Abstract
Botulinum neurotoxins (BoNTs) are among the most poisonous substances known to man, but paradoxically, BoNT-containing medicines and cosmetics have been used with great success in the clinic. Accidental BoNT poisoning mainly occurs through oral ingestion of food contaminated with Clostridium botulinum. BoNTs are naturally produced in the form of progenitor toxin complexes, which are high molecular weight (up to ~900 kDa) multi-protein complexes composed of BoNT and several non-toxic neurotoxin-associated proteins (NAPs). NAPs protect the inherently fragile BoNTs against the hostile environment of the gastrointestinal tract and help BoNTs pass through the intestinal epithelial barrier before they are released into the general circulation. These events are essential for ingested BoNTs to gain access to motoneurons, where they inhibit neurotransmitter release and cause muscle paralysis. In this review, we discuss the structural basis for assembly of NAPs and BoNT into the progenitor toxin complexes that protect BoNT and facilitate its delivery into the bloodstream.
1 Introduction
Botulinum neurotoxins (BoNTs) are produced by the anaerobic, Gram-positive, spore-forming bacteria Clostridium botulinum. Seven serotypes of BoNTs (termed A–G) have been identified, which cause botulism, a severe neurological disease associated with a life-threatening flaccid paralysis that affects both humans and animals (Binz et al. 1990; Hatheway 1990; Montecucco and Schiavo 1995; Sugiyama 1980).
Following toxin ingestion, there are two major points at which the interaction between BoNT and host cells is pivotal. To date, most attention has been paid to the BoNT-neuron interaction. Here, BoNTs act as sequence-specific endopeptidases that cleave SNAREs (soluble N-ethylmaleimide sensitive factor attachment protein receptors), blocking the release of acetylcholine at neuromuscular junctions (NMJs) and thus paralyzing the affected muscles (Blasi et al. 1993a; Schiavo et al. 1992). However, there is a large gap in our understanding of the critical events that precede the BoNT–neuron interaction, the most important of which is how the inherently fragile BoNTs survive the harsh environment (low pH, protease-rich) of the gastrointestinal (GI) tract and cross the intestinal epithelial barrier to enter the bloodstream.
The mechanism by which this is accomplished involves the secret weapons of BoNTs; the progenitor toxin complexes (PTC). PTCs are high molecular weight (up to ~900 kDa) multi-protein complexes produced by C. botulinum and are composed of BoNT and several non-toxic neurotoxin-associated proteins (NAPs) (Collins and East 1998). The NAPs comprise four clostridial proteins; a non-toxic non-hemagglutinin (NTNHA) protein and three hemagglutinin proteins (HA-17, HA-33, and HA-70). The naturally occurring minimally functional PTC (M-PTC) is composed of BoNT (~150 kDa) and NTNHA (~140 kDa). Other PTCs, with molecular masses ranging from 500 kDa to 900 kDa, are assembled by the addition of various combinations of HAs to the M-PTC through unknown mechanisms (Ito et al. 2011; Montecucco and Schiavo 1995).
The oral toxicity of BoNTs is markedly increased when co-assembled with NAPs into PTCs (Ohishi et al. 1977). This is achieved through two complementary processes. NTNHA directly interacts with BoNT and plays a major role in shielding it in the hostile GI environment. In addition, HAs interact with intestinal epithelial cells and play an active role in BoNT transport (Fujinaga et al. 1997; Fujinaga et al. 2004; Niwa et al. 2007).
Clearly, a detailed knowledge of the molecular architecture of the PTC is required to understand the strategic interactions between BoNT and the host that take place in the GI tract. This review focuses on the structure-function relationship of BoNT PTCs, including the mechanism by which NAPs protect BoNTs, the regulatory mechanisms underlying PTC assembly, and the structural basis by which NAPs participate in BoNT-host interactions.
2 BoNT and NAPs
2.1 Molecular architecture of BoNT
The seven BoNTs have a high degree of primary sequence conservation, although all are antigenically distinct (Lacy and Stevens 1999). BoNT is synthesized as a single polypeptide chain of ~150 kDa and post-translationally nicked by an unknown protease into a 50 kDa light chain (LC) and a 100 kDa heavy chain (HC) (Montecucco and Schiavo 1995). The LC and HC remain covalently linked through a disulfide bond until they encounter reducing conditions in the neuronal cytosol (Montecucco and Schiavo 1995). Crystal structures have been reported for full-length BoNT/A (PDB: 3BTA), BoNT/B (PDB: 1EPW), and BoNT/E (PDB: 3FFZ) (Kumaran et al. 2009; Lacy et al. 1998; Swaminathan and Eswaramoorthy 2000). All three structures are similar in that they exhibit a modular architecture comprising three domains. The LC is a protease. The HC is composed of two domains: the N-terminal domain (HN, also known as the translocation domain) mediates translocation of LC across the endosomal membrane, whereas the C-terminal domain (HC) is the cell surface receptor-binding domain.
The first point of attack at the NMJ involves a highly specific interaction between BoNT and the motoneuron. A well-accepted dual-receptor model proposes that BoNTs simultaneously bind to gangliosides, a class of glycosphingolipids enriched at nerve terminals, and to specific protein receptors (Binz and Rummel 2009; Chai et al. 2006; Jin et al. 2006; Montecucco et al. 2004; Stenmark et al. 2008) (see chapter 4 this book). The BoNT–ganglioside interaction ensures a high toxin–cell encounter rate, while the protein receptors confer cell specificity. BoNT serotypes A, E, and F recognize synaptic vesicle protein 2 (SV2) as the protein receptor, whereas BoNT/B and G utilize synaptotagmin I and II (SytI and II) (Binz and Rummel 2009; Rummel et al. 2009).
The warhead of BoNTs is the LC, which is a Zn2+-dependent endopeptidase that cleaves specific peptide bonds within the neuronal SNAREs (synaptobrevin, syntaxin, and SNAP-25) (see chapter 7 this book). BoNT/A and E cleave SNAP-25 while serotypes B, D, F, and G cleave synaptobrevin. BoNT/C is unique in that it is able to hydrolyze both syntaxin and SNAP-25 (Blasi et al. 1993b; Foran et al. 1996; Schiavo et al. 1995; Vaidyanathan et al. 1999; Williamson et al. 1996).
Once the toxin is endocytosed, the next step is to deliver LC across the intracellular membranes to its targets in the cytosol (Schiavo et al. 2000). The HN fragment has two long helices forming a coiled-coil (Fig. 1) and likely acts as both a channel and a transmembrane chaperone to ensure the successful transit of LC from the acidic endosomes into the cytosol (Fischer and Montal 2006, 2007a, b; Koriazova and Montal 2003). We speculate that HC remains bound to receptors on the endosomal membrane where it brings the N-terminus of HN in contact with the membrane prior to the HN-facilitated translocation of LC. However, the details of the translocation process are largely unknown.
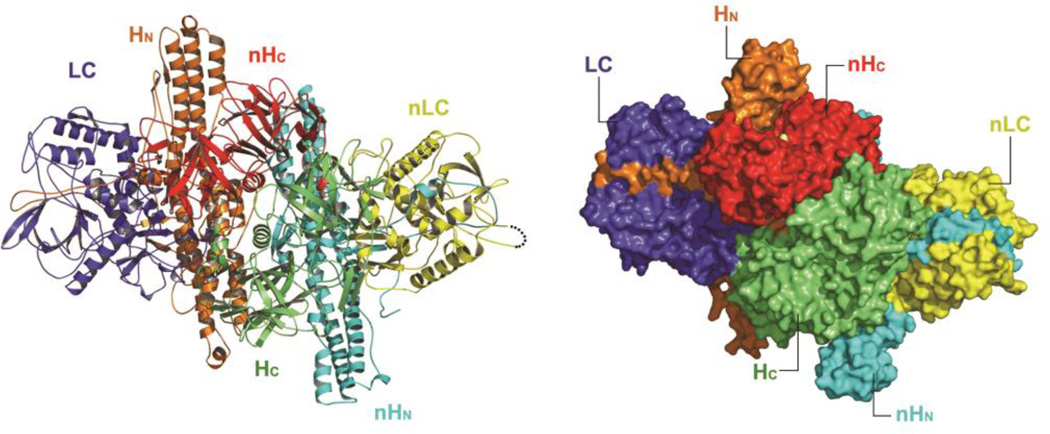
Fig. 1.
The structure of the M-PTC of BoNT/A. BoNT/A domains are blue (LC), orange (HN), and green (HC). NTNHA-A domains are yellow (nLC), cyan (nHN), and red (HC). The cartoon and surface representations of the M-PTC are shown in the left and right panels, respectively.
Before they can gain access to the motoneuron, BoNTs must traverse the GI tract and enter the bloodstream. Paradoxically, the highly sophisticated BoNTs are intrinsically vulnerable to the harsh physiological environment of the GI tract. For example, the free BoNT/A is readily degraded by digestive proteases such as trypsin and pepsin and is almost completely inactivated at pH 3 or less (Gu et al. 2012). In addition, in vitro assays have shown that trans-epithelial transcytosis of free BoNT/A is very inefficient compared with HA-containing PTCs (Fujinaga et al. 2009; Matsumura et al. 2008; Niwa et al. 2007). The responsibility for protecting BoNTs and facilitating their transit out of the GI tract falls to the NAPs.
