Abstract
Avian retroviruses have undergone intense study since the beginning of the 20th century. They were originally identified as cancer-inducing filterable agents in chicken neoplasms. Since their discovery, the study of these simple retroviruses has contributed greatly to our understanding of retroviral replication and cancer. Avian retroviruses are continuing to evolve and have great economic importance in the poultry industry worldwide. The aim of this review is to provide a broad overview of the genome, pathology, and replication of avian retroviruses. Notable gaps in our current knowledge are highlighted, and areas where avian retroviruses differ from other retrovirus are also emphasized.
Introduction
The study of avian retroviruses began with the work of two Danish researchers, Vilhelm Ellerman and Oluf Bang, more than a century ago. In a seminal 1908 publication, they showed that a form of leukemia and lymphoma that afflicts chickens, called avian leukosis, could be transferred by cell free-filtrates[1]. This was the first evidence that viruses could cause cancer, but because leukemia was not recognized as cancer, the significance of their work was not appreciated at the time. Three years later however, the American scientist Peyton Rous discovered a second virus that induced sarcomas in chickens[2]. Rous sarcoma virus (RSV) has been studied extensively and has greatly aided our current understanding of both retroviral replication and cancer. We now know that RSV is derived from the more common avian leukosis virus (ALV). These and related viruses are collectively referred to as the avian sarcoma/leukosis viruses (ASLV) and will be the focus of this review because they are the most common and well studied of the avian retroviruses. Other types of retroviruses discovered in birds include the gammaretrovirus, reticuloendotheliosis virus (REV)[3], and lymphoproliferative disease virus of turkeys (LPDV)[4,5].
ASLV Genome / Subgroups
ASLVs are members of the Alpharetrovirus genus of the family Retroviridae[6]. The gag-pol-env gene order first determined for these simple retroviruses has proven to be common to all retroviruses. However, the complex retroviruses also encode accessory proteins in overlapping reading frames. The ASLV gag gene encodes the structural proteins capsid (CA), matrix (MA), and nucleocapsid (NC), as well as the viral protease (PR), which in most retroviruses is part of pol. Thus, ASLVs synthesize more PR than other retroviruses. The ASLV pol gene encodes the reverse transcriptase (RT) and integrase (IN) enzymes, and the env gene encodes the transmembrane (TM) and surface (SU) envelope glycoproteins. The viral genome is flanked on either side by long terminal repeats (LTRs). The most commonly studied ASLVs are classified into 6 distinct subgroups (A, B, C, D, E, and J) based on differences in their envelope glycoproteins, interference patterns, and host range in chicken cells of varying phenotypes. The chicken genome also encodes many endogenous retroviruses (ERVs), a minority of which are related to exogenous ASLVs[7]. Surprisingly, the majority of the ERVs are related to beta- and gammaretroviruses, suggesting they were once the dominant type of exogenous avian retrovirus[7].
Pathology
Chickens are thought to be the natural hosts of ASLVs[8], although some wild bird species can also be infected[9]. The virus predominantly spreads horizontally by either direct or indirect contact, but can also be transmitted vertically from hen to egg.
