Abstract
Understanding and modulating the cellular response to implanted biomaterials is crucial for the field of tissue engineering and regenerative medicine. Since cells typically reside in an extracellular matrix containing nanoscale architecture, identifying synthetic nanostructures that induce desirable cellular behaviors could greatly impact the field. Using nanoimprint lithography, nanostructured patterns were generated on thin film polymeric materials. The ability of these surfaces to influence protein adsorption, fibroblast proliferation and morphology, and fibrotic markers was investigated. Nanostructured features with aspect ratios greater than five allowed for less protein adsorption, resulting in decreased fibroblast proliferation and rounded cellular morphology. These nanofeatures also induced significantly lower gene expression of collagen 1α2, collagen 3α1, and growth factors such as connective tissue growth factor, integrin linked kinase, transforming growth factor β1 (TGF-β1), and epidermal growth factor, key factors associated with a fibrotic response. The results demonstrate that select nanostructured surfaces could be used to modulate the fibrotic behavior in cells and have the potential to be used as antifibrotic architecture for medical implants or tissue engineering scaffolds.
Introduction
Implant-induced fibrosis has generated much attention in the medical community and in the field of tissue engineering. Almost all soft tissue implants undergo fibrotic encapsulation and eventual loss of functional tissue in the vicinity surrounding the implant. The fibroblast is a specific cell that synthesizes and deposits the extracellular matrix (ECM), forming the structural network for soft tissue. Although it plays a critical role in wound healing, the over-proliferation of fibroblasts and the subsequent overproduction of ECM proteins have been implicated in fibrosis. It is well known that implant fibrosis and fibrotic encapsulation can often contribute to medical device failures, ranging from breast implant contracture to biosensor inactivation.1–3 Fibrosis has also been implicated in postsurgical adhesions, contributing to the failure of gastrointestinal, gynecological, and sinus surgeries.4
To this end, new biomaterial interfaces that foster an antifibrotic environment must be developed. Previously, there has been considerable work on chemistry-based approaches for decreasing fibrosis. For example, Risbud et al. reported how biocompatible hydrogels composed of chitosan-pyrrolidone arrest capsular fibroblast growth.5 Other materials such as alginate, hyaluronic acid, and derivatives of chitin have been demonstrated to mimic fetal wound healing by selectively inhibiting fibroblast growth.6,7 Recently, however, it has been established that cells are capable of responding to nanotopographical cues found in their microenvironment. The ECM is composed of complex architectural features at the nanoscale, including pores, fibers, ridges, and protein band periodicities of ∼60 nm.8 Nanoscale features, being at the subcellular size scale, have the ability to influence cellular behaviors such as morphology, proliferation, and differentiation.9–11 Therefore, nanotopography offers the opportunity to perturb a wide range of cellular responses. A better understanding of the cell–material interface on the nanoscale enables the exploration of a spectrum of interactions that are crucial to designing advanced medical devices and implants. Herein we report how nanostructured biomaterials can be used to generate an antifibrotic environment for cells.
To investigate how nanotopography influences cellular behavior, nanostructures must be fabricated with a high level of repeatability and precision. Current advancements in nano- and microtechnology offer new possibilities of probing cell–material interactions to better understand biological functions.12,13 Ranging from microcontact printing to photolithography, there are various techniques for systematically controlling topographical features. One such fabrication technique to achieve nanofeatures is nanoimprint lithography (NIL). This technique is a stamping process capable of generating nanometer length patterns as small as 10 nm.14,15 In contrast to conventional photolithography, nanofeatures are generated by the mechanical deformation of a thermoplastic material using a mold with nanofeatures. Molds are fabricated using electron beam lithography to overcome the diffraction limit of light and to produce features on the nanoscale.
The design of the nanostructured substrates was inspired by the ECM that surrounds soft tissue implants. Mimicking the ECM involved rationally designing structures that were similar to collagen, the most abundant ECM protein in the body. Collagen consists of staggered arrays of tropocollagen molecules that bind together to form fibrils. These collagen fibrils have diameters that range from 100 to 500 nm and lengths up to the millimeter length scale.16 Despite significant research efforts over the past two decades, the effect of collagen fibril geometry on wound healing and fibrosis remains largely unknown. Herein we designed surfaces with arrays of nanopillars that have diameters ranging from 200 to 800 nm to capture the full breadth of collagen fibril diameters that are found in nature. Moreover, collagen and many other components of the ECM are hierarchical structures from the molecular length scale to the macroscopic length scale. Therefore, one of the substrates was designed to mimic the complex hierarchical structure of collagen by containing micron-sized features with two levels of nanosized features. All four of the nanostructured substrates were designed to emulate the in vivo cross section of severed collagen fibers that results from soft tissue injury as a two-dimensional array of nanopillars. To this end, we utilized NIL to generate various nanostructured surfaces in two commonly used polymeric materials as a platform to examine how nanotopographical cues influence fibroblast behavior. Since cellular attachment and proliferation are mediated by protein adsorption to the underlying substrate, we hypothesized that nanostructured topography would influence protein adsorption, thereby affecting the fibrotic response.
