial–temporal PET features accurately and precisely predicted pathologic tumor response to CRT in esophageal cancer.
INTRODUCTION
Esophageal cancer remains one of the most lethal malignancies, with a 5-year relative survival rate of only 17% (1) despite continued advances in therapy. In the United States, it is estimated that 17,460 patients were diagnosed with esophageal cancer and 15,070 died from the disease in 2012 (1). The preferred primary treatment strategy for locally advanced esophageal cancer has been transitioning from surgery (esophagectomy) to trimodality therapy, which consists of concurrent neoadjuvant chemoradiotherapy (CRT) followed by surgery (2). Recently, it was suggested that not all patients benefit from surgery after induction CRT and that definitive CRT (CRT alone) could also become an option (3). Evidence suggests that surgery after CRT can significantly improve local control (4, 5). These improvements in local control, however, have been tempered by the increased mortality (9%–12%) and morbidity (30%) compared to CRT alone (mortality, 0.8%–3.5%). Several studies have shown that tumor response to CRT remains an important predictor of both local control and overall survival (3–5). Complete responders to CRT appear to have superior outcomes, regardless of whether they undergo surgical resection. These data also support that the addition of resection can improve outcomes for patients who are discovered to have residual tumor following completion of CRT. Given the added mortality and morbidity of surgery after CRT, as well as the high local failure rate for CRT alone, it is critical to accurately identify patients who respond to CRT so that surgery may be safely deferred. It is equally important to accurately identify patients who do not respond to CRT so that early surgical salvage can be initiated.
Recent studies have emerged suggesting that spatial PET/CT features, including tumor volume (6), tumor shape (7), total glycolytic volume (8), and spatial patterns (texture features) (9), are more informative than the traditional response measure with maximum standardized uptake values (SUVmax) in various tumors. The authors demonstrated that comprehensive spatial–temporal 18F-FDG PET features were useful predictors of pathologic tumor response to CRT in esophageal cancer (10). The diversity of the new features suggest that it would be advantageous to combine multiple features in evaluation of tumor response (11) instead of traditional PET response criteria that are based on cutoff values of a single measure (8, 12). The objective of this study is to construct sophisticated tumor response models using comprehensive tumor features to accurately and precisely predict pathologic tumor response to CRT in patients with esophageal cancer.
MATERIALS AND METHODS
Patients
This retrospective study was approved by the IRB. The cohort included 20 consecutive patients (median age, 64 years) with esophageal cancer, who underwent trimodality therapy from 2006 to 2009 and had PET/CT scans both before and after CRT (Table 1). Staging was according to AJCC Cancer Staging Manual sixth edition (13), where M1a is extensive local–regional lymph node disease without distant metastasis.
TABLE 1.
Patient Characteristics (n = 20)
| Characteristic | No. of patients |
|---|---|
| Sex | |
| Male | 18 |
| Female | 2 |
| Primary site | |
| Proximal | 0 |
| Distal | 20 |
| Mid | 0 |
| Throughout | 0 |
| Histology | |
| Squamous cell carcinoma | 3 |
| Adenocarcinoma | 17 |
| Histologic grade | |
| Well differentiated | 3 |
| Moderately differentiated | 10 |
| Poorly differentiated | 5 |
| Unknown | 2 |
| TNM stage | |
| T1 | 0 |
| T2 | 2 |
| T3 | 18 |
| T4 | 0 |
| N0 | 6 |
| N1 | 14 |
| M0 | 18 |
| M1a | 2 |
| Pathologic response | |
| Pathologic complete response | 5 |
| Microscopic residual disease | 4 |
| Gross residual disease | 11 |
PET/CT Imaging, Chemoradiotherapy
Pre-CRT PET/CT imaging was performed 3–5 weeks before the beginning of CRT. Post-CRT PET/CT was performed 4–6 weeks after completion of CRT but before surgery. All PET/CT studies were acquired on the same scanner following an institutional standard protocol (10). All patients were treated with external-beam radiotherapy to the same total dose of 50.4 Gy with concurrent Cisplatin or Carboplatin chemotherapy (14).