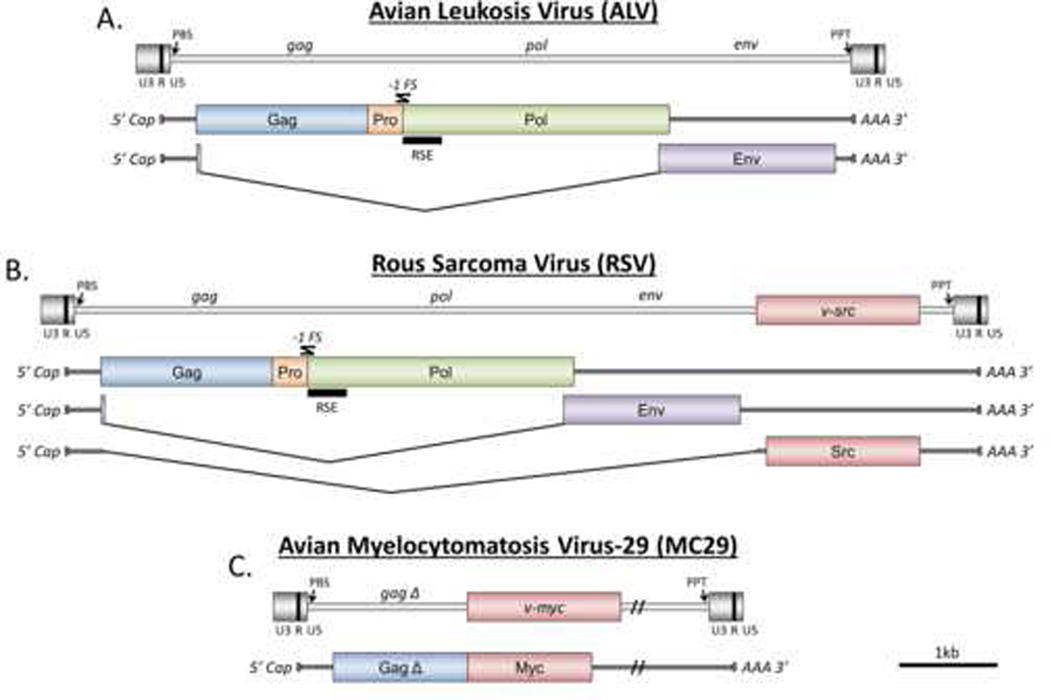
ASLVs are divided broadly into two classes: slowly transforming and acutely transforming. ALVs (Fig. 1A) make up the slow transforming class, causing tumors within a relatively long time frame of several weeks to months. ALV induces tumors by integrating within or near cancer-related genes, perturbing their expression or function. This process, first discovered with ALV, is called viral insertional mutagenesis and is common to other oncogenic simple retroviruses. Initial experiments detected ALV integration in the myc gene locus in tumors, causing deregulated expression of normal Myc protein from the ALV promoter[10,11]. Similar integrations in the myc locus were seen with REV-induced lymphomas [12]. Subsequent work also detected common integrations into a locus called bic[13], which we now know is the precursor for oncogenic microRNA-155. Recently, clonal ALV integrations into the telomerase reverse transcriptase (TERT) promoter were seen in B-cell lymphomas in chickens[14]. In addition, ALV induces erythroblastosis by insertion into the EGF receptor (c-erbB) locus in certain chickens lines[15].
Figure 1.
The ASLV proviruses and the transcripts of ALV (A), RSV (B), and MC29 (C) are shown. The location of viral oncogenes within the proviral genome are indicated when they exist. Protein coding portions of the transcripts are represented by colored rectangles. Locations of primer binding sites (PBS), RNA stability elements (RSE), polypurine tracts (PPT), and −1 framshift sites (−1 FS) are indicated. Polyprotein cleavage sites are not shown. This figure was adapted from [62].
ALV-A is the most common ASLV found in commercial flocks. It most often induces lymphoid leukosis, a B-cell lymphoma that begins in the bursa of Fabricius and metastasizes to the spleen, liver, and occasionally other organs. Although the virus is common, only a small fraction of infected birds in commercial flocks develop neoplasms. In addition, selective breeding has had some success limiting viral spread, reducing the economic impact of the virus[16].
In contrast, a new virus with a new subgroup, ALV-J was discovered in the late 1980’s. It has since spread rapidly resulting in great economic loss, especially in China. ALV-J is believed to have originated from a recombination event between ALV and an endogenous retroviral element[17–19]. The virus induces a different spectrum of tumors than ALV-A, primarily myeloid leukosis and hemangiomas[20,21]. The molecular basis of oncogenesis in these tumors is not well understood.
Acutely transforming viruses are distinct from ALV in that they contain one or two viral oncogenes in their genome, and induce neoplastic transformation more rapidly, within days or weeks. These viral oncogenes are derived from host cellular genes that have been incorporated into the virus by gene capture and are over-expressed by the strong viral promoters. For example, RSV picked up the v-src oncogene, a mutated version of the c-src tyrosine kinase gene found in host cells (Fig. 1b)[22,23]. Many acutely transforming ASLV strains have been isolated, carrying a wide variety of viral oncogenes, including myc, myb, fps, yes, jun, ets and erbB. With the exception of some strains of RSV, acutely transforming avian retroviruses are replication deficient(Fig. 1C), requiring a helper virus to replicate[24].