Materials and Methods
Fabrication of nanostructured surfaces
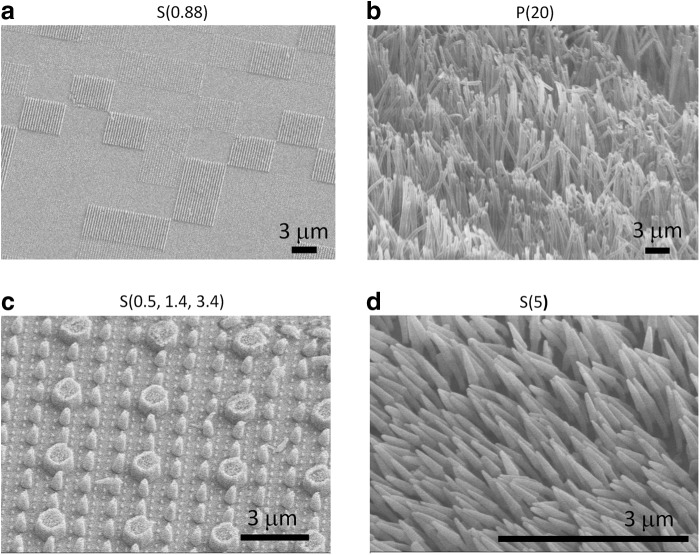
Fabrication of a mold for the nanostructured thin films was performed using electron beam lithography (JEOL JBX-9300FS EBL; Jeol Ltd., Tokyo, Japan). A nanostructured pattern was generated on a polymethylmethacrylate (PMMA) resist that had been spin casted onto an underlying silicon substrate. After developing away the PMMA resist, anisotropic reactive ion etching was employed to precisely etch the underlying silicon substrate, resulting in the nanofeatured molds shown in Figure 1. The mold was stamped into FDA-approved polypropylene or polystyrene thin films (25.4 mm; Premier Lab Supply, Inc., Port St. Lucie, FL). Briefly, the polystyrene or polypropylene film was placed in contact with the silicon mold and exposed to T=170°C and P=2 GPa using an Obducat 6-inch nanoimprint lithography system (Obducat, Lund, Sweden). Afterward, the mold was removed to reveal well-defined nanofeatures on the thin film.
FIG. 1.
Nanoimprint lithography was employed to imprint the nanofeatures from nanofeatured molds into polystyrene or polypropylene thin films. Scanning electron microscopy (SEM) images reveal (a) a polystyrene thin film with features having an aspect ratio of 0.88, (b) a polypropylene thin film with features having an aspect ratio of 20, (c) a polystyrene thin film with hierarchical nanofeatures having aspect ratios of 0.5, 1.4, and 3.4, and (d) a polystyrene thin film with features having an aspect ratio of 5.
Contact angle measurements
Water contact angle measurements were performed with a goniometer to measure the wettability of the nanostructured surfaces. The nanostructured surfaces were thoroughly cleaned in a 99% isopropyl alcohol bath and then fully dried. A single drop of deionized (DI) water was placed on each nanostructured surface. The contact angle of the water droplet is the angle at which the liquid–vapor interface meets the solid–liquid interface and is the resultant between the adhesive and cohesive thermodynamic forces. As a droplet spreads over a surface, the contact angle decreases. Therefore, the contact angle is an inverse measure of the wettability of a surface. A contact angle greater than 90 indicates that the wetting of the surface is thermodynamically unfavorable, so the fluid will minimize the surface contact and form a liquid droplet. Using this analysis, the contact angles of DI water were measured.
Protein adsorption to nanostructured surfaces
Nanostructured surfaces were cut into circular shapes with a 6-mm diameter biopsy punch and glued to the bottom of a 24-well plate (VWR, Brisbane, CA) with a silicone medical adhesive glue (Silastic Medical Adhesive Silicone Type A; Dow Corning, Midland, MI). After the glue dried, the thin films were thoroughly cleaned with 70% ethanol, followed by DI water overnight, and dried with compressed air. Solutions of FITC-IgG (Sigma-Aldrich, St. Louis, MO), FITC-BSA (Sigma-Aldrich), and fibrinogen-Alexa Fluor 568 (Life Technologies, Grand Island, NY), (each at a concentration of 0.1 mg/mL) were incubated on the nanostructured surface for 2 h. The thin films were rinsed with DI water and sonicated (Ultrasonic Bath Sonicator Model 75T; VWR) for 10 min at the default frequency. They were then mounted for visualization with a spinning disk confocal microscope (Ti-E Microscope; Nikon Instruments, Inc., Melville, NY, with a CSU-22 spinning disk confocal; Yokogawa Electric Corporation, Newnan, GA).
Culture of cells on topographic nanostructured surfaces
NIH 3T3 mouse fibroblasts were cultured in the DMEM high glucose (Gibco; Life Technologies) supplemented with 10% fetal bovine serum (HyClone, Logan, UT) and 100 IU/mL penicillin–streptomycin (Sigma-Aldrich). The cultures were incubated in an atmosphere containing 5% CO2 and 90% relative humidity at 37°C. Growth media were exchanged every third day. The cells were subcultured at 90% confluency by trypsinization with 0.25% trypsin-EDTA. Cells from passage numbers between 160 and 170 were used for the experiments.
Cyquant proliferation assay
Six millimeter biopsy punches were used to cut the nanostructured thin films into a circular shape. The thin films were then glued to the bottom of the wells of 96-well plates (VWR) using the same glue as previously described. The films were disinfected with 70% ethanol overnight and air-dried in a CII safety cabinet. Fibroblasts were seeded at a density of 2000 cells/well and cultured over a 4-day time period. At 24, 48, 72, and 96 h, the media was aspirated and the plate was frozen at −80°C overnight. A cell pellet of 1 million cells was used for quantifying the number of cells that proliferated on the substrates. The cell pellet was serially diluted to generate a standard curve that ranged from 50 to 50,000 cells. The green fluorescent Cyquant dye was used to detect the number of cells for each time point (Cyquant Cell Proliferation Assays; Life Technologies). The signal was read on a fluorometer (SpectraMAX 190; Molecular Devices, Sunnyvale, CA) at a wavelength of 495 nm.