Pathologic Assessment
Surgical resection was performed 1–7 weeks after the post-CRT PET/CT, and 6–13 weeks after CRT. The resected surgical specimen was submitted to the same pathologist for evaluation. The specimen was microscopically examined, and semi-quantitatively categorized into 1 of 3 groups: pathologic complete response (pCR), microscopic residual disease (mRD), or gross residual disease (gRD), according to the amount of residual viable carcinoma observed in relation to areas of fibrosis (15). In this study, the primary tumor alone was considered, and both pCR and mRD were considered as “responders,” because they have been shown to have similar survival rates (14), while gRD was considered as “non-responder”.
Spatial–Temporal PET Features Extraction
Details of the spatial–temporal PET feature extraction have been reported (10). Post-CRT PET/CT scans were rigidly registered to pre-CRT PET/CT scans. A tumor volume of interest (VOI) was semi-automatically delineated on the PET image using a region-growing method with a threshold of SUV ≥ 2.5. The result was reviewed and manually edited by a radiologist. The resulting VOI represented the entire hypermetabolic tumor volume and was denoted as VOI_SUV2.5. Another VOI_SUVpeak was defined as the 3×3×3-voxel cube centered at the SUVmax point, representing the peak metabolically active part of the tumor. Finally, comprehensive spatial–temporal PET features were extracted to characterize the tumor SUV intensity distribution, spatial variations (texture), geometry, and their associated changes resulting from CRT. For each image and each VOI, 9 intensity features (25), 8 Haralick texture features (26), 15 geometry features (27, 28), and 1 volume-intensity feature were extracted (Table 2). After incorporating temporal changes and excluding quantitatively identical features, a total of 137 features were obtained for each tumor.
TABLE 2.
Spatial-Temporal 18F FDG-PET Features
| Intensity features |
| Minimum |
| Maximum |
| Mean |
| Standard deviation |
| Sum |
| Median |
| Skewness |
| Kurtosis |
| Variance |
| Texture features |
| Energy |
| Entropy |
| Correlation |
| Inverse difference moment |
| Inertia |
| Cluster shade |
| Cluster prominence |
| Haralick correlation |
| Geometry features |
| Volume |
| Major diameter |
| Minor diameter |
| Eccentricity |
| Elongation |
| Oriented bounding box volume |
| Bounding box volume |
| Roundness |
| Region ratio |
| Orientation |
| Feret diameter |
| Number of lines |
| Perimeter |
| Physical size |
| Flatness |
| Geometry–intensity feature |
| Total glycolytic volume |
Clinical Parameters and Demographic Features
Sixteen clinical parameters and demographic features were extracted from patients’ charts. Clinical parameters included differentiation, stage, T stage, N stage, M stage, distant metastasis, type of chemotherapy, radiotherapy dose, treatment with concomitant boost, location of tumor, tumor involves gastroesophageal junction, histology, total extent of disease, and extent of disease >4 cm. Demographic features included age and sex.
Feature Selection and Response Modeling
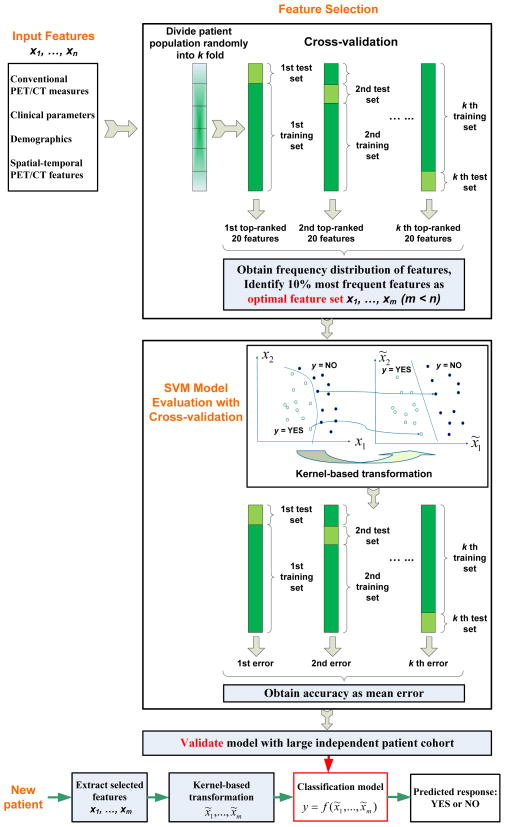
Four groups of tumor features were examined: (1) 16 conventional PET/CT response measures (Table 3); (2) 16 clinical parameters and demographics as described above; (3) 137 spatial–temporal PET features (Table 2); and (4) all 169 combined features. Figure 1 illustrates the flow of feature selection and model construction, with cross-validations to remove bias in feature selection and to prevent over-fitting of the model, respectively. The pathologic tumor response of the surgical specimen was used as the ground truth. The 20 patients were randomly partitioned into a training set (for constructing the model) and a testing set (for cross-validating the model and assessing its accuracy). 10-, 5- and 2-fold cross-validations (randomly leaving 10%, 20%, and 50% patients out in the testing set) were repeatedly used for both feature selection and model accuracy evaluation. Our goal was to model pathologic tumor response to CRT as a function (f) of each of the 4 groups of tumor features so that:
| Eq. 1 |
TABLE 3.
