Abstract
SMAD4 is a common mediator of the TGF-beta signaling pathway. One of the members of this pathway, TGF-beta 1, has an important role in controlling gut inflammation in relation to the continuous stimulation of the intestinal microbiota. SMAD4 haploinsufficiency in humans has been linked to juvenile polyposis hereditary hemorrhagic telangiectasia syndrome (JP/HHT; OMIM#17505). Hematochezia and colonic mucosal inflammation suggestive of inflammatory bowel diseases (IBD) have been reported in JP/HHT. Stimulated by recent experience with two affected pediatric patients presented here, we explored the potential role of Smad4 haploinsufficiency in a murine model of colonic inflammation.
Smad4+/− mice were maintained on a mixed C57/129SvEv background. Chronic colitis was induced with repeated administration of dextran sulfate sodium (DSS) in drinking water. The colonic mucosal microbiota was interrogated by massively parallel pyrosequencing of the bacterial 16S rRNA gene.
66.7% of Smad4+/− mice were sensitive to DSS colitis compared to 14.3% of wild type (Chi-Square p=0.036). The augmented colitis was associated with microbiota separation in the Smad4+/− mice. Enterococcus and Enterococcus faecalis specifically was increased in abundance in the colitis-prone animals.
Smad4 haploinsufficiency can associate with increased susceptibility to large bowel inflammation in mammals with variable penetrance in association with the colonic mucosal microbiota. These findings may reveal implications not only towards colonic inflammation in the setting of SMAD4 haploinsufficiency, but for colorectal cancer as well.
Keywords: colitis, SMAD4, mouse, microbiota, inflammatory bowel disease
Introduction
The transforming growth factor (TGF)-beta signaling pathway orchestrates a variety of biological processes, such as cell proliferation, growth regulation, cell cycle control, apoptosis, and adhesion, by a wide range of cytokines [1, 2]. The TGF-beta signaling pathway is composed of cell surface receptors and intracellular effectors (receptor-mediated SMADs; R-SMADs). Upon the binding of a ligand to a cell surface receptor, the R-SMADs are phosphorylated by the receptors and associate with the common SMAD, SMAD4. This complex accumulates in the nucleus and modulates transcription through a fine tuned interplay with different transcriptional co-factors. SMAD4, encoded by the SMAD4 gene, is the only identified mammalian co-SMAD. Therefore, SMAD4 plays a crucial role in the downstream regulation of the TGF-beta pathway.
Abnormalities in genes encoding members of the TGF-beta signaling pathway are linked to different clinical entities, including juvenile polyposis (JP, OMIM#174900) and hereditary hemorrhagic teleangiectesia (HHT or Osler-Weber-Rendu disease; OMIM#187300). Juvenile polyposis is an autosomal dominant hamartomatous polyposis syndrome with multiple juvenile polyps throughout the GI tract and an increased risk for GI malignancies. HHT is also an autosomal dominant disease, characterized by vascular dysplasia resulting in arteriovenous malformations of multiple organ systems, including skin (teleangiectasias), central nervous system, lung, and liver. These malformations carry the possibility of life-threatening vascular accidents. The affected genes are SMAD4 and BMPR1A (ALK3) in JP, and ENG, ACVRL1 (ALK1), and SMAD4 in HHT, respectively. ALK1, ALK3, and ENG encode receptors involved in the TGF-beta family signaling pathways. Gallione and colleagues recognized that mutations in SMAD4 can cause the combined phenotype of JP and HHT and delineated such cases as a separate entity: juvenile polyposis with hereditary hemorrhagic telangiectasia syndrome (JP/HHT; OMIM#17505) [3].
TGF-beta has an important role in the down-regulation of the inflammatory response and in the maintenance of normal gut environment. It is expressed in mucosal epithelial cells, stromal cells, and mononuclear cells in the lamina propria. Disruption of the TGF-beta signaling cascade can results in an inflammatory condition [4]. Faulty phosphorylation of the downstream mediator SMAD3 is associated with diminished TGF-beta response [4]. Interestingly, the phosphorylation of SMAD3 is defective in the affected bowel in ulcerative colitis (UC) and Crohn disease (CD) patients due to the overexpression of the inhibitory SMAD, SMAD7 [5, 6]. It is possible that haploinsufficiency in other downstream mediators such as SMAD4 causes a similar phenotype. In accordance with this possibility, a JP/HHT patient has been reported to have UC as an associated condition [7].