Immunofluorescence staining of F-actin and cell area quantification
Cells were fixed in 3.7% paraformaldehyde for 15 min and permeabilized with Triton X (0.1%) for 20 min. The F-actin was stained with phalloidin conjugated to Alexa Fluor 568 and diluted 1:40 in phosphate-buffered saline (PBS) for 30 min at 25°C (Life Technologies). The nucleus was stained with Hoechst 33258 at a dilution of 1:2000 for 30 min (Life Technologies). Samples were washed three times for 5 min in 1 mL PBS and mounted on a glass cover slip with hard mounting media (Vectashield; Vector Laboratories, Burlingame, CA) before inspection with spinning disk confocal microscopy. Images were analyzed with ImageJ software that was downloaded from the National Institutes of Health (Image J, http://rsb.info.nih.gov/ij/). The images were imported into the software as an 8-bit grayscale sequence and then smoothed with brightness and contrast functions to enhance the features relative to the background. The threshold function was applied to create a binary image for particle analysis. ImageJ software detected the cell based on the binary image and calculated parameters such as circularity, cell area, perimeter, and aspect ratio. The data obtained from ImageJ were imported to Microsoft Excel for calculating means and standard deviations for circularity (0–1). Circularity was calculated using the equation, 4π(area/perimeter2). A circularity value of 1.0 indicates a perfect circle and a value that approaches 0.0 is an increasingly elongated polygon, suggesting a more spread out cellular morphology.
Scanning electron microscopy
Fibroblasts were seeded onto the nanostructured thin films. On day 2, they were fixed with 3% glutaraldehyde in 0.1 M sodium cocadylate (pH 7.4), containing 0.1 M sucrose. After 45 min of incubation, the cells were rinsed with the cacodylate-sucrose buffer for 10 min. Next, the samples were dehydrated by gently adding solutions of ethanol in a graded series of concentrations (35%, 50%, 75%, 95%, 100%, and 100%) for 10 min each. The last step was to replace the 100% ethanol solution with HMDS, and then air-dried in the fume hood for 30 min. The samples were then sputter coated with 200 angstroms of gold–palladium and then imaged on a Hitachi S-5000 scanning electron microscope.
Quantitative polymerase chain reaction methods
The following films were cut into 6-mm circular shapes and glued as previously described: the homogeneous low aspect ratio substrate S(0.5, 1.4, 3.4), the highest aspect ratio substrate P(20), P(flat), S(flat), or tissue culture polystyrene (TCPS). 3T3 cells were seeded at a density of 5000 cells/cm2 in a 96-well tissue culture plate. After 72 h, cells were lysed, reverse transcription was performed, and quantitative polymerase chain reaction (qPCR) (StepONEPlus; Applied Biosystems, Life Technologies) was performed using the Fast SYBR Green cells to CT kit in accordance with the manufacturer's instructions (Life Technologies). The experiment was carried out in triplicate (n=3) and the evaluation of mRNA expression by qPCR was also run in triplicate. The expression of GAPDH, collagen type 1 alpha 2, collagen type 3 alpha 1, connective tissue growth factor (CTGF), epidermal growth factor (EGF), integrin linked kinase (ILK), and transforming growth factor β1 (TGF-β1) were analyzed. The real-time polymerase chain reaction (PCR) results were analyzed using the Livak method and normalized to GAPDH transcript levels.17 The primer sequences that were used are as follows: the GAPDH forward was 5′-CTC TCT GCT CCT CCT GTT CG-3′ and GAPDH reverse was 5′-GCC CAA TAC GAC CAA ATC C-3′. The collagen type 1 alpha 2 primer sequences in the forward and reverse directions were AAG GGT GCT ACT GGA CTC CC and TTG TTA CCG GAT TCT CCT TTG G, respectively. The collagen type 3 alpha 1 primer sequences in the forward and reverse directions were CTGTAACATGGAAACTGGGGAAA and CCATAGCTGAACTGAAAACCACC, respectively. The ILK primer sequences in the forward and reverse directions were CACGGCAATGTGCCACTTC and GCTCACAAGAGCCCCGTTAG, respectively. The EGF primer sequences in the forward and reverse directions were AGCATCTCTCGGATTGACCCA and CCTGTCCCGTTAAGGAAAACTCT, respectively. The TGF-β1 primer sequences in the forward and reverse directions were CTTCAATACGTCAGACATTCGGG and GTAACGCCAGGAATTGTTGCTA, respectively. And finally, the CTGF primer sequences in the forward and reverse directions were GGGCCTCTTCTGCGATTTC and ATCCAGGCAAGTGCATTGGTA, respectively.
Statistical analysis
Data are reported as mean value±standard deviation. Multiple comparisons were analyzed with a one-way ANOVA test followed by the Holm-Sidak t-test. p-values of less than 0.05 were considered statistically significant. For the proliferation data, each group had an n=6, and for the gene expression data, each group had an n=3 with a technical replicate of n=3.
Results
Nanofabrication
NIL was utilized to generate four unique nanostructured topographies as shown in Figure 1. The different substrates consisted of a polypropylene film with high aspect ratio nanopillars [height (H)=16 μm, diameter (D)=800 nm, aspect ratio (AR)=20, surface roughness=850 nm]; a polystyrene film with high aspect ratio nanopillars (H=1 μm, D=200 nm, AR=5, surface roughness=73 nm); a polystyrene film with a patchwork pattern of nanostructures (H=175 nm, D=200 nm, AR=0.88, surface roughness=9 nm); and finally, a polystyrene film with three different nanopillars combined in a hierarchical pattern with the largest pillar diameter of 1 μm having an average height of 520 nm, the medium pillar diameter of 400 nm having an average height of 560 nm, and the smallest pillar diameter of 200 nm having an average height of 680 nm. The ARs for the three structures were 0.5, 1.4, and 3.4, and the average surface roughness was 145 nm. All films will be referred to as either P or S for polypropylene or polystyrene, respectively, followed by the nanostructure aspect ratio in parentheses [e.g., “P(20)” encodes for the polypropylene film with a nanostructure aspect ratio of 20].