Conventional PET/CT Measures Used in Tumor Response Evaluation*
| SUV features |
| SUVmean (post, VOI_SUVpeak) |
| SUVmax (post, VOI_SUV2.5) |
| SUVmean Decrease (VOI_SUVpeak) |
| SUVmean (pre, VOI_SUVpeak) |
| SUVmax (pre, VOI_SUV2.5) |
| SUVmean ratio (pre, post, VOI_SUVpeak) |
| SUVmax decrease (pre, post, VOI_SUV2.5) |
| SUVmax ratio (pre, post, VOI_SUV2.5) |
| Volume and length features |
| Major diameter (post, VOI_SUV2.5) |
| Major diameter Ratio (pre, post, VOI_SUV2.5) |
| Volume (post, VOI_SUV2.5) |
| Volume ratio (pre, post, VOI_SUV2.5) |
| Major diameter (pre, VOI_SUV2.5) |
| Major diameter decrease (pre, post, VOI_SUV2.5) |
| Volume (pre, VOI_SUV2.5) |
| Volume decrease (pre, post, VOI_SUV2.5) |
Parentheses denote VOI and images from which a feature was extracted.
pre = Pre-CRT SUV, post = Post-CRT SUV, diff = pre-CRT SUV – post-CRT SUV.
FIGURE 1.
Workflow diagram illustrating feature selection and model construction with cross-validations for prediction of tumor response.
Feature Ranking and Selection
Since there are many features, to avoid model over-fitting to the training set, a feature selection process with cross-validation (Fig. 1) was applied first within each feature group for each model. A support vector machine–based feature ranking (SVMFR) method (16) was used to rank all features by recursively removing features and testing the predictive ability of the remaining features for each patient partition. A frequency distribution of the top-ranked 20 features for each of the k partitions was obtained. The optimal feature set was then identified as the 10% most frequently selected features over all k partitions from the frequency distribution. In this manner, the feature selection bias was removed.
Predictive Model Construction
We used 2 machine learning (ML) models, logistic regression (LR) and SVM, to obtain function f in Eq. 1. Figure 1 illustrates the modeling process using SVM with all tumor features as an example.
SVM with sequential minimal optimization
In our SVM, a polynomial kernel was used to transform the input tumor features from x1, … xm space into x̃1, … x̃m space, so that the 2 classes (“responders” and “non-responders”) became linearly separable (Fig. 1). The hyperplane (or the straight line in the simplified 2D illustration) that represents the largest separation between the closest members (support vectors) of the two classes was determined, providing a classification rule or model y = f (x̃1,…, x̃m), which classified a new or testing patient as a “responder” or a “non-responder” based on its y value.
Logistic regression
For comparison, we also implemented the widely used LR model. LR first transformed the response variable into Pr[YES | x1,…, xm], a probability variable corresponding to the “responders” given tumor features x1,…, xm. A logit transformation, log{Prp[YES | x1,…, xm]/(1−Pr[YES | x1,…, xm])}, was then applied so that the resulting variable lies between negative infinity and positive infinity. The transformed variable was approximated using a linear function of input tumor features (linear regression). The resulting model was Pr[YES | x1,…, xm] = 1/[1+exp(−w0 −w1x1 –…– wm xm)] with weights w. The weights were obtained by fitting the model to the training set using maximum log-likelihood estimation.
The outputs of the ML models were the predicted pathologic response represented as a binary variable (yes or no), which corresponded to “responder” or “non-responder”, respectively (Fig. 1). To test over-fitting of each model, 10-, 5- and 2-fold cross-validations were again used in model evaluation (Fig. 1). The model accuracy was calculated as the mean accuracy over all partitions.