Viral entry
The surface of all retroviral virions are studded with Env proteins, which bind to specific receptors on the target cell surface, beginning the infection cycle. Four distinct cellular receptors have been identified that mediate ASLV entry. The tva, tvc, and chNHE1[25] genes confer susceptibility to ASLV of subgroup A, C, and J respectively, while tvb encodes the host cell receptor for subgroups B, D, and E. Two different alleles of tvb have been identified, tvbs1 and tvbs3, which confer susceptibility to ASLV subgroups B and D, and B, D, and E respectively[26–28]. ALV has a broad cellular tropism, able to replicate in a range of tissues and organs in the chicken[29,30].
Once bound to its receptor on the host cell, virion/cell fusion is activated through a two-step mechanism. First, the envelope glycoprotein undergoes a conformational change at the cell surface that mediates viral uptake and endosomal trafficking. Then, the acidic environment of the endosome activates hemifusion and release of the capsid into the cytoplasm[28]. Once in the cytoplasm, reverse transcription of the viral RNA can begin. Reverse transcription is an intricate multi-step process in which cellular dNTPs and packaged tryptophan tRNAs are utilized by the reverse transcriptase to convert viral RNA into a double stranded DNA provirus. Detailed reviews of the reverse transcription process are available elsewhere[31].
Nuclear Entry/Integration
Prior to nuclear entry, the DNA copy of the viral genome associates with integrase and other viral and cellular proteins to form the preintegration complex (PIC). The ability of the PIC to gain access to nuclear DNA varies among retroviruses. Gammaretroviruses can only integrate after nuclear envelope disassembly in mitosis, while lentiviruses are able to infect non-dividing cells via active transport into the nucleus[32]. For decades it was thought that alpharetroviruses, like gammaretroviruses, were only able to infect dividing cells[33]. Contrary to this idea, it was observed that cells infected with avian sarcoma virus (ASV) upon the release of serum starvation, could be stably integrated into host genomic DNA prior to mitosis[34], suggesting that nuclear envelope breakdown is not necessary for ASV integration. It is now thought that ASV can infect a variety of non-cycling cells[35–38], and the ASV integrase contains a nuclear localization signal (NLS) that can mediate active transport of the ASV integrase through the nuclear pore[39]. This work, together with earlier observations[40], suggests a block at reverse transcription rather than nuclear import may be responsible for the low infectivity of ASLV in non-dividing cells[38].
Once in the nucleus, the PIC mediates integration of the provirus into the host genomic DNA. Integration site selection varies among retroviruses, with some showing strong selection for certain areas of the genome. ASLV integration is more indiscriminant that most retroviruses, only slightly preferring areas of active transcription. This difference in target site selection is true regardless of whether the viruses are infecting chicken or human cells[41]. The reason for this pattern of integration targeting has not been completely elucidated[42].
Transcription/Splicing/Nuclear Export
Transcription of the ASLV provirus is directed by the viral LTRs, which contain strong promoter and enhancer sequences. Because ASLVs do not encode transcriptional transactivators, they rely entirely on host transcription factors, which bind to the U3 region of the LTR and drive RNA Polymerase II transcription of the provirus. All viral transcripts have an m7G cap at the 5’ end and undergo 3’ end cleavage and polyadenylation by cellular machinery prior to export from the nucleus.
All replication-competent ASLVs produce a single primary RNA transcript. This full-length, unspliced viral RNA serves as mRNA for translation of gag and pol genes, as well as the genomic material that is packaged into new virions. In addition, a fraction of these primary transcripts are spliced to generate the subgenomic env mRNA. In the case of RSV, a second spliced transcript is generated which encodes the v-src oncogene(Fig. 1B). Replication-deficient ASVs such as myelocytomatosis virus (MC29)(Fig. 1C) typically produce only a single unspliced gag-onc fusion transcript.
The full-length viral RNA transcript (~7–9 kb) is an aberration in the cellular context, in which intron-containing transcripts are not usually exported from the nucleus for translation. To ensure successful replication, ASLV has evolved a variety of RNA elements that aid in the efficient transcription, export, and translation of these long unspliced viral RNAs.