Contact angle measurements and protein adsorption
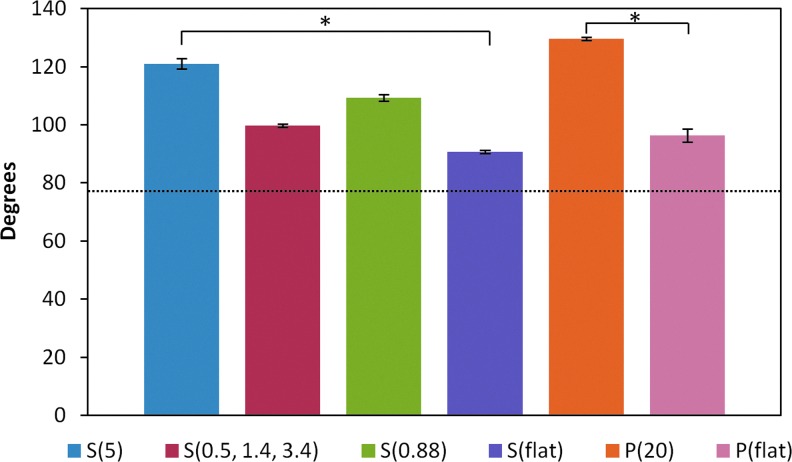
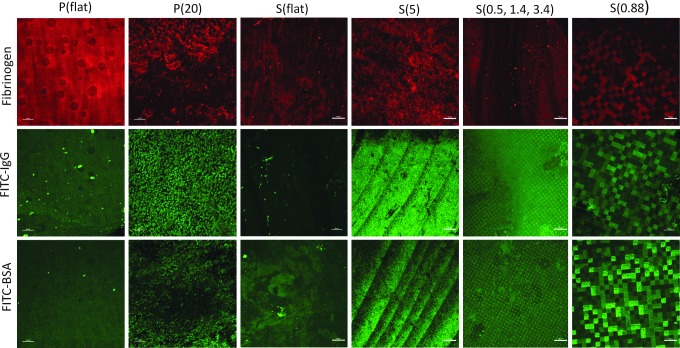
Characterization of hydrophobicity was performed through static water contact angle measurements. Figure 2 shows the contact angles in degrees for each nanostructured substrate. P(20) resulted in a higher contact angle compared with the P(flat) control. Similarly, the polystyrene nanostructured S(5), S(0.5, 1.4, 3.4), and S(0.88) resulted in higher contact angles than the S(flat) control. Next, we tested the hypothesis that higher contact angles correlated with elevated protein adsorption levels. To test this hypothesis, we incubated the nanostructured substrates in solutions composed of proteins that are commonly found in blood to mimic the in vivo environment of blood-contacting medical device implants and biosensors. The blood protein solutions were composed of either FITC-BSA, FITC-IgG, or Alexa Fluor 568-Fibrinogen.18 Next, the nanostructured substrates were imaged with confocal microscopy as shown in Figure 3. The nanotopography dramatically affects the way proteins adsorb to the various surfaces. For example, the higher aspect ratio features [S(5) and P(20)] seem to adsorb FITC-BSA, FITC-IgG, and fibrinogen only to the tips of the nanopillars. Interestingly, FITC-BSA and FITC-IgG adsorb to the tops of the micron-sized features as well as to the nanopillar tips of the hierarchical S(0.5, 1.4, 3.4) nanostructured thin film. However, fibrinogen appears to adsorb nonpreferentially on the S(0.5, 1.4, 2.4) surface. Furthermore, the heterogeneously patterned S(0.88) surface induced preferential protein adsorption in a quilt-like pattern for all three proteins.
FIG. 2.
The water contact angle measurements demonstrate that the presence of nanofeatures on the polystyrene and polypropylene thin films increases the contact angles. This result translates into a more globally hydrophobic surface. *p<0.05. Color images available online at www.liebertpub.com/tea
FIG. 3.
Alexa Fluor-Fibrinogen, FITC-IgG, and FITC-BSA were physisorbed onto the nanostructured and control surfaces. The proteins adsorbed to the surfaces in different patterns. Color images available online at www.liebertpub.com/tea
Fibroblast proliferation
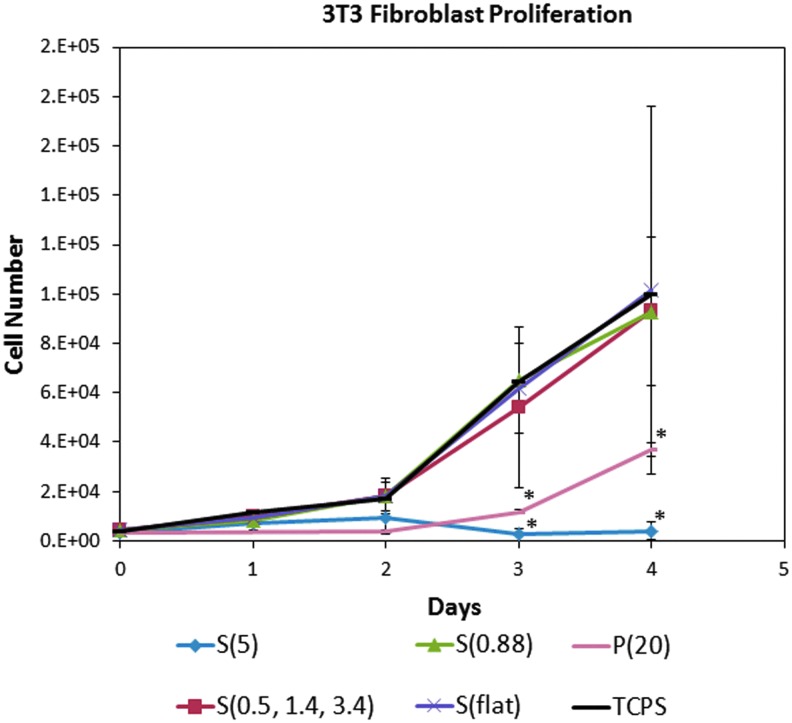
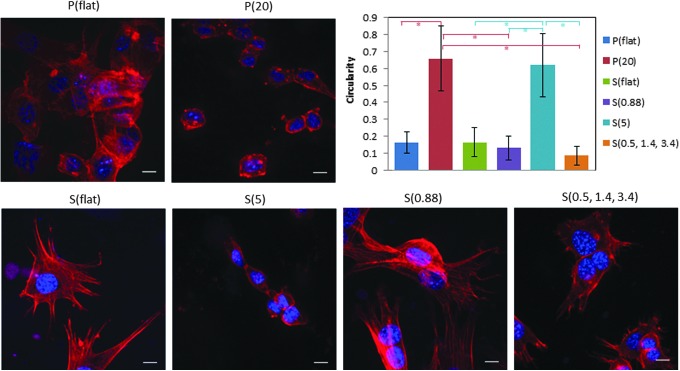
Fibroblasts were cultured on the nanostructured substrates for 4 days and measured with the Cyquant cell proliferation assay at different time points to generate a cell proliferation curve. Figure 4 shows that fibroblasts grown on P(20) and S(5), the substrates with the highest aspect ratio features, seem to grow at the slowest rate. Additionally, observation of F-actin in the immunofluorescence staining showed different cell morphologies on the nanostructured surfaces compared to flat controls. As presented in Figure 5, the fibroblasts were well spread with many stress fibers on the