2.2 NAPs: The bodyguards of BoNTs
All serotypes of BoNT are produced by bacteria in their native form as stable and non-covalent complexes with NAPs to form PTCs. The PTC protects the toxin sufficiently to reduce the oral lethal dose by 10- to 100-fold compared with the free BoNT (Cheng et al. 2008; Ohishi et al. 1977; Sugii et al. 1977). In general, BoNT and the four NAPs (NTNHA and three HAs) are encoded in a gene cluster: the downstream segment containing bont and ntnha and the upstream segment containing the three HA genes, ha-17, ha-33, and ha-70. This gene arrangement is defined as the ha70-ha17-ha33-botR-ntnha-bont cluster, or the ha cluster (Hill et al. 2009) (see chapter 1 this book). Interestingly, the HA genes are absent from BoNT/E and BoNT/F strains, as well as some BoNT/A strains. Instead, they contain the orfX3-orfX2-orfX1-(botR)-p47-ntnha-bont complex genes (abbreviated as orfX cluster) (East et al. 1998; East and Collins 1994; Fujii et al. 1993; Hill et al. 2009) (see chapter 1 this book). The expression and function of the orfX proteins are unknown.
Many different forms of PTC have been identified, with molecular masses ranging from ~300 kDa to ~900 kDa. It remains largely unknown how and why different PTCs exist. The M-PTC is ~300 kDa and is composed of BoNT and NTNHA, while other PTCs (e.g., 490, 610, 680, and 900 kDa) are assembled by the addition of various combinations of HAs to the M-PTC (Ito et al. 2011; Montecucco and Schiavo 1995). Historically, the three major forms of PTC are defined as M-PTC (~300 kDa), L-PTC (~500 kDa), and LL-PTC (~900 kDa), which are also termed the 12S, 16S, and 19S complex, respectively. The serotype A BoNTs are complexed in all three forms and BoNT/B, C, D, and G in two forms, L-PTC and M-PTC. In contrast, bacterial serotypes E and F, which do not have the HA genes, may only produce BoNT complexed in the M-PTC form.
We now know that NTNHA directly interacts with BoNT and plays a major role in shielding it in the hostile GI environment. Furthermore, NTNHA binding and protection are exquisitely regulated to allow BoNT to be released upon entry into the circulation (Gu et al. 2012). The three HA NAPs interact with intestinal epithelial cells and play an active role in BoNT transport (Fujinaga et al. 1997; Fujinaga et al. 2004; Niwa et al. 2007). Strong support for the functional role of HAs in this regard is provided by the finding that HAs directly bind to E-cadherin and disrupt the intercellular epithelial barrier (Ito et al. 2011; Matsumura et al. 2008; Sugawara et al. 2010) (see also chapter 3). These functions of NTNHA and the HAs will be discussed in more detail later.
The NAP proteins clearly play crucial roles in the early stages of BoNT oral poisoning. Therefore, a better understanding of the structure and function of NAPs as well as their interplay with BoNT will help us find a novel approach to prevent BoNT entry into the general circulation.
3 Structure of the M-PTC
3.1 A recombinant expression system to dissect the PTCs
Our limited knowledge of PTCs is in part due to the challenge associated with recombinant protein production. Most of the current research-grade BoNT and NAPs as well as BoNT-containing medicines are partially purified from natural sources. Many studies on in vitro assembly of PTCs have employed native BoNT PTC, where individual NAPs had first to be stripped from the PTC using harsh conditions such as guanidine hydrochloride. A common problem with this approach is that the purified PTC components may be contaminated by other PTC components and/or other clostridial proteins, which can complicate the data interpretation and may even account for some of the discrepancies in experimental observations. Furthermore, as a Category A reagent, the extremely high toxicity of BoNTs prevents most laboratories from performing detailed biochemical and biophysical analyses, which require large quantities of toxin. Therefore, there is an urgent need to establish a safe and efficient recombinant expression system to enable comprehensive characterization of PTCs. To this end, several laboratories have developed novel approaches based on overexpression of recombinant BoNT and individual NAPs in E. coli, which allows their purification to high homogeneity (Gu et al. 2012; Miyata et al. 2009; Rummel et al. 2004). The following discussion will focus on BoNT/A1, which is one of the major causes of botulism in humans and the most commonly used medicine among the BoNTs.
In a recent paper, a genetically modified, catalytically inactive recombinant BoNT/A1 (termed BoNT/Ai), which carries three mutations in the catalytic site of LC; E224Q/R363A/Y366F, was produce in E. coli (Gu et al. 2012). It is known that either double mutation E224Q/Y366F or R363A/Y366F is sufficient to reduce the catalytic activity of BoNT/A-LC to an undetectable level (Binz et al. 2002; Breidenbach and Brunger 2004; Fu et al. 2006; Li et al. 2000). Consistent with this, BoNT/Ai has no detectable neurotoxicity in the mouse phrenic nerve toxicity assay when tested at 30,000-fold higher concentrations than active BoNT/A (Binz et al. 2002; Gu et al. 2012; Li et al. 2000). The crystal structure of BoNT/Ai was determined at a resolution of 5 Å, which confirmed that the triple mutations do not alter the overall structure of BoNT/A (Gu et al. 2012).
In addition to the recombinant BoNT/Ai, the corresponding full-length NTNHA-A (NTNHA-A1, ~140 kDa) was also produced in E. coli (Gu et al. 2012). NTNHA-A shares low sequence identity (~20%) with BoNT/A, but its structure is unknown. During long-term storage, NTNHA-A slowly self-processes into two polypeptide chains (~125 and 15 kDa) that remain non-covalently linked. N-terminal amino acid sequencing identified the cleavage site between Lys133 and Lys134. This is consistent with the known spontaneous nicking of native NTNHA in the homologous area, which occurs through an unknown mechanism (Fujita et al. 1995; Inoue et al. 1996; Miyata et al. 2009; Sagane et al. 2002). Recombinant forms of HA proteins for various BoNT serotypes have also been produced in several laboratories (Inoue et al. 2003; Nakamura et al. 2009; Nakamura et al. 2008).
3.2 NTNHA-A forms a tight protective complex with BoNT/A
The wild-type PTCs are stable at pH values of 6.25 or less but release of BoNTs occurs at pH values above 7.0 (Boroff et al. 1966; DasGupta and Boroff 1968; Eisele et al. 2011). The recombinant BoNT/Ai and NTNHA-A form a tight complex at acidic pH (pH 6.0) with Kd (disassociation constant) of ~30.8 nM and a stoichiometry of 1:1, as quantified by isothermal titration calorimetry and analytic ultracentrifugation (Gu et al. 2012). No interaction could be detected at pH 7.5. Thus, the recombinant M-PTC fully recapitulates the pH-dependent assembly of the wild-type PTC. This also suggests that BoNT and NTNHA, not the HA proteins, are mainly responsible for the pH-dependent assembly of PTC.
It is well known that the oral toxicity of free BoNTs is significantly less than that of PTCs (Cheng et al. 2008; Ohishi et al. 1977; Sugii et al. 1977). In vitro studies also showed that free BoNTs are much more easily detoxified than those in PTC by pepsin and pancreatin, and by gastric and intestinal juices (Ohishi et al. 1977; Sugii et al. 1977; Sugiyama 1980). To reproduce the physiological conditions faced by BoNT in the GI tract, the response of free BoNT/A and the M-PTC to low pH and digestive proteases was examined. It is found that free BoNT/A was almost completely inactivated by incubation at pH 3 or less, but remained fully active at pH 4–8. Similarly, the free BoNT/A and NTNHA-A were easily degraded into short peptides by digestive proteases, as is observed with most other proteins (Gu et al. 2012). In contrast, the in vitro-reconstituted M-PTC fully protected BoNT/A from protease- and low pH-induced inactivation, mimicking the behavior of the wild-type PTC. No such mutual protection was observed at neutral pH, which induced dissociation of the M-PTC. Thus, NTNHA plays a major role in protecting and stabilizing BoNTs under the pH and protease conditions prevailing in the GI tract, whereas HA proteins likely play a minor role in this process.
3.3 Architecture of the M-PTC
To understand the molecular mechanisms underlying BoNT/A protection and pH-dependent assembly of the M-PTC, we set out to determine the crystal structure of the M-PTC. Prior to this work, the best structural information available on a PTC was a projection map of the BoNT/A LL-PTC at 30 Å resolution (Burkard et al. 1997) and a negative stain transmission microscopic image of BoNT/D L-PTC (Hasegawa et al. 2007).
Not surprisingly, large molecular mass multi-protein complexes are notoriously difficult to crystallize, and if they do, their diffraction is often limited to low resolution. After years of effort and numerous crystals screened at synchrotrons, the structure of the M-PTC composed of the full-length BoNT/Ai and NTNHA-A could be determined at 3.9 Å resolution. Despite the high quality of the structure model, which was built based on the unbiased experimental phase, it could not provide the detailed information needed to interrogate the extensive protein interactions between BoNT/Ai and NTNHA-A.
The breakthrough came only with the successful nanobody-facilitated crystallization of the M-PTC. Nanobodies are the smallest antigen-binding fragments (~12–15 kDa) of naturally occurring heavy-chain-only antibodies (VHH) present in camelids (Hamers-Casterman et al. 1993; Muyldermans et al. 2001) and they have been exploited as inhibitors for BoNTs (Conway et al. 2010; Dong et al. 2010; Mukherjee et al. 2012; Tremblay et al. 2010). VHH provided additional sites to facilitate a more compact crystal packing (Korotkov et al. 2009; Lam et al. 2009), which significantly improved the diffraction quality of crystals to 2.7 Å resolution. Although the free and the VHH-bound M-PTC were crystallized in different crystal forms under different crystallization conditions, the structures of the M-PTC are indistinguishable, demonstrating that this structure is a faithful representation of the physiological conformation of the M-PTC, independent of VHH binding and crystal packing.