For example, ASLV employs several mechanisms to protect these transcripts from splicing, such as suboptimal 3’ splice sites[43], and cis-acting RNA elements. One cis-acting element is the negative regulator of splicing (NRS). The NRS acts as a faux 5’ splice site by interacting with the 3’ splice site and recruiting components of the spliceosome. This sequesters the 3’ splice site away from the 5’ splice site and further reduces the efficiency of splicing[44].
Unlike complex retroviruses, which encode accessory proteins that mediate nuclear export, ASLV relies on a 100 nt direct repeat (DR) RNA sequence. ALV employs a single DR in its 3’ untranslated region (UTR), while RSV has two sequences, located on either side of the src oncogene. The DRs must form a highly stable stem loop structure to mediated nuclear export, which is dependent on the nuclear export factor Tap, although the DR does not appear to bind Tap directly[45,46].
The full length virus faces yet another obstacle during translation. Because the viral RNA has a stop codon at the end of the gag gene, the region downstream appears to be a long 3’UTR, which often targets transcripts for degradation by the nonsense mediated decay (NMD) machinery. In order to avoid degradation, ASLV has evolved a 400 nt element, named the RNA stability element (RSE)[47]. Positioned immediately downstream of the gag termination codon, the RSE protects the full length viral RNA from NMD mediated degradation. Work is currently underway to determine the mechanism by which the RSE functions.
Translation
The full length viral transcript acts as template for synthesis of two different polyproteins, Gag-Pro and Gag-Pro-Pol. Because the virion is composed of more Gag structural proteins than Pol proteins, all retroviruses have developed means to synthesize more Gag than Pol, despite being encoded on the same transcript. ASLV accomplishes this with a short A-U rich “slippery sequence” just upstream of the gag termination codon, followed by an RNA pseudoknot[48]. This pauses the ribosome over the slippery sequence, occasionally (~5% of the time) causing it to slip backwards a single nucleotide before continuing forward. This “−1 frameshift” places the gag termination codon out of frame and allows the ribosome to read through to the pol termination codon, generating the Gag-Pro-Pol polyprotein.
During synthesis, ASLV Gag undergoes further modification. Most retroviral Gag proteins undergo low levels of phosphorylation and myristylation at their N-termini. ASLV Gag does undergo low level phosphorylation[49], but is not myristylated. Instead ASLV Gag is acetylated at its N-terminus[50]. The function of this acetylation is currently unknown.
The Env polyprotein is synthesized from a separate spliced transcript. Unique among retroviruses, the ASLV env splice donor resides within gag, just downstream of the gag start codon. This appends the first six amino acids of Gag onto the beginning of each Env polyprotein[49,51,52]. The Env polyprotein undergoes additional processing and modification in the endoplasmic reticulum. During this process, the Env polyprotein is proteolytically cleaved into three fragments, is glycosylated, undergoes folding, forms a trimer[53], and is then exported to the cell surface. Once the trimer has formed, Env is competent to bind its cognate receptor which is also processed through the secretory pathway. This forms the basis of superinfection resistance. By binding the receptor in the ER and remaining bound as it is presented at the cell surface, infection of the cell with any virus that uses that receptor is prevented.
In addition to the canonical translation products, three small peptides are synthesized from upstream open reading frames (uORFs) adjacent to the gag gene. These uORFs are conserved among all ASLVs and may play a role in translation and packaging[54,55].
Virion Assembly/Budding
The assembly and budding process is mediated in large part by Gag. Following synthesis, the RSV Gag protein is transiently imported to the nucleus where the Gag NC domain interacts with a packaging sequence (Ψ) on the viral genomic RNA[56,57]. After Gag dimerization, a nuclear export signal within the p10 domain of Gag then mediates export of the ribonucleoprotein (RNP) complex by the CRM1 Pathway[57,58].
The RNP complex then undergoes phosphoinositide-dependent[59] trafficking from the nucleus and stably associates with the plasma membrane via a membrane binding domain(MBD) at the N-terminus of Gag[60]. The degree to which assembly occurs prior to plasma membrane localization is not well understood. At the plasma membrane the viral polyproteins, processed Env proteins, two covalently linked viral genomic RNAs and tryptophan tRNAs coalesce and the virion buds from the cell surface. Various domains of Gag, and multiple host proteins have been shown to be important in this process[49,61]. The viral PR mediates cleavage of the polyproteins, and the virion obtains a mature morphology capable of infection shortly after.