control flat substrates. However, the fibroblasts grown on P(20) and S(5) substrates were rounder and had fewer stress fibers. Quantitative analysis of cell morphology as measured by circularity confirmed what was observed qualitatively. Fibroblasts grown on the high aspect ratio P(20) and S(5) surfaces have statistically significantly (p<0.001) higher circularity values than those of the cells grown on low aspect ratio substrates, P(flat), S(flat), S(0.88), and S(0.5, 1.4, 3.4).
FIG. 4.
3T3 fibroblast proliferation was quantified with the Cyquant assay. Over 4 days, the fibroblasts grew significantly more slowly on the P(20) and the S(5) nanostructured surfaces compared to the other surfaces. Color images available online at www.liebertpub.com/tea
FIG. 5.
The fibroblast morphology indicates that the higher aspect ratio nanofeatures induce a rounded cellular morphology compared to the lower aspect ratio nanofeatures and the flat controls. The scale bars are 10 μm. The circularity values for the fibroblasts grown on high aspect ratio nanostructured surfaces [P(20) and S(5)] are significantly higher than those of the low aspect ratio nanostructured surfaces [P(flat), S(flat), S(0.88), S(0.5, 1.4, 3.4)]. The * symbol indicates p<0.001. This result indicates that the fibroblasts are unable to attach and spread on the high aspect ratio nanofeatures. Color images available online at www.liebertpub.com/tea
Cell morphology
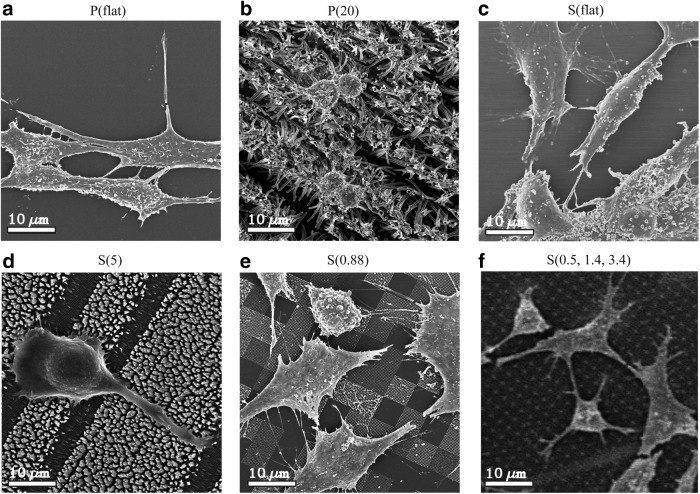
Scanning electron microscopy (SEM) was employed to directly image the interactions of the filopodial projections with the nanofeatures. Fibroblasts grown on flat polypropylene (Fig. 6a) showed a typical, spread fibroblast shape. In contrast, fibroblasts grown on P(20) films (Fig. 6b) were more rounded. As seen with flat polypropylene, fibroblasts grown on flat polystyrene (Fig. 6c) were spread out with many filopodia. Fibroblasts on S(5) films (Fig. 6d) were more rounded like those on the P(20) film. In contrast, fibroblasts grown on the low aspect ratio polystyrene films, S(0.88) and S(0.5, 1.4, 3.4) showed a more typical fibroblast morphology (Fig. 6e, f). Interestingly, fibroblasts growing on the S(0.88) substrates preferentially grow on certain nanopatterned features. The fibroblasts exhibit star-like filopodial projections that adhere to the nanopatterned square patches. On S(0.5, 1.4, 3.4) films, fibroblasts seem to preferentially extend filopodia to taller structures (Fig. 1c).
FIG. 6.
SEM images demonstrate how the fibroblasts are interacting directly with the nanostructures. (a) Fibroblasts grown on P(flat). (b) Fibroblasts grown on P(20). (c) Fibroblasts grown on S(flat). (d) Fibroblasts grown on S(5). (e) Fibroblasts grown on S(0.88). (f) Fibroblasts grown on S(0.5, 1.4, 3.4). Similarly to Figure 5, the P(20) and S(5) nanostructured surfaces induce a rounded morphology, indicating an antifibrotic environment. The scale bars are 10 μm.