Statistical Analysis
The accuracy of using each feature group and each model to predict the pathologic tumor response was quantified using the AUC, defined as the area under receiver operating characteristic (ROC) curves. In addition, the sensitivity and specificity of each model were calculated and compared using the unpaired t test at a significance level of 0.05. Model precision was evaluated with the 95% confidence intervals (CI).
RESULTS
Because LR and SVM are 2 distinct models, our feature selection process resulted in different optimal feature sets for each model (Table 4). The optimal feature set for SVM contained the optimal feature set for LR, except when applied to clinical parameters and demographics, where histology was the only feature selected for SVM. This was in agreement with a larger study of 164 patients by Koshy et al. (14) that showed histology was the most and only predictive individual clinical parameter.
TABLE 4.
Optimal Feature Set Resulting from Feature Selection*
| Metrics/Features | Logistic regression | Support vector machine |
|---|---|---|
| Conventional PET/CT measures | Major diameter (pre, VOI_SUV2.5) | Major diameter (pre, VOI_SUV2.5) Volume (pre, VOI_SUV2.5) SUVmax Decline (pre, post, VOI_SUV2.5) SUVmax Ratio (pre, post, VOI_SUV2.5) |
| Clinical parameters and demographics | Histology Tumor involves gastroesophageal junction Sex T stage |
Histology |
| Spatial–temporal PET features | Inertia (diff, VOI_SUVpeak) Orientation (post, VOI_SUV2.5) Variance (post, VOI_SUVpeak) Cluster prominence (post, VOI_SUVpeak) Inverse difference moment (post, VOI_SUV2.5) |
Inertia (diff, VOI_SUVpeak) Orientation (post, VOI_SUV2.5) Variance (post, VOI_SUVpeak) Cluster prominence (post, VOI_SUVpeak) Inverse difference moment (post, VOI_SUV2.5) Energy (post, VOI_SUV2.5) Entropy (post, VOI_SUV2.5) Haralick correlation (pre, VOI_SUV2.5) Median (post, VOI_SUV2.5) Inverse difference moment (Diff, VOI_SUVpeak) Median (post, VOI_SUVpeak) Entropy (diff, VOI_SUVpeak) Roundness (post, VOI_SUV2.5) Mean (post, VOI_SUV2.5) Elongation (pre, VOI_SUVpeak) Total glycolytic volume (diff, VOI_SUV2.5) |
| All features | Orientation (post, VOI_SUV2.5) Tumor involves gastroesophageal junction Inertia (diff, VOI_SUVpeak) Energy (post, VOI_SUV2.5) Entropy (post, VOI_SUV2.5) Skewness (diff, VOI_SUVpeak) |
Orientation (post, VOI_SUV2.5) Tumor involves gastroesophageal junction Inertia (diff, VOI_SUVpeak) Energy (post, VOI_SUV2.5) Entropy (post, VOI_SUV2.5) Skewness (diff, VOI_SUVpeak) Inverse difference moment (post, VOI_SUV2.5) Cluster prominence (post, VOI_SUVpeak) Inverse difference moment (diff, VOI_SUVpeak) Inertia (post, VOI_SUV2.5) Volume (post, VOI_SUV2.5) Elongation (pre, VOI_SUVpeak) T stage Variance (post, VOI_SUVpeak) Haralick correlation (pre, VOI_SUV2.5) Median (post, VOI_SUV2.5) Total glycolytic volume (diff, VOI_SUV2.5) |
Parentheses denote VOI and images from which a feature was extracted.
pre = Pre-CRT SUV, post = Post-CRT SUV, diff = pre-CRT SUV – post-CRT SUV.