We present two JP/HHT cases where the possibility of inflammatory bowel disease (IBD) as a co-morbid condition complicated the clinical picture. Based on these data, we examined the response to chemically-induced colitis in Smad4+/− mice.
Cases
Patient 1
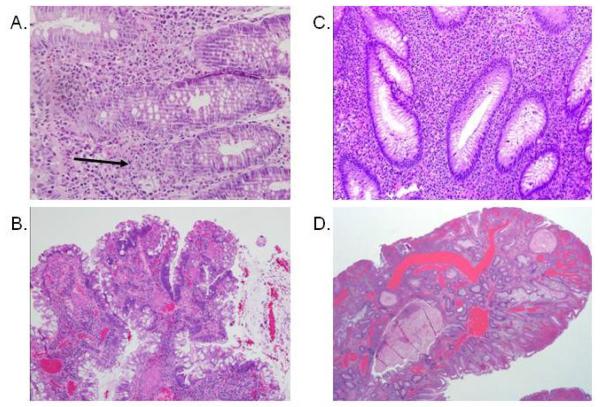
The 7-year-old white female presented with a chief complaint of rectal bleeding. She had congenital arachnoid cyst and VP shunt as an infant. Family history was suggestive of an inherited condition; her mother had rectal bleeding but had never been evaluated, and her grandmother had an ileostomy, but the reason was unclear. Colonoscopy grossly showed active colitis that resembled IBD with colonic ulcerations, and multiple colonic polyps were also observed. The histology showed mild chronic inflammation with limited architectural distortion (Figure 1A), and juvenile polyps. She was treated with mesalamine and became symptom-free. At about 9-10 years of age she developed some chest pains, intermittent shortness of breath, lip and nail bed cyanosis, and clubbing. Ultimately, she was discovered to have multiple bilateral pulmonary arteriovenous malformations. The most prominent of these, in the right lower lobe, was embolized via coils at the age of 12. After this procedure, her pulmonary symptoms resolved and her energy level improved. Genetic testing of SMAD4 was performed, and a pathogenic deletion was detected (1588delC), which was reported by Calva-Cerqueira et al. (JP16) [8]. She continued to have intermittent rectal bleeding, and colonoscopies at the age of 14 and 15 years revealed one juvenile polyp each, with adenomatous changes (Figure 1B). At that time she was starting to complain of shortness of breath again, and recommendations were made to follow-up with her pulmonary doctor. She has been lost to follow-up since that time.
Figure 1.
Mild chronic colitis and juvenile polyp with adenomatous changes in Patient 1 with JP/HHT: A. Mixed inflammatory infiltrate in the lamina propria, focally invading the glandular epithelium (arrow). Mild architectural distortion, tortuosity of the glands was present. B. Juvenile polyp with focal adenomatous changes characterized by hyperchromasia and pseudostratification of the epithelial nuclei. Patient 2 with JP/HHT: C. Dense mixed inflammation including numerous eosinophils in the lamina propria. Mild architectural distortion with glandular trifurcation was seen. D. Juvenile polyp with ectatic vessels from colectomy specimen.
Patient 2
The white male born at 34 weeks gestation developed recurrent epistaxis beginning at 5 years of age. He subsequently was diagnosed with HHT based on the family history of his mother who had pulmonary hemorrhage and a stroke. Genetic testing of the two most common genes (ENG and ALK1) was negative. He had multiple episodes of epistaxis requiring blood transfusions. Secondary to hematochezia between 11 and 12 years of age, a colonoscopy was performed, which revealed a juvenile polyp with mucosal inflammation. A subsequent colonoscopy was interpreted as chronic inflammatory bowel disease in active phase with pseudopolyps. Biopsies from both colonoscopies showed mixed inflammation with mild architectural distortion (Figure 1C). He was treated with prednisone without response. Based on the clinical, endoscopic, and histologic findings, genetic testing for SMAD4 was performed, and a previously recognized pathogenic mutation (1081C>T) [7] was detected. Secondary to continued GI blood loss a colectomy with creation of an ileostomy was performed. There were numerous polyps in the resected specimen, histologically consistent with juvenile polyps, but with prominent vascular ectasia (Figure 1D). Video capsule endoscopy showed telangiectasias in the small bowel, but no active bleeding. Over the next months his hemoglobin stabilized and he has remained well.