Gene expression
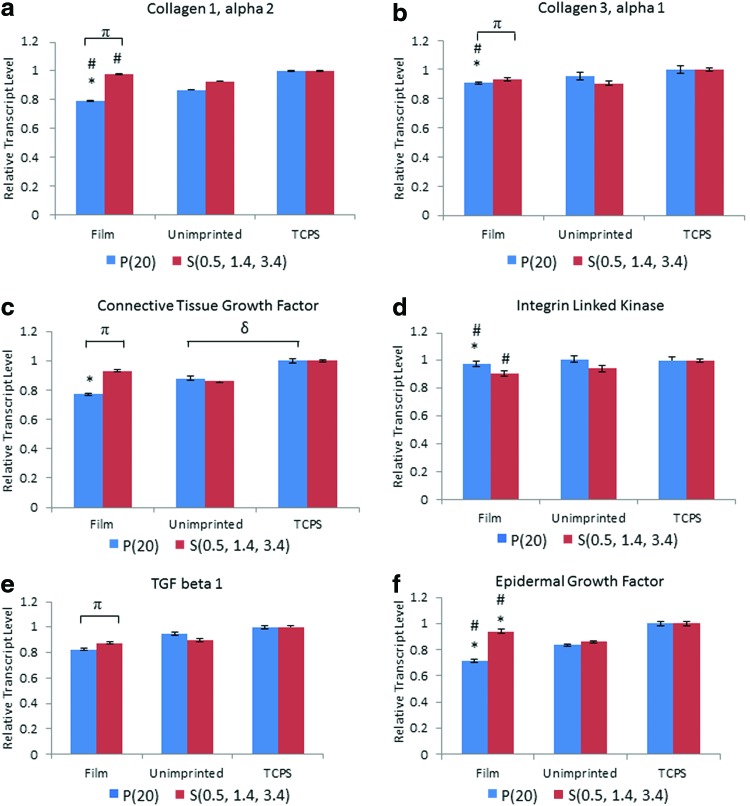
Expression of the following six genes was measured by qPCR: collagen 1α2 (COL1α2), collagen 3α1 (COL3α1), CTGF, ILK, transforming growth factor β1 (TGF-β1), and EGF. The results indicate that P(20) elicits a significant down-regulation in fibrotic markers as shown in Figure 7. For example, the fibroblast gene expression of both COL1α2 (Fig. 7a) and COL3α1 (Fig. 7b) on P(20) are significantly downregulated compared to the lower aspect ratio S(0.5, 1.4, 3.4), unimprinted control, and TCPS control, suggesting an overall decrease in collagen production with high aspect ratio nanostructures. Similarly, CTGF, TGF-β1, and EGF growth factors responsible for the proliferative behavior of fibroblasts are substantially downregulated in the presence of the P(20) nanostructured surface.
FIG. 7.
Gene expression studies for (a) collagen 1α2, (b) collagen 3α1, (c) connective tissue growth factor, (d) integrin linked kinase, (e) transforming growth factor β1, and (f) epidermal growth factor are displayed for P(20) and S(0.5, 1.4, 3.4). The * refers to a significant difference of p<0.05 between the film and the tissue culture polystyrene (TCPS) control, while the # corresponds to a significant difference of p<0.05 between the film and the unimprinted control group. π and δ refer to a significant difference of p<0.05 between P(20) and S(0.5, 1.4, 3.4) films and unimprinted control and TCPS control, respectively. Color images available online at www.liebertpub.com/tea
Discussion
The ECM is a hierarchical environment containing microscale to nanoscale features ranging from microns to tens of nanometers in length. Fibroblasts in a wound site interact with clusters of severed collagen fibrils in an inhomogeneous pattern, and they may perceive this environment as surfaces containing nano- and microgrooves, ridges, and pillars with variable densities and roughnesses. A better understanding of the role that nanotopographical cues have on the fibrotic phenotype is important to the development of implanted materials.
In this study, we demonstrated how nanostructure-mediated cues elicit a dramatic effect on protein adsorption, cell shape, proliferation, and gene expression, with potential applications for decreasing fibrosis in vivo. We tested four different nanostructured surfaces and discovered that surfaces with higher aspect ratio features above five fostered an antifibrotic microenvironment. This result may be directly related to the surface structure or indirectly related to the way the surface topography influences ECM protein recruitment. The longer and thinner nanofeatures may resist protein adsorption due to steric hindrance and entropic repulsive forces.19 Although others have observed differential cellular responses to nanotopography, most research has focused on cell–surface interactions and have not investigated the intermediate effect that nanostructures have on protein adsorption.9,10,20–22 Unlike microtopography, whereby the cells are mechanically constrained between features, nanotopography may work by influencing how proteins are presented to cells. The adsorption of proteins to biomaterial surfaces is a dynamic process whereby proteins bind, rearrange, and detach. This process plays a crucial role in influencing cell growth, proliferation, and the overall incorporation of the biomaterial into the body. Depending on the shape of the nanofeature, the new surface energy may result in a more hydrophilic or hydrophobic environment, which can lead to protein resistance or even protein denaturation in the extreme case. The results from Figure 2 indicate that regardless of material chemistry, nanotopography increases the contact angles of polystyrene and polypropylene, and therefore increases the hydrophobicity and surface energy of the thin films. P(20) and S(5) both have higher contact angles compared to the other surfaces. Perhaps, this superhydrophobic effect from the nanostructures may induce conformational denaturation of the adsorbed proteins, ultimately affecting the cell–materials interactions.23 Without suitable protein presentation on the surface, the fibroblasts are less capable of attaching and maintaining healthy phenotypes.24
As we can see from Figure 3, the three blood proteins have drastically different adsorption patterns on the different surfaces. This is a striking result because it suggests that the nanostructure of the underlying surface influences the way proteins adsorb, which could be relevant to the surface properties of medical device implant materials. Interestingly, fibrinogen appears to adsorb to the same surface in a different way than FITC-IgG and FITC-BSA. For many of the surfaces, there are patches of no protein adsorption, particularly for P(20) and S(5). Fibrinogen is a much larger protein than IgG and BSA with a Stokes radius of 10.7 nm, twice the size of IgG (5.5 nm) and three times the size of BSA (3.4 nm).25 Therefore, it may be difficult for the relatively larger fibrinogen molecules to conform to surfaces with high roughness such as in P(20) and S(5).