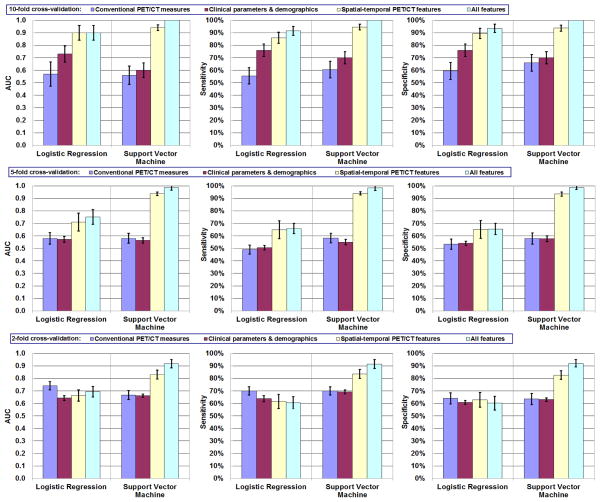
Figure 2 shows the model accuracy (AUC) and precision (95% CI) obtained from repeating the 10-, 5-, and 2-fold cross-validations. The best prediction was obtained using the SVM model with 17 features from all combined features (SVMall). All patients within the testing set were correctly classified during the repetition of 10-fold cross-validations, resulting in a mean AUC of 1.00 (100% sensitivity, 100% specificity). Table 4 indicates that SVMall contained 1 conventional PET/CT measure, “residual metabolic tumor volume (i.e., SUV ≥ 2.5) post-CRT”; 2 clinical parameters, “whether tumor involves gastroesophageal junction” and “T stage”; and 14 spatial–temporal PET (3 intensity, 8 texture, 2 geometry, and 1 volume-intensity) features, suggesting that all 3 groups of tumor features and all 4 categories of spatial-temporal PET features contained useful predictors of response. The model performance was stable when leaving more patients out with 5- and 2-fold cross-validations compared to 10-fold cross-validation. Only a small reduction in mean AUC (from 1.00 to 0.99 and 0.92) was observed.
FIGURE 2.
Response modeling accuracy and reliability: mean AUC, sensitivity, and specificity of 10-, 5-, 2-fold cross-validations. Error bars indicate 95% confidence intervals.
Figure 2 also shows the sensitivity and specificity obtained from each model using different groups of features. When the SVM model was used with 10-fold cross validation, significantly higher sensitivity was achieved by using all features including spatial–temporal PET features (100%) than by using conventional PET/CT measures (60%) or clinical parameters and demographics (70%) alone (P < 0.001). Significantly higher specificity (P < 0.001) was achieved as well. Similar results were obtained when the LR model was used (92% sensitivity, 94% specificity; both P < 0.001).
For 10-fold cross-validation, the differences between SVM and LR models were not significant when using any of the four groups of features (P > 0.06). However, when using selected spatial–temporal PET features or using all features in 5- and 2-fold cross-validations, the SVM models demonstrated significantly better results than the LR models (P < 0.0001 and P < 0.0002, respectively).
DISCUSSION
18F-FDG PET has shown promising results in predicting pathologic response to CRT and long-term prognosis in esophageal cancer (12, 17). Westerterp et al. (18) and Swisher et al. (19) showed that PET had the highest accuracy (76%) among PET, endoscopic ultrasonography, and CT for predicting pathologic response to CRT with sensitivity ranges of 71%–100% and specificity ranges of 55%–100%. Levien et al. (20) showed that PET can be useful for predicting pathologic response with sensitivity of 61.3% and specificity of 60.0%. Reviewing 20 studies, Kwee (12) found that the sensitivity and specificity of PET for predicting pathologic response to neoadjuvant therapy ranged from 33% to 100% and from 30% to 100%, respectively. Although promising, the accuracy of PET is still low and none of these studies achieved both high sensitivity and high specificity.
Almost all published 18F-FDG PET studies quantify therapeutic response in tumors with SUVmax (21, 22). Changes in SUVmax and sometimes SUVmax before (pre-) or after (post-) CRT are correlated to post-CRT pathologic response or survival. However, SUVmax is a single-point estimate, whereas most solid tumors show significant heterogeneity in both degree and distribution of FDG uptake. Heterogeneity in FDG uptake has been associated with important biological and physiologic parameters (9, 23) and has been shown to be a prognostic factor in many cancers (9, 24). In our previous work, comprehensive spatial–temporal 18F-FDG PET features were extracted and found to be useful predictors of pathologic tumor response (10).
In this work, a predictive model generally included multiple tumor features. The best model achieved significantly higher accuracy (AUC 1.00) than the best individual response measure (inertia post-CRT, AUC 0.85, (10)). Both prediction accuracy and precision were significantly improved by using selected spatial–temporal PET features instead of conventional PET/CT measures or clinical parameters and demographics. This was the case for both the SVM and LR models. These results suggested that spatial–temporal PET features provided richer information than the other feature groups.