The M-PTC structure has revealed a wealth of information and shows many unanticipated features (Fig. 1). First, BoNT/Ai adopts a distinct quaternary arrangement in the M-PTC. Pair-wise structural comparisons with the free BoNT/A (Lacy et al. 1998) yield root-mean-square deviations (r.m.s.d) of 0.6, 1.1, and 1.3 Å for LC, HN, and HC, respectively, suggesting the existence of conserved structures within each domain. Unexpectedly, HC in the M-PTC differs from HC in free BoNT/A in that it rotates about 140° though a linker between HN and HC. Second, NTNHA-A is remarkably similar to BoNT/A. The three domains of NTNHA-A, when pair-wise compared with LC, HN, or HC, yield r.m.s.d. of 2.2, 2.3, and 1.9 Å, respectively, and thus were named nLC, nHN and nHC (Figs. 1–2). BoNT/Ai and NTNHA-A form an interlocked compact complex, reminiscent of a handshake. In the center of this molecular complex is the HC fragment that is embraced by all three domains of NTNHA-A (Fig. 1). Interestingly, LC does not directly contact NTNHA-A.
Fig. 2.
Structures of BoNT/A and NTNHA-A in the M-PTC. The domain colors are as described for Figure 1.
3.4 The structure of NTNHA-A
Despite an overall similarity in domain organization and individual domain structure to BoNT/A, NTNHA has several unique features and has clearly lost most of BoNT’s features necessary to fulfill the neuron-specific attack (Fig. 2).
The protease activity of LC is absolutely dependent on a bound Zn2+ whereas nLC does not bind zinc. This has been carefully verified by inspecting an anomalous difference electron density map and is consistent with the fact that no protease activity has been observed for nLC. Primary sequence analysis of LC revealed an “HELIH+E” signature motif in all BoNTs (Ile is highly conserved except in BoNT/C and D where it is replaced by Asn and Thr, respectively). The His and the second Glu residues coordinate a Zn2+, while the first Glu coordinates a water molecule for hydrolysis. This motif is found in a variety of Zn2+-dependent metalloproteases such as thermolysin, which suggests that LCs may utilize a similar enzymatic mechanism (Binz et al. 2002; Kurazono et al. 1992). However, such a motif is not found in NTNHA-A or any other serotype. Interestingly, a structure-based sequence alignment shows that this motif is replaced by a highly conserved “KCLIK” motif in NTNHAs (Gu et al. 2012).
A second difference between the two molecules is that a long loop connecting LC and HC in BoNT/A is missing in NTNHA. This loop is cleaved post-translationally to activate BoNT/A, while LC and HC remain linked via an essential disulfide bond (Cys430–Cys454 in BoNT/A) that is conserved in all BoNTs (de Paiva et al. 1993). In all available crystal structures of the full-length BoNTs, most of this loop (Ile435–Ala449 in BoNT/A) has no visible electron density, indicating a highly flexible structure in this area. It is suggested that this loop keeps LC attached to HC via the conserved disulfide bond during transportation of LC, and then releases LC upon reduction in the cytosol. This disulfide bond and the homologous loop in BoNT/A are not found in any serotype of NTNHA.
The structure of NTNHA also suggests that nHC is unlikely to have neuron-binding capacity due to the many deletions of surface exposed loops in HC. Notably, none of the GT1b-binding residues of BoNT/A-HC are conserved in NTNHA-A, including the highly conserved ganglioside-binding motif present in many BoNT serotypes (Rummel et al. 2009; Rummel et al. 2004; Stenmark et al. 2008). Accordingly, NTNHA-A did not bind to immobilized ganglioside mixtures (Rummel unpublished results).
Despite these differences, NTNHA does retain an intriguing structural feature of BoNTs; the so-called translocation domain belt. The belt region (Asn493–Asp546 in BoNT/A) is a mostly extended loop that wraps around LC and is suggested to be a pseudosubstrate inhibitor of the LC protease, and/or a chaperone during translocation of LC into the cytosol (Brunger et al. 2007). NTNHA-A maintains the homologous belt region (Asp451–Asn496, referred to as the nBelt) as part of nHN, which wraps around nLC in an extended conformation. The function of the nBelt remains to be discovered.
It is worth noting that NTNHA has a structural feature that is not present in the equivalent part of BoNT/A, which is a large insert in nLC. Sequence analysis shows that a 33-residue fragment in this area (Gly116–Ala148 in NTNHA-A, termed the nLoop), is conserved in NTNHA serotypes A1, B, C, D, and G, but is missing in serotypes A2, E, and F. Many of the known spontaneous nicking sites in NTNHA are located in the nLoop (Fujita et al. 1995; Inoue et al. 1996; Miyata et al. 2009; Sagane et al. 2002). The whole nLoop has no visible electron density in the M-PTC structure, indicating high flexibility in this area. It has been reported that the nicking sites in NTNHA are masked in the HA-bound PTC, and that the M-PTC that contains the nicked NTNHA can no longer assemble with HAs (Kouguchi et al. 2002; Sagane et al. 2002). These data suggest that the nLoop of NTNHA is likely part of the binding site for HAs, and thus participates in assembly of the larger sized PTCs (Gu et al. 2012). Consistent with this hypothesis, NTNHA-A2, E, and F, which do not have the nLoop, also lack the accompanying HA proteins and only form the HA-negative M-PTC (East and Collins 1994; Fujii et al. 1993; Lin et al. 2010).
Collectively, these findings corroborate the hypothesis that BoNT/A and NTNHA-A might have evolved from a common ancestor by gene duplication, which has resulted in a similar general architecture but distinct biological functions. On the basis of amino acid sequence alignment, there is high sequence identity among the different serotypes of NTNHA. For example, NTNHA-A shows 82.9%, 65.8%, 65.9%, 65.8%, 74.9%, and 72.2% identity with NTNHA of serotypes B, C, D, E, F, and G, respectively (Krebs and Lebeda 2008). This high sequence identity suggests that the overall structural architecture of NTNHA should be conserved across the different serotypes. On the other hand, the sequence identity is much lower among the different serotypes of BoNTs: BoNT/A shares only 39.4%, 32.5%, 33.1%, 40.4%, 39.0%, and 39.6% identity with BoNT/B, C, D, E, F, and G, respectively (Krebs and Lebeda 2008) (see chapter 1). This raises several interesting questions for future studies. Why are the amino acid sequences of NTNHAs relatively static compared with those of the BoNTs? Are interactions between BoNT and NTNHA serotype-specific? Does NTNHA have functions other than protecting BoNT?
3.5 A novel conformation of BoNT/A in the M-PTC
BoNT/Ai adopts a novel conformation in the M-PTC. Specifically, HC rotates about 140° through a linker between HN and HC, which brings the receptor-binding site on HC into the vicinity of the C-terminus of HN (Fig. 3). In contrast, the LC-HN moiety of complexed BoNT/Ai maintains the same structure as that observed in the free BoNT/A, BoNT/B, and a truncated BoNT/A composed of LC-HN (Masuyer et al. 2009). The HC reorientation of BoNT/Ai is likely induced by NTNHA-A rather than pH because the same conformation is adopted by all structures of free BoNT/A or BoNT/B crystallized at pHs ranging from 5.0 to 7.0 (Chai et al. 2006; Eswaramoorthy et al. 2004; Lacy et al. 1998; Swaminathan and Eswaramoorthy 2000).
Fig. 3.
The HC of BoNT/A can adopt multiple conformations. Surface representations of the structures of BoNT/A in the M-PTC (a), free forms of BoNT/A (BoNT/B adopts an identical conformation) (b), and free BoNT/E (c). (d) Cartoon model showing the three conformations of the HC fragment depicted in (a)–(c). The receptor-binding site in HC is indicated by a red star. (e) Sequence alignment of the HN-HC linker. Residues that adopt an α-helix or β-sheet are in orange or green, respectively.
The conformational changes of BoNT/A in the M-PTC are caused mostly by rotation of HC, with the LC-HN moiety as the base, via a linker composed of residues Leu845–Thr876 in BoNT/A. In the free BoNT/A, this linker contains an α-helix (Gln860–Lys871) and two 310-helices (Leu853–Lys855 and Ile873–Thr876) and is structurally conserved in the free BoNT/B. However, the linker adopts a different structure in the complex form, comprising an α-helix (Leu863–Ile874) and two 310-helices (Leu853–Lys855 and Asp858–Gln860) (Fig. 3e). Interestingly, the equivalent linker in BoNT/E (Leu819–Ser850) has no rigid secondary structure, which could explain the flexible domain organization of BoNT/E revealed by the electron microcopy and crystal structures (Fischer et al. 2008; Kumaran et al. 2009).
Mutagenesis studies suggested that the correct reorientation of HC is crucial for effective HN-mediated translocation (Gu et al. 2012). It was speculated that the flexible HN–HC linker may play a role in coordinating HC-mediated receptor binding and HN-mediated translocation, given that the membrane-anchored receptors impose some geometric restrictions on the position of HC with respect to the membrane surface (Jin et al. 2006), and the long helical HN needs to strategically orientate on the membrane to achieve efficient translocation of LC to the cytosol (Baldwin et al. 2007; Kumaran et al. 2009; Montal 2010). This is consistent with the hypothesis that the more rapid translocation of LC to the cytosol observed with BoNT/E than with BoNT/A is due to the more flexible BoNT/E-like linker that allows an “easier” reorientation of HC to achieve the optimal conformation for translocation (Fischer et al. 2008; Keller et al. 2004; Kumaran et al. 2009; Wang et al. 2008).