Conclusion
Since the discovery of avian retroviruses more than a century ago, much has been learned about their replication and life cycle. ASLV has proven to be very adaptable. Its ability to capture and use cellular genes, evolve to use a variety of cellular receptors, and recombine to form ALV-J attests to this. Though our understanding of the virus has increased, there are still many aspects of ASLV replication that warrant further study.
Highlights.
Overview of the genome, pathology, and replication of avian retroviruses
Areas where avian retroviruses differ from other retrovirus are emphasized
Gaps in current knowledge are highlighted
Acknowledgements
We acknowledge support from NIH grants RO1CA48746 and RO1CA124596 to KLB. JJ was supported in part by NIH training grant T32GM007231.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest have been highlighted as:
* of special interest
** of outstanding interest
- 1.Ellermann V, Bang O. Experimentelle leukämie bei hühnern. Zentralbl Bakteriol Parasitenkd Infekt. Hyg Abt Orig. 1908 [no volume]. [Google Scholar]
- 2.Rous P. A sarcoma of the fowl transmissible by an agent separable from the tumor cells. J. Exp. Med. 1911;13:397–411. doi: 10.1084/jem.13.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Theilen GH, Zeigel RF, Twiehaus MJ. Biological studies with RE virus (strain T) that induces reticuloendotheliosis in turkeys, chickens, and Japanese quail. J. Natl. Cancer Inst. 1966;37:731–743. [PubMed] [Google Scholar]
- 4.Biggs PM, McDougall JS, Frazier JA, Milne BS. Lymphoproliferative disease of turkeys 1. Clinical aspects. Avian Pathol. 1978;7:131–139. doi: 10.1080/03079457808418265. [DOI] [PubMed] [Google Scholar]
- 5.Payne LN. Retrovirus-induced disease in poultry. Poult. Sci. 1998;77:1204–1212. doi: 10.1093/ps/77.8.1204. [DOI] [PubMed] [Google Scholar]
- 6.King AM, Lefkowitz E, Adams MJ, Carstens EB, editors. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier; 2011. [Google Scholar]
- 7. Bolisetty M, Blomberg J, Benachenhou F, Sperber G, Beemon K. Unexpected diversity and expression of avian endogenous retroviruses. MBio. 2012;3:e00344–e00312. doi: 10.1128/mBio.00344-12. This paper shows that most endogenous avian retroviruses are not related to ASLVs (alpha retroviruses) but are beta-like, gamma-like, and alphabeta hybrids. About 20% of the 500 chicken ERVs are transcribed and translated.
- 8.Payne LN. Epizootiology of Avian Leukosis Virus Infections. In: Boer GFD, editor. Avian Leukosis. Springer US; 1987. pp. 47–75. [Google Scholar]
- 9.Li D, Qin L, Gao H, Yang B, Liu W, Qi X, Wang Y, Zeng X, Liu S, Wang X, et al. Avian leukosis virus subgroup A and B infection in wild birds of Northeast China. Vet. Microbiol. 2013;163:257–263. doi: 10.1016/j.vetmic.2013.01.020. [DOI] [PubMed] [Google Scholar]
- 10. Hayward WS, Neel BG, Astrin SM. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature. 1981;290:475–480. doi: 10.1038/290475a0. This is the first demonstration in any species of retroviral activation of a cellular oncogene by insertional mutagenesis.
- 11.Payne GS, Bishop JM, Varmus HE. Multiple arrangements of viral DNA and an activated host oncogene in bursal lymphomas. Nature. 1982;295:209–214. doi: 10.1038/295209a0. [DOI] [PubMed] [Google Scholar]
- 12.Noori-Daloii MR, Swift RA, Kung HJ, Crittenden LB, Witter RL. Speciic integration of REV proviruses in avian bursal lymphomas. Nature. 1981;294:574–576. doi: 10.1038/294574a0. [DOI] [PubMed] [Google Scholar]
- 13.Clurman BE, Hayward WS. Multiple proto-oncogene activations in avian leukosis virus-induced lymphomas: evidence for stage-specific events. Mol. Cell. Biol. 1989;9:2657–2664. doi: 10.1128/mcb.9.6.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang F, Xian RR, Li Y, Polony TS, Beemon KL. Telomerase reverse transcriptase expression elevated by avian leukosis virus integration in B cell lymphomas. Proc. Natl. Acad. Sci. U. S. A. 2007;104:18952–18957. doi: 10.1073/pnas.0709173104. Activation of TERT is common in B-cell lymphomas induced by ALV infection of chick embryos.