When using the same feature group and comparing the performance of LR and SVM models, the results varied from group to group. SVM achieved significantly higher accuracy than LR when using spatial–temporal PET feature group. The reason is that this group contained more candidate features, whose complimentary relationship for response prediction is hard to identify with LR. On the other hand, SVM has been proven to be able to extract complex relationships among a large number of features (25). Because the candidate feature group of the conventional PET/CT measures or clinical parameters and demographics contained only 16 features and because only 1 or 4 features were selected into the optimal subset, LR resulted in better results than SVM. The reason is that with this small number of selected features it would be difficult for SVM to achieve high accuracy.
When utilizing many candidate features for classification, it is important to identify an optimal, smaller feature set to prevent model over-fitting. Our feature selection process removed redundant features that introduce colinearity and noise into the models (26). Furthermore, feature selection bias was eliminated by collecting frequency distribution of the selected features over all patient partitions. The feature selection resulted in 16 features for the SVM using spatial–temporal PET features alone. When using all combined features, the number of selected features was 17. 1 conventional PET/CT measure and 2 clinical parameters replaced 2 spatial-temporal PET features, resulting in increased prediction accuracy (AUC increased from 0.94 to 1.00). This means features from conventional PET/CT measures and clinical parameters provided some complementary information to the spatial–temporal PET features.
Another important aspect of constructing predictive models is to avoid model over-fitting. To test this, we used different number of patients to train and test our models, namely 10-, 5- and 2-fold cross-validations. When leaving more patients out of training set, the prediction accuracy decreased. However, the AUC was still above 0.90 for SVMall model suggesting that it was not notably affected by over-fitting. LR model was not as stable as SVM model in this case (AUC dropped from 0.90 to below 0.70).
The authors provided an explanation as to why spatial–temporal PET features were useful predictors of tumor response (10). In addition to the spatial–temporal PET features, 1 conventional PET/CT measure (residual metabolic tumor volume) and 2 clinic parameters (whether tumor involves gastroesophageal junction and T stage) were included in the optimal feature set for the SVMall model. Evidence suggests that smaller residual metabolic tumor volume (27) and lower T stage (13) are associated with better tumor response. The effects of tumor involvement of the gastroesophageal junction, however, are not yet clear and are under investigation at our institution using a larger dataset.
There were variations in the interval between CRT and surgery (6–13 weeks) and the interval between post-CRT PET/CT and surgery (1–7 weeks). Several studies suggest that in rectal cancer delaying surgery after CRT may increase the pCR rate. However, based on our knowledge, three studies in esophageal cancer suggest that the timing of surgery did not affect the pCR rate (28–30). Examination of our data using Z-test suggested that neither interval affected the pCR rate (both P = 0.09). The results were not affected by variations in the intervals.
One limitation of this proof-of-principle study is that pathologic response in the primary tumor alone was examined. A pCR on the primary tumor site may not completely exclude lymph node invasion. An extension of the study to lymph node is under investigation. Another limitation is that it is a retrospective analysis of a small patient cohort. Although 10-, 5- and 2-fold cross-validations showed that the model was not notably affected by over-fitting, the predictive accuracy and stability of the models should be validated in a larger, independent patient cohort as shown in Fig. 1. Validation is also needed to confirm that the selected features are indeed meaningful measures and important for response evaluation in esophageal cancer. When the model is validated, it can be used to more appropriately select patients for surgery, thus avoiding the mortality and morbidity of surgery in responders for whom surgery can be safely deferred. The methodology can also be applied to evaluate response during CRT, which will provide the opportunity for early adjustments on treatment strategies including: giving a higher dose in definitive CRT to responders, changing the type of chemotherapy or performing surgery earlier to non-responders.
CONCLUSION
The SVM model using all features including spatial–temporal PET features accurately and precisely predicted pathologic tumor response to CRT in 20 patients with esophageal cancer. It has the potential to be used to safely defer surgery or to give a higher dose in definitive CRT for patients who respond to CRT. This will ultimately improve patient’s quality of life while reducing costs.
Summary.
This work constructed sophisticated predictive models using multiple, comprehensive tumor response measures, including conventional PET/CT measures, clinical parameters and demographics, and spatial–temporal PET features. The models achieved very high accuracy (100% sensitivity, 100% specificity) for prediction of pathologic tumor response to chemoradiotherapy in 20 patients with esophageal cancer.
Acknowledgments
This work was supported in part by National Cancer Institute Grant R21 CA131979 and R01 CA172638. Shan Tan was supported in part by the National Natural Science Foundation of China 60971112 and 61375018, and by Fundamental Research Funds for the Central Universities 2012QN086.