Mechanistically, HC-mediated receptor binding is the earliest step during neuron invasion, and likely one of the most crucial, because damage to HC will jeopardize the enrichment of BoNT/A on the neuron membrane (Montecucco 1986). Structural modeling shows that the ganglioside-binding pocket in HC is mostly unchanged upon binding to NTNHA (Fig. 4) (Stenmark et al. 2008). The potential binding site for the protein receptor, predicted to be homologous to the Sytbinding site in BoNT/B (Jin et al. 2006), is also exposed on the molecular surface of the M-PTC (Fig. 4). It is thus reasonable to predict that BoNT/A will still be capable of binding its dual receptors in the context of the M-PTC.
Fig. 4.
NTNHA binding does not directly block the receptor-binding sites in BoNT/A. (a) The structure of HCA in the M-PTC (complex, green) is superimposed with an HCA–GT1b (apo, orange) complex (PDB code: 2VU9). Key residues that interact with GT1b (purple sticks) are shown in stick representation. Syt-II (ribbon in purple) is modeled based on HCB–Syt-II complex (PDB code: 2NM1). (b) The structures in panel (a) are shown in the context of the M-PTC with NTNHA-A in a surface representation (gray).
4 Dynamic association of the M-PTC
4.1 BoNT/A and NTNHA-A interactions
The molecular handshake of BoNT/Ai and NTNHA-A buries an unusually large solvent-accessible area (~3,200 Å2 per molecule), resulting in a tightly bound complex at pH 6.0. The binding is predominantly driven by enthalpy (Kd = 30.8 nM, ΔH = −17.6 kcal mol−1, and ΔS = − 24.8 cal mol−1 K−1). It is interesting to note that most of the intra-complex interactions are mediated by HC, which forms extensive interfaces with all three domains of NTNHA-A. The unique M-PTC architecture is consistent with a comprehensive antibody mapping study, which showed that all epitopes on BoNT/A that are masked in the PTC are mapped to HC, whereas epitopes in LC are all exposed in the PTC (Chen et al. 1997).
To investigate the interactions of BoNT/A within the M-PTC, we took advantage of the recombinant expression system to perform a systematic domain truncation study. We found that the binding behavior of the full-length BoNT/A could largely be replicated by the isolated HC, which binds to NTNHA-A with a high affinity at pH 6.0 (Kd = 48.3 nM, ΔH = −12.3 kcal mol−1, and ΔS = −8.0 cal mol−1 K−1) but not at pH 7.5. Consistent with this, a truncated BoNT/A lacking HC no longer binds NTNHA-A. Somewhat surprisingly, contributions to the association of the complex are not distributed evenly across the large interacting interface. A single point mutation (e.g. HC-K1000A) could significantly decrease the binding affinity of BoNT/A for NTNHA-A by up to 7-fold and HC-K1000A/K1039A/K1121A virtually disrupted the M-PTC (Gu et al. 2012). Thus, there may be “hot spots” within the intra-complex interactions that are crucial to maintain a stable M-PTC.
The clamp-like binding of HC by NTNHA-A is reminiscent of a diverse array of protein chaperones (Stirling et al. 2006). However, NTNHA-A differs from the conventional role of chaperones, which facilitate protein folding, by providing large and multivalent binding surfaces for BoNT/A to shield it from the hostile gut environment. Interestingly, it has been reported that HC of the free BoNT is more susceptible to proteolytic cleavage than are the LC and HN domains, suggesting that protection of HC is likely critical (Chen et al. 1997; Shone et al. 1985). Future studies should be guided by the structure of the M-PTC to determine how, or if, the apparent “biased” protection towards HC has evolved as an optimal strategy to protect BoNT/A.
4.2 pH-dependent association of the PTC
One of the most fascinating features of the M-PTC is that BoNT/A can be released from the fully armored complex by a simple pH change. It was reasoned that the pH-dependent assembly of the M-PTC may be regulated by specific residues (referred to as pH sensors) that adopt pH-dependent conformational changes. Attempting to identify pH sensors in a 300 kDa protein complex is a daunting task. Fortunately, the structure of the M-PTC has prompted to focus on HC, which is responsible for most of the intra-complex interactions and maintains the pH-sensing feature observed in the full-length BoNT/A.
Which residues could serve as pH sensors? Titratable residues in the acidic environment are histidine (His), glutamate (Glu), and aspartate (Asp), which have typical side chain pKa values (where Ka is the acid dissociation constant) around 6.1, 4.3, and 3.9, respectively. Protonation of these residues has been reported to be involved in low pH-sensing in the pH-gated urea channel, chloride channel, histidine kinases, and viral membrane fusion proteins (Mueller et al. 2008; Perez and Groisman 2007; Stroffekova et al. 1998; Weeks and Sachs 2001).
Using structure-based mutagenesis and in vitro binding assays, Glu982 and Asp1037 of BoNT/A were identified as two key residues that mediate the pH-dependent binding between BoNT/A and NTNHA-A. A surface electrostatic potential analysis showed that Glu982 and Asp1037 are located in a generally positively charged surface in HC, whereas the opposing NTNHA surface is negatively charged. Thus, the negative charge of the deprotonated Glu982 and Asp1037 would potentially weaken the local complementary electrostatic interactions in a pH-dependent manner.
The pKa describes the tendency of a titratable residue to give up a proton. For internal ionizable groups in proteins in different microenvironments, their pKa values and charged states could be substantially different from their intrinsic values (Pace et al. 2009). Based on the crystal structures, the pKa of Glu982 and Asp1037 were calculated to increase to neutral or mild alkaline pH when in the complex form compared to the free BoNT/A (Li et al. 2005). Clearly, the assembly of the M-PTC is a finely tuned dynamic process. It can be hypothesized that when the BoNT/A and NTNHA-A approach each other to form the transient complex, the pKa of Glu982 and Asp1037 of BoNT/A will gradually increase, partly because of the desolvation effect when they are buried in the complex. In an acidic environment, pH sensors, including Glu982 and Asp1037 of BoNT/A, will be protonated and subsequently stabilize complex assembly. In contrast, the pH sensors will be deprotonated in neutral or alkaline environments, resulting in repulsive charge interactions that destabilize the transient complex.
It is worth noting that we are just beginning to understand the pH-sensing mechanism underlying M-PTC assembly. Other pH-sensing components in addition to Glu982 and Asp1037 must exist in BoNT/A. Strong evidence for this has come from the thermodynamic studies on the mutated HC-E982A, which is able to bind NTNHA-A at pH 7.5. Nevertheless, a significant loss of enthalpy (ΔΔH ~6.3 kcal mol−1) was observed for binding at pH 7.5 compared to that at pH 6.0.
This suggests that the network of protein-protein interactions between HC and NTNHA-A and solvation of the HC surface, contributed by the combined surface pH-sensing elements, are different at pH 7.5 and pH 6.0. To add another layer of complexity, the key residues in BoNT/A that mediate BoNT–NTNHA interactions, including Glu982 and Asp1037, are not conserved in other BoNT serotypes. The structure of BoNT/A M-PTC should provide a framework for future studies to fully dissect the comprehensive pH-sensing mechanism across different serotypes.
5 HA proteins
5.1 The structure of the HA proteins
The HA proteins account for up to 60% of the molecular mass of PTC, but we have a poor understanding of their role in the pathogenesis of BoNTs. The crystal structures of the isolated HAs have been reported for selected serotypes, including HA-17 (BoNT/D), HA-33 (BoNT/A, C, and D), and HA-70 (BoNT/C) (Arndt et al. 2005; Hasegawa et al. 2007; Inoue et al. 2003; Nakamura et al. 2009; Nakamura et al. 2008). However, it is largely unknown how the HAs assemble with each other and subsequently with BoNT and NTNHA.
HA-70 of BoNT/C consists of two domains, HA-70a and HA-70b, which are proteolytically cleaved from the full-length HA-70. The HA-70a is a single domain fragment with three α-helices and eight β-strands. The HA-70b subcomponent consists of three subdomains (I, II and III). Domain I of HA-70b is very similar to HA-70a. Domains II and III adopt a similar jelly-roll–like β-sandwich structure (Nakamura et al. 2009). Intriguingly, HA-70 forms a three-bladed propeller-like trimer in crystals, with a pore located at the center of the trimer (Fig. 5). The pore, estimated to be 33 Å in length and 21 Å in diameter, is a compact cylindrical β-barrel composed of 12 β- strands from HA-70a and the domain I of HA-70b (Nakamura et al. 2009).
Fig. 5.
A model of the assembly of the PTC. The structures are shown in a cartoon representation: HA-17 (salmon, BoNT/D, PDB code 2E4M), HA-33 (green, BoNT/D, PDB code 2E4M), HA-70 (cyan/yellow/gray, BoNT/C, PDB code 2ZS6), BoNT (gold, BoNT/A1, PDB code 3V0A), and NTNHA (blue, BoNT/A1, PDB code 3V0A).
HA-33 has a dumbbell-like shape, composed of two β-trefoil domains linked by an α-helix (Fig. 5). The β-trefoil fold is a common structural module found in many proteins, including many lectins, where it acts as an oligosaccharide-binding unit. The β-trefoil domains of HA-33 from BoNT/A, C, and D are highly similar. However, the domains are capable of twisting against each other by as much as 10°, via the helical linker, suggesting significant inter-domain conformational plasticity (Arndt et al. 2005; Hasegawa et al. 2007; Inoue et al. 2003). HA-17 seems to tightly associate with HA-33, forming a complex that remains bound even in the presence of high concentrations of guanidine hydrochloride (Kouguchi et al. 2002). The crystal structure shows that HA-17 of BoNT/D also adopts a β-trefoil fold, with each HA-17 associating with two HA-33 molecules by binding to their N-terminal β-trefoil domains (Hasegawa et al. 2007).