The various protein adsorption patterns strongly correlate to changes in phenotypic behaviors. For example, the nanotopographies of the surfaces may contribute to the different fibroblast morphologies that were observed by immunofluorescence and SEM. Interestingly, fibroblasts grown on P(20) and S(5) exhibited more rounded morphologies as quantified by circularity in Figure 5. In contrast, cells grown on S(0.5, 1.4, 3.4), S(0.88), and the flat control surfaces displayed well-spread morphologies. Numerous studies in the literature have described a common trend of decreased cellular adhesion and spreading with an increase in nanofeature height.26,27 This phenomenon is thought to be associated with perturbed focal adhesion formation, as well as an inhibition of protein adsorption. In our studies, we observed differential protein adsorption patterns on the various surfaces. The nanostructures that induced rounded cellular morphologies also had protein adsorption localized only to the tips of the nanopillars. This would suggest that the cellular filopodia could not find suitable sites of attachment due to the restriction of protein adsorption to the tips. Furthermore, this would indicate that the higher aspect ratio nanofeatures restrict integrin adhesion formation to the apexes of the nanopillars.
In this study, there are two mechanisms of action on the nanoscale that contribute to the fibroblast response to nanotopography: protein adsorption and substrate mechanics. In Figure 6, it is apparent that the low aspect ratio nanofeatures promote more interaction with the cells. S(0.5, 1.4, 3.4) has relatively low aspect ratio features, and we observed filopodia projections extending to the tips of the nanopillars. Filopodia tips are ∼100 nm and are the main sensory tools for spatial information.28 Since the S(0.5, 1.4, 3.4) nanostructures have larger diameters that range from 200 nm to 1 μm, the fibroblasts were able to form intact focal adhesions to this surface.7 Similarly, S(0.88) is another nanostructured surface with low aspect ratio features. Fibroblasts on this surface were well spread, and each filopodia appears to terminate on a region that has a high protein density. In contrast, it is apparent that the fibroblasts on P(20) and S(5) are pinned to the top of the nanopillars and unable to spread. It is well established that fibroblasts proliferate and form organized F-actin and stress fibers on higher moduli substrates.29,30 To the perspective of a fibroblast, a low aspect ratio feature is locally stiffer than a high aspect ratio feature due to the higher critical buckling load of a shorter and wider feature. These observations are consistent with the well-established fact that anchorage-dependent cells must attach and spread on an underlying substrate with a sufficient mechanical stiffness. Additionally, our study suggests that protein adsorption patterns to nanofeatures contribute to the cellular spreading as well.
Since focal adhesions link the cytoskeleton to the ECM through integrin clustering, it was hypothesized that the ability of the fibroblast to adhere and spread will alter cell signaling and mechanotransduction processes.32 A potential explanation for the observed nanotopographical effects on fibroblast behavior could be the model of tensional integrity (tensegrity) developed by Ingber et al. in 1993.31 In this model, integrins act as mechanoreceptors that transmit mechanical signals through the tensionally integrated cytoskeleton. The higher aspect ratio nanostructured substrates such as P(20) and P(5) have higher surface roughnesses, which may correspond to fewer cellular contact points for integrin engagement. Therefore, nanostructured surfaces that induce fewer and weaker focal adhesions may be associated with lower ECM production, proliferation, and gene expression since the reduced tension will lead to lower signal transduction to the nucleus.25 Consistent with this hypothesis, P(20), the nanostructure with the highest aspect ratio features, induced lower gene expression levels for COL1α2, COL3α1, CTGF, TGF-β1, and EGF compared to the lower aspect ratio substrate, S(0.5, 1.4, 3.4). This decreased gene expression is in agreement with the significant reduction in cell growth observed on the same surface. COL1α2 and COL3α1 are significant components of scar tissue, so a downregulation in these genes suggests that this higher aspect ratio nanostructured surface could act as an antifibrotic interface.
Taken together, these results demonstrate that the aspect ratio and organization of nanotopographical cues influence fibroblast proliferation, cellular morphology, and expression of key fibrotic markers compared to flat control surfaces of the same material chemistry. The observed phenotypic effects are most likely due to altered protein adsorption to the nanofeatures. The proposed mechanism is through decreased focal adhesion formation and cell spreading. This work has broad applications for decreased scar formation around implanted biomaterials.
Acknowledgments
We would like to thank Professor Ronald Fearing and the University of California, San Francisco Nikon center for their valuable insight and advice, Devin Brown and Nicole Devlin at the Georgia Tech Microelectronics Research Center for generating the NIL molds, Sunland Biotechnology for ELISA testing, and American Preclinical Services for the animal studies. Funding for this work was kindly provided by the Kimberly-Clark Corporation and the National Institutes of Health (NIH).