Footnotes
Conflict of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.American Cancer Society. [Accessed on May 13, 2013..];Cancer Facts & Figures. 2012 Available from: www.cancer.org/Research/CancerFactsFigures/CancerFactsFigures/cancer-facts-figures-2012.
- 2.Tepper J, Krasna MJ, Niedzwiecki D, et al. Phase III trial of trimodality therapy with cisplatin, fluorouracil, radiotherapy, and surgery compared with surgery alone for esophageal cancer: CALGB 9781. J Clin Oncol. 2008;26:1086–1092. doi: 10.1200/JCO.2007.12.9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monjazeb AM, Riedlinger G, Aklilu M, et al. Outcomes of patients with esophageal cancer staged with [(18)F]fluorodeoxyglucose positron emission tomography (FDG-PET): can postchemoradiotherapy FDG-PET predict the utility of resection? J Clin Oncol. 2011;28:4714–4721. doi: 10.1200/JCO.2010.30.7702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stahl M, Stuschke M, Lehmann N, et al. Chemoradiation with and without surgery in patients with locally advanced squamous cell carcinoma of the esophagus. Journal of Clinical Oncology. 2005;23:2310–2317. doi: 10.1200/JCO.2005.00.034. [DOI] [PubMed] [Google Scholar]
- 5.Bedenne L, Michel P, Bouche O, et al. Chemoradiation followed by surgery compared with chemoradiation alone in squamous cancer of the esophagus: FFCD 9102. J Clin Oncol. 2007;25:1160–1168. doi: 10.1200/JCO.2005.04.7118. [DOI] [PubMed] [Google Scholar]
- 6.Prasad SR, Jhaveri KS, Saini S, et al. CT tumor measurement for therapeutic response assessment: comparison of unidimensional, bidimensional, and volumetric techniques initial observations. Radiology. 2002;225:416–419. doi: 10.1148/radiol.2252011604. [DOI] [PubMed] [Google Scholar]
- 7.O’Sullivan F, Roy S, O’Sullivan J, et al. Incorporation of tumor shape into an assessment of spatial heterogeneity for human sarcomas imaged with FDG-PET. Biostatistics. 2005;6:293–301. doi: 10.1093/biostatistics/kxi010. [DOI] [PubMed] [Google Scholar]
- 8.Wahl RL, Jacene H, Kasamon Y, Lodge MA. From RECIST to PERCIST: Evolving Considerations for PET response criteria in solid tumors. J Nucl Med. 2009;50:122S–150S. doi: 10.2967/jnumed.108.057307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tixier F, Le Rest CC, Hatt M, et al. Intratumor heterogeneity characterized by textural features on baseline 18F-FDG PET images predicts response to concomitant radiochemotherapy in esophageal cancer. J Nucl Med. 2011;52:369–378. doi: 10.2967/jnumed.110.082404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reference blinded for review.
- 11.Vaidya M, Creach KM, Frye J, et al. Combined PET/CT image characteristics for radiotherapy tumor response in lung cancer. Radiotherapy and Oncology. 2012;102:239–245. doi: 10.1016/j.radonc.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 12.Kwee RM. Prediction of tumor response to neoadjuvant therapy in patients with esophageal cancer with use of 18F FDG PET: a systematic review. Radiology. 2010;254:707–717. doi: 10.1148/radiol.09091324. [DOI] [PubMed] [Google Scholar]
- 13.American Joint Committee on Cancer. AJCC Cancer Staging Manual. 6. New York, NY: Springer; 2002. [Google Scholar]
- 14.Koshy M, Greenwald BD, Hausner P, et al. Outcomes After Trimodality Therapy for Esophageal Cancer: The Impact of Histology on Failure Patterns. Am J Clin Oncol. 2010 doi: 10.1097/COC.0b013e3181e841ce. [DOI] [PubMed] [Google Scholar]
- 15.Mandard AM, Dalibard F, Mandard JC, et al. Pathologic assessment of tumor regression after preoperative chemoradiotherapy of esophageal carcinoma. Clinicopathologic correlations. Cancer. 1994;73:2680–2686. doi: 10.1002/1097-0142(19940601)73:11<2680::aid-cncr2820731105>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 16.Guyon I, Weston J, Barnhill S, et al. Gene Selection for Cancer Classification using Support Vector Machines. Mach Learn. 2002;46:389–422. [Google Scholar]