Several different structural models of the fully assembled PTC have been proposed (Hasegawa et al. 2007; Inoue et al. 1996; Kouguchi et al. 2002; Lietzow et al. 2008; Mutoh et al. 2003; Suzuki et al. 2005). The discrepancies among the models are largely due to the lack of systematic biochemical and biophysical characterization of HAs in solution. For example, we do not yet know if the trimeric HA-70 or the HA-17/33 complex that were observed in crystal structures represent the physiologically relevant conformations in solution. Furthermore, future studies should address whether and how HA-17/33 interacts with HA-70, and how BoNT and/or NTNHA may interact with the HA proteins (Fig. 5).
5.2 HAs mediate toxin-host interactions
HA proteins appear to play an important role in absorption of PTC by interacting with oligosaccharides on intestinal epithelial cells (Fujinaga et al. 1997; Fujinaga et al. 2004; Inoue et al. 2001; Kojima et al. 2005; Nakamura et al. 2007) (also see chapter 3). One N-acetylneuraminic acid (Neu5Ac) binding site has been reported in HA-70b of BoNT/C, which is located in a cleft formed by domains II and III (Nakamura et al. 2009). The HA-70–Neu5Ac interaction was not well defined in the reported crystal structure, likely due to the low binding affinity of a single Neu5Ac to HA-70. Improved crystal structures are reported recently, which show that HA-70 of BoNT/C can recognize both α2-3- and α2-6-sialylated oligosaccharides, and that most of the HA-sugar interactions are contributed by Neu5Ac (Yamashita et al. 2012). A binding assay of HA-70 of BoNT/C against α2-3-sialyllactosamine- or α2-6-sialyllactosamine-conjugated BSA suggests that it might have a higher affinity for α2-3-sialylated oligosaccharide (Yamashita et al. 2012). However, the crystal structures show that α2-6-sialyllactosamine forms more hydrogen bonds with HA-70 compared with α2-3-sialyllactosamine.
HA-33 of BoNT/C has been reported to have three binding sites that could interact with several sugars, including Neu5Ac, galactose (Gal), and N-acetylgalactosamine (GalNAc) (Nakamura et al. 2008). This finding is largely derived from crystal-soaking experiments, where HA-33 crystals were soaked in high-concentration sugar solutions prior to collection of diffraction data for structure determination. It is worth noting that all three sugars examined in this study bind differently to the two identical HA-33 molecules in an asymmetric unit in the crystal, suggesting there may be crystallographic artifacts. Furthermore, inconsistent results have been reported regarding the GalNAc-binding site(s) in HA-33 of BoNT/C (Nakamura et al. 2008; Nakamura et al. 2011). Thus, the physiological role of HA-33–sugar interactions awaits clarification in future studies.
One of the most exciting recent breakthroughs is the discovery that E-cadherin (E-cad), an epithelial cell surface adhesion molecule, is a host receptor for HA proteins. It was suggested that HA proteins directly bind to and disrupt E-cad–mediated cell-to-cell adhesion, thus facilitating the absorption of BoNT through the intestinal epithelium via a paracellular route (Ito et al. 2011; Matsumura et al. 2008; Sugawara et al. 2010) (also see chapter 3). Consistent with this hypothesis, the interaction is highly specific: HAs bind only to E-cad and not to N-cad or VE-cad (Sugawara et al. 2010). Furthermore, the HA–E-cad interaction is serotype- and species-specific. For example, the HAs of BoNT/A or B efficiently bind to human, bovine, or mouse E-cad, but not to chicken E-cad; whereas the HA of BoNT/C does not recognize human E-cad (Sugawara et al. 2010). These observations correlate well with the epidemiology of botulism. For instance, birds are more resistant to oral intoxication of BoNT/A (Notermans et al. 1980) and there are few reports of human botulism caused by BoNT/C (Coffield et al. 1997).
Collectively, these observations suggest that the host susceptibility to botulism is likely determined in part by the interplay between HA proteins and host receptors in the GI tract. Future studies should focus on dissecting the poorly understood HA-mediated interplay between toxin and host, including the identity of the host carbohydrate and protein receptors. A better understanding of the molecular mechanisms controlling these steps will facilitate the development of novel strategies to block the early stages of oral intoxication by preventing BoNT entry into the general circulation.
6 Conclusions
We have observed an unprecedented growth in our knowledge of the structure and function of botulinum neurotoxin progenitor complexes, due in large part to worldwide collaborations among scientists from different fields with complementary expertise. Among the crucial findings, the first crystal structure of the minimal progenitor toxin complex of BoNT/A has been reported recently (Gu et al. 2012), which has rationalized a large set of genetic and biochemical observations.
BoNT/A and NTNHA-A form an interlocked handshake-like complex, which lends both proteins extraordinary stability against low pH and digestive proteases. Interestingly, the non-toxic NTNHA-A has low sequence identity to BoNT/A but adopts a similar architecture. Despite this, NTNHA-A is devoid of the characteristic structural features of BoNT/A that are crucial to its biological functions (e.g., endopeptidase activity, host receptor binding). The M-PTC structure also helps pinpoint several pH-sensing residues that are key players in balancing the seemingly contradictory needs of BoNT/A and NTNHA-A: strong binding for protection in the gut and timely release upon gaining entry to the general circulation.
The fascinating mode of BoNT/A protection and assembly should also facilitate mechanistic studies on other BoNT serotypes and other bacterial toxins. However, many questions remain to be addressed. How does the M-PTC assemble with the HA proteins? Does the ~500 kDa HA complex further protect BoNT/A against low pH and proteases? How does the HA complex interact with host receptors and facilitate translocation across the epithelium? How does the PTC disassemble upon reaching the relative safety of the bloodstream? The answers to these questions await rigorous structural and functional analyses in the future.
On another front, a better understanding of the protection and transportation functions of the PTC may help identify the Achilles’ heel of this complex and expedite the design of novel approaches to counteract BoNT intoxication. This could be accomplished, for example, by small molecules that promote premature disassembly and destruction of the PTC in the GI tract, or which disrupt HA-mediated toxin–host recognition in the small intestine to prevent BoNT entry into the circulation. Such preventive countermeasures for oral BoNT intoxication will be crucial in situations such as during an outbreak of botulism, and could be a strong deterrent against the use of BoNT as a bioterrorism weapon.
Finally, the unique features of the complex involved in BoNT protection, absorption, and dynamic assembly suggest that PTC-based vehicles could be engineered to shield proteinaceous drugs from GI destruction and thus allow their oral administration. Furthermore, re-targeted BoNT with altered cell tropism could be constructed that would allow tissue- and organ-specific drug delivery. If successful, this would be an enormous step forward in the development of therapeutic biologics.