Materials and Methods
Mice
The murine Smad4 null allele (Smad4tm1.1Rob), designed as Smad4− , has been described [9]. Mice were maintained on a C57BL/6J;129S7/SvEvBrd mixed genetic background (abbreviated C57/129SvEv throughout text). Smad4 haploinsufficient (Smad4+/−) animals and WT littermate controls (Smad4+/+) were generated by crossing Smad4+/− to C57/129SvEv F1 hybrid mice. Smad4−/− mice do not gastrulate leading to early embryonic death, [9] therefore only heterozygous and WT offspring were generated. Weanling mice were genotyped by tail genomic DNA using PCR as described, [9] and tagged for later identification. Adult male mice, 18 weeks of age, were used in this study. The haploinsufficient and WT mice were maintained in mixed litters (3 out of 4 cages had both Smad4 haploinsufficient and WT animals for the dextran sodium sulfate [DSS] studies, for example). The Smad4+/− to WT microbiota comparisons were performed on non-DSS treated mice separately. All experimental animals were maintained in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory animals, and approved by the Institutional Review Board of the Baylor College of Medicine.
Chronic DSS colitis
3% (wt/vol) dextran sodium sulfate (DSS) (MW=36,000-50,000, MP Biomedicals, LLC, Solon, OH) was dissolved in the drinking water of the animals and provided ad libitum for 7-8 days, followed by 14 days of regular drinking water in 3 cycles of DSS exposure according to Liu ES et al. [10]. This molecular weight of DSS has been shown to induce colonic inflammation in previous work [11]. Mice were weighed daily when on DSS and intermittently between exposures. At 66 days, the animals were asphyxiated and colonic lengths were measured (n=14), except for mice that lost greater than 25% of their starting weight (n=2) during the experiment. The latter animals were euthanized early according to the approved animal protocol and designated as the “mortality group”. The colons were placed on ice, transected longitudinally, cleansed from feces, washed with ice cold normal saline, and followed by collection of colonic mucosa by scraping with a glass microscope slide [12] (excluding the cecum). The mucosal scrapings were flash frozen on dry ice, and stored at −80 °C as earlier described [13].
DNA Extraction for Microbial Studies
Mucosal scrapings were thawed, centrifuged at 14,000 rpm for 30 seconds, and re-suspended in 500μl RLT buffer (Qiagen, Valencia, CA) (with β- mercaptoethanol). Sterile 5mm steel beads (Qiagen, Valencia, CA) and 500μl sterile 0.1mm glass beads (Scientific Industries, Inc., NY, USA) were added for complete bacterial lysis in a Qiagen TissueLyser (Qiagen, Valencia, CA), and the mixture was run at 30Hz for 5 minutes in Eppendorf tubes. Samples were centrifuged briefly and 100μl of 100% ethanol was added to a 100μl aliquot of the sample supernatant. This mixture was added to a DNA spin column, and DNA recovery protocols were followed as instructed in the QIAamp DNA Mini Kit (Qiagen, Valencia, CA) starting at step 5 of the Tissue Protocol. DNA was eluted and diluted to a final concentration of 20ng/μl.
Massively parallel bTEFAP
Bacterial taxonomic examination within the colonic mucosal samples was performed by bacterial tag-encoded FLX-Titanium amplicon pyrosequencing (bTEFAP) as described previously [14]. bTEFAP utilizes Titanium reagents and procedures, and a one-step PCR with a mixture of Hot Start and HotStar high fidelity taq polymerases to generate DNA amplicons originating from the V1-V3 region numbered in relation to E. coli 16SrRNA gene. The sequence information from the variable regions of 16SrRNA can enable the bacterial taxonomic annotation of each individual read (i.e. operational taxonomic unit [OTU]) based on publicly available databanks (for more detail see Bacterial diversity data analysis). The bTEFAP procedures were performed at the Research and Testing Laboratory (Lubbock, TX) (www.researchandtesting.com).