Disclosure Statement
The authors confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
References
- 1.Chegini N.Peritoneal molecular environment, adhesion formation and clinical implication. Front Biosci 7,e91, 2002 [DOI] [PubMed] [Google Scholar]
- 2.Hunter L.W., Lieske J.C., Tran N.V., and Miller V.M.The association of matrix Gla protein isomers with calcification in capsules surrounding silicone breast implants. Biomaterials 32,8364, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Giurgiutiu V., et al. Electromechanical impedance sensor for in vivo monitoring the body reaction to implants. J Invest Surg 17,257, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Schippers E., Tittel A., Ottinger A., and Schumpelick V.Laparoscopy versus laparotomy: comparison of adhesion-formation after bowel resection in a canine model. Dig Surg 15,145, 1998 [DOI] [PubMed] [Google Scholar]
- 5.Risbud M., Hardikar A., and Bhonde R.Growth modulation of fibroblasts by chitosan-polyvinyl pyrrolidone hydrogel: Implications for wound management? J. Biosci 25,25, 2000 [DOI] [PubMed] [Google Scholar]
- 6.Li Y., Rodrigues J., and Tomas H.Injectable and biodegradable hydrogels: gelation, biodegradation and biomedical applications. Chem Soc Rev 41,2193, 2012 [DOI] [PubMed] [Google Scholar]
- 7.Mast B.A., Diegelmann R.F., Krummel T.M., and Cohen I.K.Hyaluronic acid modulates proliferation, collagen and protein synthesis of cultured fetal fibroblasts. Matrix 13,441, 1993 [DOI] [PubMed] [Google Scholar]
- 8.Yim E.K.F.et al. Nanopattern-induced changes in morphology and motility of smooth muscle cells. Biomaterials 26,5405, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pennisi C.P., et al. Nanoscale topography reduces fibroblast growth, focal adhesion size and migration-related gene expression on platinum surfaces. Colloids Surf B Biointerfaces 85,189, 2011 [DOI] [PubMed] [Google Scholar]
- 10.Kim D.-H., et al. Mechanosensitivity of fibroblast cell shape and movement to anisotropic substratum topography gradients. Biomaterials 30,5433, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karuri N.W., Porri T.J., Albrecht R.M., Murphy C.J., and Nealey P.F.Nano- and microscale holes modulate cell-substrate adhesion, cytoskeletal organization, and -beta1 integrin localization in SV40 human corneal epithelial cells. IEEE Trans Nanobioscience 5,273, 2006 [DOI] [PubMed] [Google Scholar]
- 12.Qian T., and Wang Y.Micro/nano-fabrication technologies for cell biology. Med Biol Eng Comput 48,1023, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma G., et al. Polymer particle shape independently influences binding and internalization by macrophages. J Control Release 147,408, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou S.Y., Krauss P.R., and Renstrom P.J.Imprint lithography with 25-nanometer resolution. Science 272,85, 1996 [Google Scholar]
- 15.Chou S.Y., and Krauss P.R.Imprint lithography with sub-10 nm feature size and high throughput. Microelectron Eng 35,237, 1997 [Google Scholar]
- 16.Gautieri A., Vesentini S., Redaelli A., and Buehler M.J.Hierarchical structure and nanomechanics of collagen microfibrils from the atomistic scale up. Nano Lett 11,757, 2011 [DOI] [PubMed] [Google Scholar]
- 17.Livak K.J., and Schmittgen T.D.Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25,402, 2001 [DOI] [PubMed] [Google Scholar]
- 18.Mabuchi K., et al. Changes with respect to time in the in vivo adsorption of plasma proteins onto artificial heart blood pumps. ASAIO J 38,M536, 1992 [DOI] [PubMed] [Google Scholar]
- 19.Rixman M.A., Dean D., Macias C.E., and Ortiz C.Nanoscale intermolecular interactions between human serum albumin and alkanethiol self-assembled monolayers. Langmuir 19,6202, 2003 [Google Scholar]
- 20.Wood M.A., Bagnaninchi P., and Dalby M.J.The beta integrins and cytoskeletal nanoimprinting. Exp Cell Res 314,927, 2008 [DOI] [PubMed] [Google Scholar]
- 21.Dalby M.J.Cellular response to low adhesion nanotopographies. Int J Nanomedicine 2,373, 2007 [PMC free article] [PubMed] [Google Scholar]
- 22.Dalby M.J., Yarwood S.J., Johnstone H.J., Affrossman S., and Riehle M.O.Fibroblast signaling events in response to nanotopography: a gene array study. IEEE Trans Nanobioscience 1,12, 2002 [DOI] [PubMed] [Google Scholar]
- 23.Roach P., Farrar D., and Perry C.C.Interpretation of protein adsorption: surface-induced conformational changes. J Am Chem Soc 127,8168, 2005 [DOI] [PubMed] [Google Scholar]
- 24.Misra R.D.K., Girase B., Nune V.K.C., and Xu W.Cellular interactions and modulated osteoblasts functions mediated by protein adsorption. Adv Eng Mater 14,B247, 2012 [Google Scholar]
- 25.Axelsson I.Characterization of proteins and other macromolecules by agarose gel chromatography. J Chromatogr A 152,21, 1978 [Google Scholar]
- 26.Sjöström T., et al. Fabrication of pillar-like titania nanostructures on titanium and their interactions with human skeletal stem cells. Acta Biomater 5,1433, 2009 [DOI] [PubMed] [Google Scholar]
- 27.Lee J., Chu B.H., Chen K.-H., Ren F., and Lele T.P.Randomly oriented, upright SiO2 coated nanorods for reduced adhesion of mammalian cells. Biomaterials 30,4488, 2009 [DOI] [PubMed] [Google Scholar]
- 28.Yang C., and Svitkina T.Filopodia initiation: focus on the Arp2/3 complex and formins. Cell Adh Migr 5,402, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Discher D.E., Janmey P., and Wang Y.Tissue cells feel and respond to the stiffness of their substrate. Science 310,1139, 2005 [DOI] [PubMed] [Google Scholar]
- 30.Hadjipanayi E., Mudera V., and Brown R.A.Close dependence of fibroblast proliferation on collagen scaffold matrix stiffness. J Tissue Eng Regen Med 3,77, 2009 [DOI] [PubMed] [Google Scholar]
- 31.Wang N., Butler J.P., and Ingber D.E.Mechanotransduction across the cell surface and through the cytoskeleton. Science 260,1124, 1993 [DOI] [PubMed] [Google Scholar]
- 32.Dalby M.J., et al. Nanotopographical stimulation of mechanotransduction and changes in interphase centromere positioning. J Cell Biochem 100,326, 2007 [DOI] [PubMed] [Google Scholar]