References
- Arndt JW, Gu J, Jaroszewski L, Schwarzenbacher R, Hanson MA, Lebeda FJ, Stevens RC. The structure of the neurotoxin-associated protein HA33/A from Clostridium botulinum suggests a reoccurring beta-trefoil fold in the progenitor toxin complex. J Mol Biol. 2005;346:1083–1093. doi: 10.1016/j.jmb.2004.12.039. [DOI] [PubMed] [Google Scholar]
- Baldwin MR, Kim JJ, Barbieri JT. Botulinum neurotoxin B-host receptor recognition: it takes two receptors to tango. Nat Struct Mol Biol. 2007;14:9–10. doi: 10.1038/nsmb0107-9. [DOI] [PubMed] [Google Scholar]
- Binz T, Bade S, Rummel A, Kollewe A, Alves J. Arg(362) and Tyr(365) of the botulinum neurotoxin type a light chain are involved in transition state stabilization. Biochemistry. 2002;41:1717–1723. doi: 10.1021/bi0157969. [DOI] [PubMed] [Google Scholar]
- Binz T, Kurazono H, Wille M, Frevert J, Wernars K, Niemann H. The complete sequence of botulinum neurotoxin type A and comparison with other clostridial neurotoxins. J Biol Chem. 1990;265:9153–9158. [PubMed] [Google Scholar]
- Binz T, Rummel A. Cell entry strategy of clostridial neurotoxins. J Neurochem. 2009;109:1584–1595. doi: 10.1111/j.1471-4159.2009.06093.x. [DOI] [PubMed] [Google Scholar]
- Blasi J, Chapman ER, Link E, Binz T, Yamasaki S, De Camilli P, Sudhof TC, Niemann H, Jahn R. Botulinum neurotoxin A selectively cleaves the synaptic protein SNAP-25. Nature. 1993a;365:160–163. doi: 10.1038/365160a0. [DOI] [PubMed] [Google Scholar]
- Blasi J, Chapman ER, Yamasaki S, Binz T, Niemann H, Jahn R. Botulinum neurotoxin C1 blocks neurotransmitter release by means of cleaving HPC-1/syntaxin. Embo J. 1993b;12:4821–4828. doi: 10.1002/j.1460-2075.1993.tb06171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroff DA, Townend R, Fleck U, DasGupta BR. Ultracentrifugal analysis of the crystalline toxin and isolated fractions of Clostridium botulinum type A. J Biol Chem. 1966;241:5165–5167. [PubMed] [Google Scholar]
- Breidenbach MA, Brunger AT. Substrate recognition strategy for botulinum neurotoxin serotype A. Nature. 2004;432:925–929. doi: 10.1038/nature03123. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Breidenbach MA, Jin R, Fischer A, Santos JS, Montal M. Botulinum neurotoxin heavy chain belt as an intramolecular chaperone for the light chain. PLoS Pathog. 2007;3:e113. doi: 10.1371/journal.ppat.0030113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard F, Chen F, Kuziemko GM, Stevens RC. Electron density projection map of the botulinum neurotoxin 900-kilodalton complex by electron crystallography. J Struct Biol. 1997;120:78–84. doi: 10.1006/jsbi.1997.3910. [DOI] [PubMed] [Google Scholar]
- Chai Q, Arndt JW, Dong M, Tepp WH, Johnson EA, Chapman ER, Stevens RC. Structural basis of cell surface receptor recognition by botulinum neurotoxin B. Nature. 2006;444:1096–1100. doi: 10.1038/nature05411. [DOI] [PubMed] [Google Scholar]
- Chen F, Kuziemko GM, Amersdorfer P, Wong C, Marks JD, Stevens RC. Antibody mapping to domains of botulinum neurotoxin serotype A in the complexed and uncomplexed forms. Infect Immun. 1997;65:1626–1630. doi: 10.1128/iai.65.5.1626-1630.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LW, Onisko B, Johnson EA, Reader JR, Griffey SM, Larson AE, Tepp WH, Stanker LH, Brandon DL, Carter JM. Effects of purification on the bioavailability of botulinum neurotoxin type A. Toxicology. 2008;249:123–129. doi: 10.1016/j.tox.2008.04.018. [DOI] [PubMed] [Google Scholar]
- Coffield JA, Bakry N, Zhang RD, Carlson J, Gomella LG, Simpson LL. In vitro characterization of botulinum toxin types A, C and D action on human tissues: combined electrophysiologic, pharmacologic and molecular biologic approaches. J Pharmacol Exp Ther. 1997;280:1489–1498. [PubMed] [Google Scholar]
- Collins MD, East AK. Phylogeny and taxonomy of the food-borne pathogen Clostridium botulinum and its neurotoxins. J Appl Microbiol. 1998;84:5–17. doi: 10.1046/j.1365-2672.1997.00313.x. [DOI] [PubMed] [Google Scholar]
- Conway JO, Sherwood LJ, Collazo MT, Garza JA, Hayhurst A. Llama single domain antibodies specific for the 7 botulinum neurotoxin serotypes as heptaplex immunoreagents. PLoS ONE. 2010;5:e8818. doi: 10.1371/journal.pone.0008818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta BR, Boroff DA. Separation of toxin and hemagglutinin from crystalline toxin of Clostridium botulinum type A by anion exchange chromatography and determination of their dimensions by gel filtration. J Biol Chem. 1968;243:1065–1072. [PubMed] [Google Scholar]
- de Paiva A, Poulain B, Lawrence GW, Shone CC, Tauc L, Dolly JO. A role for the interchain disulfide or its participating thiols in the internalization of botulinum neurotoxin A revealed by a toxin derivative that binds to ecto-acceptors and inhibits transmitter release intracellularly. J Biol Chem. 1993;268:20838–20844. [PubMed] [Google Scholar]
- Dong J, Thompson AA, Fan Y, Lou J, Conrad F, Ho M, Pires-Alves M, Wilson BA, Stevens RC, Marks JD. A single-domain llama antibody potently inhibits the enzymatic activity of botulinum neurotoxin by binding to the non-catalytic alpha-exosite binding region. J Mol Biol. 2010;397:1106–1118. doi: 10.1016/j.jmb.2010.01.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East AK, Bhandari M, Hielm S, Collins MD. Analysis of the botulinum neurotoxin type F gene clusters in proteolytic and nonproteolytic Clostridium botulinum and Clostridium barati. Curr Microbiol. 1998;37:262–268. doi: 10.1007/s002849900376. [DOI] [PubMed] [Google Scholar]
- East AK, Collins MD. Conserved structure of genes encoding components of botulinum neurotoxin complex M and the sequence of the gene coding for the nontoxic component in nonproteolytic Clostridium botulinum type F. Curr Microbiol. 1994;29:69–77. doi: 10.1007/BF01575751. [DOI] [PubMed] [Google Scholar]
- Eisele KH, Fink K, Vey M, Taylor HV. Studies on the dissociation of botulinum neurotoxin type A complexes. Toxicon. 2011;57:555–565. doi: 10.1016/j.toxicon.2010.12.019. [DOI] [PubMed] [Google Scholar]
- Eswaramoorthy S, Kumaran D, Keller J, Swaminathan S. Role of metals in the biological activity of Clostridium botulinum neurotoxins. Biochemistry. 2004;43:2209–2216. doi: 10.1021/bi035844k. [DOI] [PubMed] [Google Scholar]
- Fischer A, Garcia-Rodriguez C, Geren I, Lou J, Marks JD, Nakagawa T, Montal M. Molecular architecture of botulinum neurotoxin E revealed by single particle electron microscopy. J Biol Chem. 2008;283:3997–4003. doi: 10.1074/jbc.M707917200. [DOI] [PubMed] [Google Scholar]
- Fischer A, Montal M. Characterization of Clostridial botulinum neurotoxin channels in neuroblastoma cells. Neurotox Res. 2006;9:93–100. doi: 10.1007/BF03033926. [DOI] [PubMed] [Google Scholar]
- Fischer A, Montal M. Crucial Role of the Disulfide Bridge between Botulinum Neurotoxin Light and Heavy Chains in Protease Translocation across Membranes. J Biol Chem. 2007a;282:29604–29611. doi: 10.1074/jbc.M703619200. [DOI] [PubMed] [Google Scholar]
- Fischer A, Montal M. Single molecule detection of intermediates during botulinum neurotoxin translocation across membranes. Proc Natl Acad Sci U S A. 2007b;104:10447–10452. doi: 10.1073/pnas.0700046104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foran P, Lawrence GW, Shone CC, Foster KA, Dolly JO. Botulinum neurotoxin C1 cleaves both syntaxin and SNAP-25 in intact and permeabilized chromaffin cells: correlation with its blockade of catecholamine release. Biochemistry. 1996;35:2630–2636. doi: 10.1021/bi9519009. [DOI] [PubMed] [Google Scholar]
- Fu Z, Chen S, Baldwin MR, Boldt GE, Crawford A, Janda KD, Barbieri JT, Kim JJ. Light chain of botulinum neurotoxin serotype A: structural resolution of a catalytic intermediate. Biochemistry. 2006;45:8903–8911. doi: 10.1021/bi060786z. [DOI] [PubMed] [Google Scholar]
- Fujii N, Kimura K, Yokosawa N, Yashiki T, Tsuzuki K, Oguma K. The complete nucleotide sequence of the gene encoding the nontoxic component of Clostridium botulinum type E progenitor toxin. J Gen Microbiol. 1993;139:79–86. doi: 10.1099/00221287-139-1-79. [DOI] [PubMed] [Google Scholar]
- Fujinaga Y, Inoue K, Watanabe S, Yokota K, Hirai Y, Nagamachi E, Oguma K. The haemagglutinin of Clostridium botulinum type C progenitor toxin plays an essential role in binding of toxin to the epithelial cells of guinea pig small intestine, leading to the efficient absorption of the toxin. Microbiology. 1997;143(Pt 12):3841–3847. doi: 10.1099/00221287-143-12-3841. [DOI] [PubMed] [Google Scholar]
- Fujinaga Y, Inoue K, Watarai S, Sakaguchi Y, Arimitsu H, Lee JC, Jin Y, Matsumura T, Kabumoto Y, Watanabe T, Ohyama T, Nishikawa A, Oguma K. Molecular characterization of binding subcomponents of Clostridium botulinum type C progenitor toxin for intestinal epithelial cells and erythrocytes. Microbiology. 2004;150:1529–1538. doi: 10.1099/mic.0.26805-0. [DOI] [PubMed] [Google Scholar]