- 15.Nilsen TW, Maroney PA, Goodwin RG, Rottman FM, Crittenden LB, Raines MA, Kung HJ. c-erbB activation in ALV-induced erythroblastosis: novel RNA processing and promoter insertion result in expression of an amino-truncated EGF receptor. Cell. 1985;41:719–726. doi: 10.1016/s0092-8674(85)80052-0. [DOI] [PubMed] [Google Scholar]
- 16.Fadly AM, Nair V. Leukosis/sarcoma group. Diseases of Poultry. 2008:514–568. [Google Scholar]
- 17.Venugopal K. Avian leukosis virus subgroup J: a rapidly evolving group of oncogenic retroviruses. Res. Vet. Sci. 1999;67:113–119. doi: 10.1053/rvsc.1998.0283. [DOI] [PubMed] [Google Scholar]
- 18.Smith LM, Toye AA, Howes K, Bumstead N, Payne LN, Venugopal K. Novel endogenous retroviral sequences in the chicken genome closely related to HPRS-103 (subgroup J) avian leukosis virus. J. Gen. Virol. 1999;80(Pt 1):261–268. doi: 10.1099/0022-1317-80-1-261. [DOI] [PubMed] [Google Scholar]
- 19.Sacco MA, Flannery DM, Howes K, Venugopal K. Avian endogenous retrovirus EAV-HP shares regions of identity with avian leukosis virus subgroup J and the avian retrotransposon ART-CH. J. Virol. 2000;74:1296–1306. doi: 10.1128/jvi.74.3.1296-1306.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Payne LN, Gillespie AM, Howes K. Myeloid leukaemogenicity and transmission of the HPRS-103 strain of avian leukosis virus. Leukemia. 1992;6:1167–1176. [PubMed] [Google Scholar]
- 21.Cheng Z, Liu J, Cui Z, Zhang L. Tumors associated with avian leukosis virus subgroup J in layer hens during 2007 to 2009 in China. J. Vet. Med. Sci. Jpn. Soc. Vet. Sci. 2010;72:1027–1033. doi: 10.1292/jvms.09-0564. [DOI] [PubMed] [Google Scholar]
- 22.Stehelin D, Varmus HE, Bishop JM, Vogt PK. DNA related to the transforming gene(s) of avian sarcoma viruses is present in normal avian DNA. Nature. 1976;260:170–173. doi: 10.1038/260170a0. [DOI] [PubMed] [Google Scholar]
- 23.Swanstrom R, Parker RC, Varmus HE, Bishop JM. Transduction of a cellular oncogene: the genesis of Rous sarcoma virus. Proc. Natl. Acad. Sci. U. S. A. 1983;80:2519–2523. doi: 10.1073/pnas.80.9.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vogt V. Retroviral Virions and Genomes. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 25.Chai N, Bates P. Na+/H+ exchanger type 1 is a receptor for pathogenic subgroup J avian leukosis virus. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5531–5536. doi: 10.1073/pnas.0509785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brojatsch J, Naughton J, Rolls MM, Zingler K, Young JA. CAR1, a TNFR-related protein, is a cellular receptor for cytopathic avian leukosis-sarcoma viruses and mediates apoptosis. Cell. 1996;87:845–855. doi: 10.1016/s0092-8674(00)81992-3. [DOI] [PubMed] [Google Scholar]
- 27.Adkins HB, Brojatsch J, Young JA. Identification and characterization of a shared TNFR-related receptor for subgroup B, D, and E avian leukosis viruses reveal cysteine residues required specifically for subgroup E viral entry. J. Virol. 2000;74:3572–3578. doi: 10.1128/jvi.74.8.3572-3578.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnard RJO, Young JAT. Alpharetrovirus envelope-receptor interactions. Curr. Top. Microbiol. Immunol. 2003;281:107–136. doi: 10.1007/978-3-642-19012-4_3. [DOI] [PubMed] [Google Scholar]
- 29.Dougherty RM, Di Stefano HS. Sites of avian leukosis virus multiplication in congenitally infected chickens. Cancer Res. 1967;27:322–332. [PubMed] [Google Scholar]