- 17.Krause BJ, Herrmann K, Wieder H, et al. 18F-FDG PET and 18F-FDG PET/CT for assessing response to therapy in esophageal cancer. J Nucl Med. 2009;50 (Suppl 1):89S–96S. doi: 10.2967/jnumed.108.057232. [DOI] [PubMed] [Google Scholar]
- 18.Westerterp M, van Westreenen HL, Reitsma JB, et al. Esophageal cancer: CT, endoscopic US, and FDG PET for assessment of response to neoadjuvant therapy--systematic review. Radiology. 2005;236:841–851. doi: 10.1148/radiol.2363041042. [DOI] [PubMed] [Google Scholar]
- 19.Swisher SG, Maish M, Erasmus JJ, et al. Utility of PET, CT, and EUS to identify pathologic responders in esophageal cancer. Annals of Thoracic Surgery. 2004;78:1152–1160. doi: 10.1016/j.athoracsur.2004.04.046. discussion 1152–1160. [DOI] [PubMed] [Google Scholar]
- 20.Levine EA, Farmer MR, Clark P, et al. Predictive value of 18-fluoro-deoxy-glucose-positron emission tomography (F-18-FDG-PET) in the identification of responders to chemoradiation therapy for the treatment of locally advanced esophageal cancer. Annals of Surgery. 2006;243:472–478. doi: 10.1097/01.sla.0000208430.07050.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kubota K. From tumor biology to clinical PET: A review of positron emission tomography (PET) in oncology. Annals of Nuclear Medicine. 2001;15:471–486. doi: 10.1007/BF02988499. [DOI] [PubMed] [Google Scholar]
- 22.Larson SM, Erdi Y, Akhurst T, et al. Tumor Treatment Response Based on Visual and Quantitative Changes in Global Tumor Glycolysis Using PET-FDG Imaging. The Visual Response Score and the Change in Total Lesion Glycolysis. Clin Positron Imaging. 1999;2:159–171. doi: 10.1016/s1095-0397(99)00016-3. [DOI] [PubMed] [Google Scholar]
- 23.Zhao S, Kuge Y, Mochizuki T, et al. Biologic correlates of intratumoral heterogeneity in 18F-FDG distribution with regional expression of glucose transporters and hexokinase-II in experimental tumor. J Nucl Med. 2005;46:675–682. [PubMed] [Google Scholar]
- 24.Marusyk A, Polyak K. Tumor heterogeneity: causes and consequences. Biochim Biophys Acta. 2010;1805:105–117. doi: 10.1016/j.bbcan.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kotsiantis SB. Supervised Machine Learning: A Review of Classification Techniques. Informatica. 2007;31:249–269. [Google Scholar]
- 26.Hua J, Xiong Z, Lowey J, et al. Optimal number of features as a function of sample size for various classification rules. Bioinformatics. 2005;21:1509–1515. doi: 10.1093/bioinformatics/bti171. [DOI] [PubMed] [Google Scholar]
- 27.Jayachandran P, Pai RK, Quon A, et al. Postchemoradiotherapy positron emission tomography predicts pathologic response and survival in patients with esophageal cancer. Int J Radiat Oncol Biol Phys. 2012;84:471–477. doi: 10.1016/j.ijrobp.2011.12.029. [DOI] [PubMed] [Google Scholar]
- 28.Chiu CH, Chao YK, Chang HK, et al. Interval Between Neoadjuvant Chemoradiotherapy and Surgery for Esophageal Squamous Cell Carcinoma: Does Delayed Surgery Impact Outcome? Ann Surg Oncol. 2013 doi: 10.1245/s10434-013-3139-7. [DOI] [PubMed] [Google Scholar]
- 29.Kim JY, Correa AM, Vaporciyan AA, et al. Does the timing of esophagectomy after chemoradiation affect outcome? Ann Thorac Surg. 2012;93:207–212. doi: 10.1016/j.athoracsur.2011.05.021. discussion 212–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruol A, Rizzetto C, Castoro C, et al. Interval between neoadjuvant chemoradiotherapy and surgery for squamous cell carcinoma of the thoracic esophagus: does delayed surgery have an impact on outcome? Ann Surg. 2010;252:788–796. doi: 10.1097/SLA.0b013e3181fc7f86. [DOI] [PubMed] [Google Scholar]