- Fujinaga Y, Matsumura T, Jin Y, Takegahara Y, Sugawara Y. A novel function of botulinum toxin-associated proteins: HA proteins disrupt intestinal epithelial barrier to increase toxin absorption. Toxicon. 2009;54:583–586. doi: 10.1016/j.toxicon.2008.11.014. [DOI] [PubMed] [Google Scholar]
- Fujita R, Fujinaga Y, Inoue K, Nakajima H, Kumon H, Oguma K. Molecular characterization of two forms of nontoxic-nonhemagglutinin components of Clostridium botulinum type A progenitor toxins. FEBS Lett. 1995;376:41–44. doi: 10.1016/0014-5793(95)01241-5. [DOI] [PubMed] [Google Scholar]
- Gu S, Rumpel S, Zhou J, Strotmeier J, Bigalke H, Perry K, Shoemaker CB, Rummel A, Jin R. Botulinum neurotoxin is shielded by NTNHA in an interlocked complex. Science. 2012;335:977–981. doi: 10.1126/science.1214270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- Hasegawa K, Watanabe T, Suzuki T, Yamano A, Oikawa T, Sato Y, Kouguchi H, Yoneyama T, Niwa K, Ikeda T, Ohyama T. A novel subunit structure of clostridium botulinum serotype D toxin complex with three extended arms. J Biol Chem. 2007;282:24777–24783. doi: 10.1074/jbc.M703446200. [DOI] [PubMed] [Google Scholar]
- Hatheway CL. Toxigenic clostridia. Clin Microbiol Rev. 1990;3:66–98. doi: 10.1128/cmr.3.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KK, Xie G, Foley BT, Smith TJ, Munk AC, Bruce D, Smith LA, Brettin TS, Detter JC. Recombination and insertion events involving the botulinum neurotoxin complex genes in Clostridium botulinum types A, B, E and F and Clostridium butyricum type E strains. BMC Biol. 2009;7:66. doi: 10.1186/1741-7007-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Fujinaga Y, Honke K, Arimitsu H, Mahmut N, Sakaguchi Y, Ohyama T, Watanabe T, Inoue K, Oguma K. Clostridium botulinum type A haemagglutinin-positive progenitor toxin (HA(+)-PTX) binds to oligosaccharides containing Gal beta1-4GlcNAc through one subcomponent of haemagglutinin (HA1) Microbiology. 2001;147:811–819. doi: 10.1099/00221287-147-4-811. [DOI] [PubMed] [Google Scholar]
- Inoue K, Fujinaga Y, Watanabe T, Ohyama T, Takeshi K, Moriishi K, Nakajima H, Inoue K, Oguma K. Molecular composition of Clostridium botulinum type A progenitor toxins. Infect Immun. 1996;64:1589–1594. doi: 10.1128/iai.64.5.1589-1594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Sobhany M, Transue TR, Oguma K, Pedersen LC, Negishi M. Structural analysis by X-ray crystallography and calorimetry of a haemagglutinin component (HA1) of the progenitor toxin from Clostridium botulinum. Microbiology. 2003;149:3361–3370. doi: 10.1099/mic.0.26586-0. [DOI] [PubMed] [Google Scholar]
- Ito H, Sagane Y, Miyata K, Inui K, Matsuo T, Horiuchi R, Ikeda T, Suzuki T, Hasegawa K, Kouguchi H, Oguma K, Niwa K, Ohyama T, Watanabe T. HA-33 facilitates transport of the serotype D botulinum toxin across a rat intestinal epithelial cell monolayer. FEMS Immunol Med Microbiol. 2011:1–9. doi: 10.1111/j.1574-695X.2011.00779.x. [DOI] [PubMed] [Google Scholar]
- Jin R, Rummel A, Binz T, Brunger AT. Botulinum neurotoxin B recognizes its protein receptor with high affinity and specificity. Nature. 2006;444:1092–1095. doi: 10.1038/nature05387. [DOI] [PubMed] [Google Scholar]
- Keller JE, Cai F, Neale EA. Uptake of botulinum neurotoxin into cultured neurons. Biochemistry. 2004;43:526–532. doi: 10.1021/bi0356698. [DOI] [PubMed] [Google Scholar]
- Kojima S, Eguchi H, Ookawara T, Fujiwara N, Yasuda J, Nakagawa K, Yamamura T, Suzuki K. Clostridium botulinum type A progenitor toxin binds to Intestine-407 cells via N-acetyllactosamine moiety. Biochem Biophys Res Commun. 2005;331:571–576. doi: 10.1016/j.bbrc.2005.04.007. [DOI] [PubMed] [Google Scholar]
- Koriazova LK, Montal M. Translocation of botulinum neurotoxin light chain protease through the heavy chain channel. Nat Struct Biol. 2003;10:13–18. doi: 10.1038/nsb879. [DOI] [PubMed] [Google Scholar]
- Korotkov KV, Pardon E, Steyaert J, Hol WG. Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody. Structure. 2009;17:255–265. doi: 10.1016/j.str.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouguchi H, Watanabe T, Sagane Y, Sunagawa H, Ohyama T. In vitro reconstitution of the Clostridium botulinum type D progenitor toxin. J Biol Chem. 2002;277:2650–2656. doi: 10.1074/jbc.M106762200. [DOI] [PubMed] [Google Scholar]
- Krebs KM, Lebeda FJ. Comparison of the structural features of botulinum neurotoxin and NTNH, a non-toxic accessory protein of the progenitor complex. The Botulinum Journal. 2008;1:116–134. [Google Scholar]
- Kumaran D, Eswaramoorthy S, Furey W, Navaza J, Sax M, Swaminathan S. Domain organization in Clostridium botulinum neurotoxin type E is unique: its implication in faster translocation. J Mol Biol. 2009;386:233–245. doi: 10.1016/j.jmb.2008.12.027. [DOI] [PubMed] [Google Scholar]
- Kurazono H, Mochida S, Binz T, Eisel U, Quanz M, Grebenstein O, Wernars K, Poulain B, Tauc L, Niemann H. Minimal essential domains specifying toxicity of the light chains of tetanus toxin and botulinum neurotoxin type A. J Biol Chem. 1992;267:14721–14729. [PubMed] [Google Scholar]
- Lacy DB, Stevens RC. Sequence homology and structural analysis of the clostridial neurotoxins. J Mol Biol. 1999;291:1091–1104. doi: 10.1006/jmbi.1999.2945. [DOI] [PubMed] [Google Scholar]
- Lacy DB, Tepp W, Cohen AC, DasGupta BR, Stevens RC. Crystal structure of botulinum neurotoxin type A and implications for toxicity. Nat Struct Biol. 1998;5:898–902. doi: 10.1038/2338. [DOI] [PubMed] [Google Scholar]
- Lam AY, Pardon E, Korotkov KV, Hol WG, Steyaert J. Nanobody-aided structure determination of the EpsI:EpsJ pseudopilin heterodimer from Vibrio vulnificus. J Struct Biol. 2009;166:8–15. doi: 10.1016/j.jsb.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- Li L, Binz T, Niemann H, Singh BR. Probing the mechanistic role of glutamate residue in the zinc-binding motif of type A botulinum neurotoxin light chain. Biochemistry. 2000;39:2399–2405. doi: 10.1021/bi992321x. [DOI] [PubMed] [Google Scholar]
- Lietzow MA, Gielow ET, Le D, Zhang J, Verhagen MF. Subunit stoichiometry of the Clostridium botulinum type A neurotoxin complex determined using denaturing capillary electrophoresis. Protein J. 2008;27:420–425. doi: 10.1007/s10930-008-9151-2. [DOI] [PubMed] [Google Scholar]
- Lin G, Tepp WH, Pier CL, Jacobson MJ, Johnson EA. Expression of the Clostridium botulinum A2 neurotoxin gene cluster proteins and characterization of the A2 complex. Appl Environ Microbiol. 2010;76:40–47. doi: 10.1128/AEM.01882-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyer G, Thiyagarajan N, James PL, Marks PM, Chaddock JA, Acharya KR. Crystal structure of a catalytically active, non-toxic endopeptidase derivative of Clostridium botulinum toxin. Biochem Biophys Res Commun. 2009;381:50–53. doi: 10.1016/j.bbrc.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Matsumura T, Jin Y, Kabumoto Y, Takegahara Y, Oguma K, Lencer WI, Fujinaga Y. The HA proteins of botulinum toxin disrupt intestinal epithelial intercellular junctions to increase toxin absorption. Cell Microbiol. 2008;10:355–364. doi: 10.1111/j.1462-5822.2007.01048.x. [DOI] [PubMed] [Google Scholar]
- Miyata K, Yoneyama T, Suzuki T, Kouguchi H, Inui K, Niwa K, Watanabe T, Ohyama T. Expression and stability of the nontoxic component of the botulinum toxin complex. Biochem Biophys Res Commun. 2009;384:126–130. doi: 10.1016/j.bbrc.2009.04.095. [DOI] [PubMed] [Google Scholar]
- Montal M. Botulinum neurotoxin: a marvel of protein design. Annu Rev Biochem. 2010;79:591–617. doi: 10.1146/annurev.biochem.051908.125345. [DOI] [PubMed] [Google Scholar]
- Montecucco C. How do tetanus and botulinum neurotoxins bind to neuronal membranes? Trends Biochem Sci. 1986;11:314–317. [Google Scholar]
- Montecucco C, Rossetto O, Schiavo G. Presynaptic receptor arrays for clostridial neurotoxins. Trends Microbiol. 2004;12:442–446. doi: 10.1016/j.tim.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Montecucco C, Schiavo G. Structure and function of tetanus and botulinum neurotoxins. Q Rev Biophys. 1995;28:423–472. doi: 10.1017/s0033583500003292. [DOI] [PubMed] [Google Scholar]
- Mueller DS, Kampmann T, Yennamalli R, Young PR, Kobe B, Mark AE. Histidine protonation and the activation of viral fusion proteins. Biochem Soc Trans. 2008;36:43–45. doi: 10.1042/BST0360043. [DOI] [PubMed] [Google Scholar]
- Mukherjee J, Tremblay JM, Leysath CE, Ofori K, Baldwin K, Feng X, Bedenice D, Webb RP, Wright PM, Smith LA, Tzipori S, Shoemaker CB. A novel strategy for development of recombinant antitoxin therapeutics tested in a mouse botulism model. PLoS ONE. 2012;7:e29941. doi: 10.1371/journal.pone.0029941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh S, Kouguchi H, Sagane Y, Suzuki T, Hasegawa K, Watanabe T, Ohyama T. Complete subunit structure of the Clostridium botulinum type D toxin complex via intermediate assembly with nontoxic components. Biochemistry. 2003;42:10991–10997. doi: 10.1021/bi034996c. [DOI] [PubMed] [Google Scholar]