- 30.Williams SM, Fitzgerald SD, Reed WM, Lee LF, Fadly AM. Tissue tropism and bursal transformation ability of subgroup J avian leukosis virus in White Leghorn chickens. Avian Dis. 2004;48:921–927. doi: 10.1637/7196-041904R. [DOI] [PubMed] [Google Scholar]
- 31.Telesnitsky A, Goff S. Reverse Transcriptase and the Generation of Retroviral DNA. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 32.Lewis PF, Emerman M. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J. Virol. 1994;68:510–516. doi: 10.1128/jvi.68.1.510-516.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Temin HM. Studies on carcinogenesis by avian sarcoma viruses. VI. Differential multiplication of uninfected and of converted cells in response to insulin. J. Cell. Physiol. 1967;69:377–384. doi: 10.1002/jcp.1040690314. [DOI] [PubMed] [Google Scholar]
- 34.Humphries EH, Glover C, Reichmann ME. Rous sarcoma virus infection of synchronized cells establishes provirus integration during S-phase DNA synthesis prior to cellular division. Proc. Natl. Acad. Sci. U. S. A. 1981;78:2601–2605. doi: 10.1073/pnas.78.4.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hatziioannou T, Goff SP. Infection of nondividing cells by Rous sarcoma virus. J. Virol. 2001;75:9526–9531. doi: 10.1128/JVI.75.19.9526-9531.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katz RA, Greger JG, Darby K, Boimel P, Rall GF, Skalka AM. Transduction of interphase cells by avian sarcoma virus. J. Virol. 2002;76:5422–5434. doi: 10.1128/JVI.76.11.5422-5434.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greger JG, Katz RA, Taganov K, Rall GF, Skalka AM. Transduction of terminally differentiated neurons by avian sarcoma virus. J. Virol. 2004;78:4902–4906. doi: 10.1128/JVI.78.9.4902-4906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Katz RA, Greger JG, Skalka AM. Effects of cell cycle status on early events in retroviral replication. J. Cell. Biochem. 2005;94:880–889. doi: 10.1002/jcb.20358. This discusses mitosis-independent integration of ASLV and the need for dNTPs for reverse transcription.
- 39.Andrake MD, Sauter MM, Boland K, Goldstein AD, Hussein M, Skalka AM. Nuclear import of Avian Sarcoma Virus integrase is facilitated by host cell factors. Retrovirology. 2008;5:73. doi: 10.1186/1742-4690-5-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fritsch EF, Temin HM. Inhibition of viral DNA synthesis in stationary chicken embryo fibroblasts infected with avian retroviruses. J. Virol. 1977;24:461–469. doi: 10.1128/jvi.24.2.461-469.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barr SD, Leipzig J, Shinn P, Ecker JR, Bushman FD. Integration targeting by avian sarcoma-leukosis virus and human immunodeficiency virus in the chicken genome. J. Virol. 2005;79:12035–12044. doi: 10.1128/JVI.79.18.12035-12044.2005. ASLV integration is found to be near random.
- 42.Craigie R, Bushman FD. HIV DNA Integration. Cold Spring Harb. Perspect. Med. 2012;2:a006890. doi: 10.1101/cshperspect.a006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McNally MT, Beemon K. Intronic sequences and 3’ splice sites control Rous sarcoma virus RNA splicing. J. Virol. 1992;66:6–11. doi: 10.1128/jvi.66.1.6-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Giles KE, Beemon KL. Retroviral splicing suppressor sequesters a 3’ splice site in a 50S aberrant splicing complex. Mol. Cell. Biol. 2005;25:4397–4405. doi: 10.1128/MCB.25.11.4397-4405.2005. ASLV splicing is regulated by a gag RNA sequence that sequesters the 3’ splice site.