Bacterial diversity data analysis
All failed sequence reads, low quality sequence ends and tags, and sequences shorter than 300bp were removed, and any non-bacterial ribosome sequences and chimeras were depleted using custom software described previously [14] and the Black Box Chimera Check software B2C2 (described and freely available at http://www.researchandtesting.com/B2C2.html). This process provided 4303 to 8871 filtered sequences in the individual mucosal DNA samples, which were queried using a distributed BLASTn.NET algorithm [15] against a database of high quality 16S rRNA bacterial sequences derived from NCBI. Database sequences were characterized as high quality based upon similar criteria as described for RDP ver 9 (http://rdp.cme.msu.edu/) [16]. Using a .NET and C# analysis pipeline the resulting BLASTn outputs were validated using taxonomic distance methods, and compiled as described previously [14]. Sequences with identity scores greater than 97% (<3% divergence) were resolved at the species level, between 95% and 97% at the genus level, between 90% and 95% at the family level, between 85% and 90% at the order level, between 80% and 85% at the class level, and below this to the phylum (77% and 80%). Statistical tests including Principle Coordinates Analysis (PCoA) using Unifrac-based distances were evaluated using NCSS 2007 (Kaysville, UT, USA).
Results
Increased weight loss upon DSS challenge in select Smad4+/− mice
DSS in drinking water induces weight loss in mice, which has been recognized as a reliable marker of colitis severity in this experimental model [13]. In a pilot experiment of a single 5 day DSS exposure in 6 Smad4+/− and 8 WT mice, 50% of the haploinsufficient animals lost more than 20% weight compared to none in the control group (two tailed Chi-Square p=0.024).
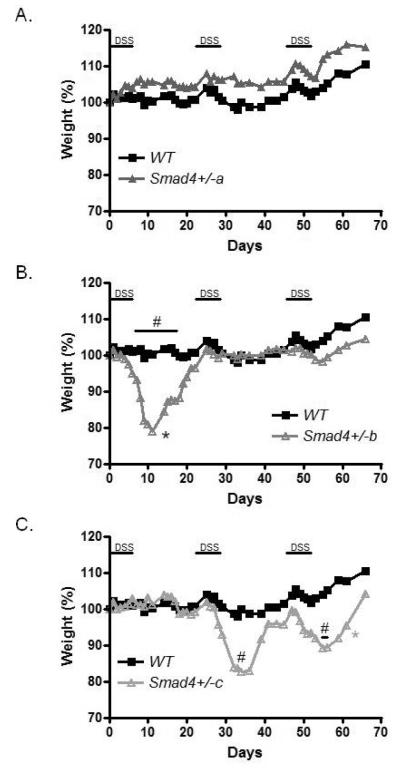
To follow up on this initial experiment, we used a chronic (3 consecutive exposures) DSS colitis trial. There were more Smad4 haploinsufficient mice that lost >12% of their original body weight (6 out of 9) than WT (1 out of 7) (Chi-Square p=0.036). As compared to WT, three distinct groups of Smad4+/− animals could be distinguished. One (group a: 3 mice) did not lose any weight during the chronic DSS challenge (Figure 2A), another (group b: 4 mice) lost weight only during the first DSS challenge (Figure 2B), and a third (group c: 2 mice) lost weight during the second and the third DSS exposure (Figure 2C). One animal from group b and one from group c had to be euthanized before the completion of the experiment for weight loss >25% according to our research protocol. Thus, mortality was only observed in the Smad4+/− animals (22%).
Figure 2.
Increased weight loss in select Smad4+/− mice during repeated exposures to dextran sulfate sodium (DSS, 3% w/v dissolved in drinking water). The graph shows average weight percentages compared to 100% on day 0 during the 66 day experiment. A. A group of Smad4+/− animals was resistant to colitis similarly to wild type (WT: n=7) (Smad4+/− a: n=3). B. A second group of Smad4+/− mice lost significant (#: p<0.05 on days 4-14) weight during the first DSS exposure (Smad4+/− b: n=4). C. A third group of Smad4 haploinsufficient mice lost significant weight during the second (#: P<0.05 on day 32) and the third (#: p<0.05 on days 55 and 56) DSS exposure (Smad4+/− c: n=2). * indicates that an animal had to be euthanized early secondary to extreme weight loss. For details see manuscript text. Error bars not shown for clarity. The WT samples are the same in all three panels, but for readability, the different Smad4+/− groups are shown as separate graphs.