- Muyldermans S, Cambillau C, Wyns L. Recognition of antigens by single-domain antibody fragments: the superfluous luxury of paired domains. Trends Biochem Sci. 2001;26:230–235. doi: 10.1016/s0968-0004(01)01790-x. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Kotani M, Tonozuka T, Ide A, Oguma K, Nishikawa A. Crystal structure of the HA3 subcomponent of Clostridium botulinum type C progenitor toxin. J Mol Biol. 2009;385:1193–1206. doi: 10.1016/j.jmb.2008.11.039. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Takada N, Tonozuka T, Sakano Y, Oguma K, Nishikawa A. Binding properties of Clostridium botulinum type C progenitor toxin to mucins. Biochim Biophys Acta. 2007;1770:551–555. doi: 10.1016/j.bbagen.2006.11.006. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Tonozuka T, Ide A, Yuzawa T, Oguma K, Nishikawa A. Sugar-binding sites of the HA1 subcomponent of Clostridium botulinum type C progenitor toxin. J Mol Biol. 2008;376:854–867. doi: 10.1016/j.jmb.2007.12.031. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Tonozuka T, Ito S, Takeda Y, Sato R, Matsuo I, Ito Y, Oguma K, Nishikawa A. Molecular diversity of the two sugar-binding sites of the beta-trefoil lectin HA33/C (HA1) from Clostridium botulinum type C neurotoxin. Arch Biochem Biophys. 2011;512:69–77. doi: 10.1016/j.abb.2011.05.012. [DOI] [PubMed] [Google Scholar]
- Niwa K, Koyama K, Inoue S, Suzuki T, Hasegawa K, Watanabe T, Ikeda T, Ohyama T. Role of nontoxic components of serotype D botulinum toxin complex in permeation through a Caco-2 cell monolayer, a model for intestinal epithelium. FEMS Immunol Med Microbiol. 2007;49:346–352. doi: 10.1111/j.1574-695X.2006.00205.x. [DOI] [PubMed] [Google Scholar]
- Notermans S, Dufrenne J, Kozaki S. Experimental botulism in Pekin ducks. Avian Dis. 1980;24:658–664. [PubMed] [Google Scholar]
- Ohishi I, Sugii S, Sakaguchi G. Oral toxicities of Clostridium botulinum toxins in response to molecular size. Infect Immun. 1977;16:107–109. doi: 10.1128/iai.16.1.107-109.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace CN, Grimsley GR, Scholtz JM. Protein ionizable groups: pK values and their contribution to protein stability and solubility. J Biol Chem. 2009;284:13285–13289. doi: 10.1074/jbc.R800080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez JC, Groisman EA. Acid pH activation of the PmrA/PmrB two-component regulatory system of Salmonella enterica. Mol Microbiol. 2007;63:283–293. doi: 10.1111/j.1365-2958.2006.05512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rummel A, Hafner K, Mahrhold S, Darashchonak N, Holt M, Jahn R, Beermann S, Karnath T, Bigalke H, Binz T. Botulinum neurotoxins C, E and F bind gangliosides via a conserved binding site prior to stimulation-dependent uptake with botulinum neurotoxin F utilising the three isoforms of SV2 as second receptor. J Neurochem. 2009;110:1942–1954. doi: 10.1111/j.1471-4159.2009.06298.x. [DOI] [PubMed] [Google Scholar]
- Rummel A, Mahrhold S, Bigalke H, Binz T. The HCC-domain of botulinum neurotoxins A and B exhibits a singular ganglioside binding site displaying serotype specific carbohydrate interaction. Mol Microbiol. 2004;51:631–643. doi: 10.1046/j.1365-2958.2003.03872.x. [DOI] [PubMed] [Google Scholar]
- Sagane Y, Watanabe T, Kouguchi H, Sunagawa H, Obata S, Oguma K, Ohyama T. Spontaneous nicking in the nontoxic-nonhemagglutinin component of the Clostridium botulinum toxin complex. Biochem Biophys Res Commun. 2002;292:434–440. doi: 10.1006/bbrc.2002.6689. [DOI] [PubMed] [Google Scholar]
- Schiavo G, Benfenati F, Poulain B, Rossetto O, Polverino de Laureto P, DasGupta BR, Montecucco C. Tetanus and botulinum-B neurotoxins block neurotransmitter release by proteolytic cleavage of synaptobrevin. Nature. 1992;359:832–835. doi: 10.1038/359832a0. [DOI] [PubMed] [Google Scholar]
- Schiavo G, Matteoli M, Montecucco C. Neurotoxins affecting neuroexocytosis. Physiol Rev. 2000;80:717–766. doi: 10.1152/physrev.2000.80.2.717. [DOI] [PubMed] [Google Scholar]
- Schiavo G, Shone CC, Bennett MK, Scheller RH, Montecucco C. Botulinum neurotoxin type C cleaves a single Lys-Ala bond within the carboxyl-terminal region of syntaxins. J Biol Chem. 1995;270:10566–10570. doi: 10.1074/jbc.270.18.10566. [DOI] [PubMed] [Google Scholar]
- Shone CC, Hambleton P, Melling J. Inactivation of Clostridium botulinum type A neurotoxin by trypsin and purification of two tryptic fragments. Proteolytic action near the COOH-terminus of the heavy subunit destroys toxin-binding activity. Eur J Biochem. 1985;151:75–82. doi: 10.1111/j.1432-1033.1985.tb09070.x. [DOI] [PubMed] [Google Scholar]
- Stenmark P, Dupuy J, Imamura A, Kiso M, Stevens RC. Crystal structure of botulinum neurotoxin type A in complex with the cell surface co-receptor GT1b-insight into the toxin-neuron interaction. PLoS Pathog. 2008;4:e1000129. doi: 10.1371/journal.ppat.1000129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling PC, Bakhoum SF, Feigl AB, Leroux MR. Convergent evolution of clamp-like binding sites in diverse chaperones. Nat Struct Mol Biol. 2006;13:865–870. doi: 10.1038/nsmb1153. [DOI] [PubMed] [Google Scholar]
- Stroffekova K, Kupert EY, Malinowska DH, Cuppoletti J. Identification of the pH sensor and activation by chemical modification of the ClC-2G Cl- channel. Am J Physiol. 1998;275:C1113–C1123. doi: 10.1152/ajpcell.1998.275.4.C1113. [DOI] [PubMed] [Google Scholar]
- Sugawara Y, Matsumura T, Takegahara Y, Jin Y, Tsukasaki Y, Takeichi M, Fujinaga Y. Botulinum hemagglutinin disrupts the intercellular epithelial barrier by directly binding E-cadherin. J Cell Biol. 2010;189:691–700. doi: 10.1083/jcb.200910119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugii S, Ohishi I, Sakaguchi G. Correlation between oral toxicity and in vitro stability of Clostridium botulinum type A and B toxins of different molecular sizes. Infect Immun. 1977;16:910–914. doi: 10.1128/iai.16.3.910-914.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama H. Clostridium botulinum neurotoxin. Microbiol Rev. 1980;44:419–448. doi: 10.1128/mr.44.3.419-448.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Watanabe T, Mutoh S, Hasegawa K, Kouguchi H, Sagane Y, Fujinaga Y, Oguma K, Ohyama T. Characterization of the interaction between subunits of the botulinum toxin complex produced by serotype D through tryptic susceptibility of the isolated components and complex forms. Microbiology. 2005;151:1475–1483. doi: 10.1099/mic.0.27801-0. [DOI] [PubMed] [Google Scholar]
- Swaminathan S, Eswaramoorthy S. Structural analysis of the catalytic and binding sites of Clostridium botulinum neurotoxin B. Nat Struct Biol. 2000;7:693–699. doi: 10.1038/78005. [DOI] [PubMed] [Google Scholar]
- Tremblay JM, Kuo CL, Abeijon C, Sepulveda J, Oyler G, Hu X, Jin MM, Shoemaker CB. Camelid single domain antibodies (VHHs) as neuronal cell intrabody binding agents and inhibitors of Clostridium botulinum neurotoxin (BoNT) proteases. Toxicon. 2010;56:990–998. doi: 10.1016/j.toxicon.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidyanathan VV, Yoshino K, Jahnz M, Dorries C, Bade S, Nauenburg S, Niemann H, Binz T. Proteolysis of SNAP-25 isoforms by botulinum neurotoxin types A, C, and E: domains and amino acid residues controlling the formation of enzyme-substrate complexes and cleavage. J Neurochem. 1999;72:327–337. doi: 10.1046/j.1471-4159.1999.0720327.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Meng J, Lawrence GW, Zurawski TH, Sasse A, Bodeker MO, Gilmore MA, Fernandez-Salas E, Francis J, Steward LE, Aoki KR, Dolly JO. Novel chimeras of botulinum neurotoxins A and E unveil contributions from the binding, translocation, and protease domains to their functional characteristics. J Biol Chem. 2008;283:16993–17002. doi: 10.1074/jbc.M710442200. [DOI] [PubMed] [Google Scholar]
- Weeks DL, Sachs G. Sites of pH regulation of the urea channel of Helicobacter pylori. Mol Microbiol. 2001;40:1249–1259. doi: 10.1046/j.1365-2958.2001.02466.x. [DOI] [PubMed] [Google Scholar]
- Williamson LC, Halpern JL, Montecucco C, Brown JE, Neale EA. Clostridial neurotoxins and substrate proteolysis in intact neurons: botulinum neurotoxin C acts on synaptosomal-associated protein of 25 kDa. J Biol Chem. 1996;271:7694–7699. doi: 10.1074/jbc.271.13.7694. [DOI] [PubMed] [Google Scholar]
- Yamashita S, Yoshida H, Uchiyama N, Nakakita Y, Nakakita SI, Tonozuka T, Oguma K, Nishikawa A, Kamitori S. Carbohydrate recognition mechanism of HA70. from Clostridium botulinum deduced from X-ray structures in complexes with sialylated oligosaccharides. FEBS Let. 2012 doi: 10.1016/j.febslet.2012.05.055. [DOI] [PubMed] [Google Scholar]