- 45.LeBlanc JJ, Uddowla S, Abraham B, Clatterbuck S, Beemon KL. Tap and Dbp5, but not Gag, are involved in DR-mediated nuclear export of unspliced Rous sarcoma virus RNA. Virology. 2007;363:376–386. doi: 10.1016/j.virol.2007.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paca RE, Ogert RA, Hibbert CS, Izaurralde E, Beemon KL. Rous sarcoma virus DR posttranscriptional elements use a novel RNA export pathway. J. Virol. 2000;74:9507–9514. doi: 10.1128/jvi.74.20.9507-9514.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weil JE, Hadjithomas M, Beemon KL. Structural characterization of the Rous sarcoma virus RNA stability element. J. Virol. 2009;83:2119–2129. doi: 10.1128/JVI.02113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jacks T, Madhani HD, Masiarz FR, Varmus HE. Signals for ribosomal frameshifting in the Rous sarcoma virus gag-pol region. Cell. 1988;55:447–458. doi: 10.1016/0092-8674(88)90031-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swanstrom R, Wills J. Synthesis, Assembly, and Processing of Viral Proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 50.Palmiter RD, Gagnon J, Vogt VM, Ripley S, Eisenman RN. The NH2-terminal sequence of the avian oncovirus gag precursor polyprotein (Pr76gag) Virology. 1978;91:423–433. doi: 10.1016/0042-6822(78)90388-4. [DOI] [PubMed] [Google Scholar]
- 51.Ficht TA, Chang LJ, Stoltzfus CM. Avian sarcoma virus gag and env gene structural protein precursors contain a common amino-terminal sequence. Proc. Natl. Acad. Sci. 1984;81:362–366. doi: 10.1073/pnas.81.2.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hunter E, Hill E, Hardwick M, Bhown A, Schwartz DE, Tizard R. Complete sequence of the Rous sarcoma virus env gene: identification of structural and functional regions of its product. J. Virol. 1983;46:920–936. doi: 10.1128/jvi.46.3.920-936.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Einfeld D, Hunter E. Oligomeric structure of a prototype retrovirus glycoprotein. Proc. Natl. Acad. Sci. U. S. A. 1988;85:8688–8692. doi: 10.1073/pnas.85.22.8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Donzé O, Damay P, Spahr PF. The first and third uORFs in RSV leader RNA are efficiently translated: implications for translational regulation and viral RNA packaging. Nucleic Acids Res. 1995;23:861–868. doi: 10.1093/nar/23.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moustakas A, Sonstegard TS, Hackett PB. Alterations of the three short open reading frames in the Rous sarcoma virus leader RNA modulate viral replication and gene expression. J. Virol. 1993;67:4337–4349. doi: 10.1128/jvi.67.7.4337-4349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scheifele LZ, Garbitt RA, Rhoads JD, Parent LJ. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. 2002;99:3944–3949. doi: 10.1073/pnas.062652199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gudleski N, Flanagan JM, Ryan EP, Bewley MC, Parent LJ. Directionality of nucleocytoplasmic transport of the retroviral gag protein depends on sequential binding of karyopherins and viral RNA. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9358–9363. doi: 10.1073/pnas.1000304107. ASLV Gag interacts with genomic RNA in the nucleus and is exported by CRM1.
- 58.Scheifele LZ, Ryan EP, Parent LJ. Detailed mapping of the nuclear export signal in the Rous sarcoma virus Gag protein. J. Virol. 2005;79:8732–8741. doi: 10.1128/JVI.79.14.8732-8741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nadaraia-Hoke S, Bann DV, Lochmann TL, Gudleski-O’Regan N, Parent LJ. Alterations in the MA and NC Domains Modulate Phosphoinositide-Dependent Plasma Membrane Localization of the Rous Sarcoma Virus Gag Protein. J. Virol. 2013;87:3609–3615. doi: 10.1128/JVI.03059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verderame MF, Nelle TD, Wills JW. The membrane-binding domain of the Rous sarcoma virus Gag protein. J. Virol. 1996;70:2664–2668. doi: 10.1128/jvi.70.4.2664-2668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pincetic A, Leis J. The Mechanism of Budding of Retroviruses From Cell Membranes. Adv. Virol. 2009;2009:6239691–6239699. doi: 10.1155/2009/623969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petropoulos C. Retroviral Taxonomy, Protein Structures, Sequences, and Genetic Maps. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]