The microbiota of colitis-prone Smad4+/− mice differs from non-colitic animals
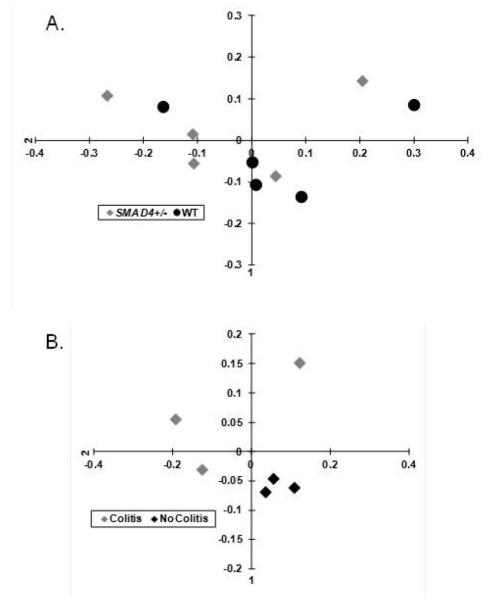
We then compared the colonic mucosal microbiota of Smad4+/− and WT animals that were not exposed to DSS. The mucosal microbiota has been proposed to being more relevant for intestinal immunomodulation than fecal content [17] and was previously examined in our earlier investigations [18, 19]. The microbiota of the haploinsufficient animals did not separate from WT (Figure 3A). However, when we compared the colonic mucosal microbiota of the Smad4+/− animals that developed significant colitis (weight loss>12%) during the first DSS exposure of the chronic colitis experiment (Figure 2, group b), to the haploinsufficient animals that did not develop colitis (Figure 2, group a) at the end of our experiment (when relative weights did not differ anymore), then a distinct partitioning could be observed between the two groups (Figure 3B).
Figure 3.
Weighted principal coordinates analysis (PCoA) of the UniFrac distance measures from the colonic mucosal samples. This analysis was performed to demonstrate relative distances (i.e. maximal differences) between the microbiota of each sample (colonic mucosa of each animal studied) at the genera level in a 2 dimensional space A. The Smad4+/− (grey diamond) and the wild type (WT; black dot) microbiota did not separate distinctly. B. The microbiota of colitis-prone (grey diamond) Smad4+/− mice segregated from the non-colitic (black diamond) Smad4+/− animals.
There was no difference between the colitic and non-colitic Smad4+/− microbiota at the phylum level. However, there were 5 genera and 6 species, which differed significantly (p<0.05 by either T test, or ANOVA) between the two groups (Table 1).
Table 1.
Genus and species level differences between colitis-prone (Colitis, n=3) and resistant (No colitis, n=3) colonic mucosal microbiota from Smad4+/− mice. The numbers were rounded to three decimals and represent abundance (%) for the no-colitis/colitis columns, and p values for the T test (two-sided, non-paired) and analysis of variance (ANOVA, two-sided) between the groups.
| Genus | No colitis | Colitis | T test | ANOVA |
|---|---|---|---|---|
| Sporichthya | 0.147 | 0.049 | 0.011 | 0.013 |
| Enterococcus | 0.033 | 0.195 | 0.012 | 0.042 |
| Paenibacillus | 0.000 | 0.040 | 0.030 | 0.081 |
| Alicyclobacillus | 0.379 | 0.163 | 0.049 | 0.040 |
| Brevibacillus | 4.288 | 13.546 | 0.053 | 0.038 |
| Species | No colitis | Colitis | T test | ANOVA |
| Clostridium cocleatum | 0.438 | 0.209 | 0.003 | 0.032 |
| Sporichthya (species unknown) | 0.147 | 0.049 | 0.011 | 0.013 |
| Enterococcus faecalis | 0.023 | 0.170 | 0.018 | 0.040 |
| Bacteroides barnesiae | 0.151 | 0.647 | 0.036 | 0.080 |
| Alicyclobacillus pohliae | 0.379 | 0.163 | 0.049 | 0.040 |
| Brevibacillus laterosporus | 4.288 | 13.546 | 0.053 | 0.038 |
Discussion
The role of the TGF-beta/SMAD signaling pathway in the pathogenesis of IBD has been studied in the past [4, 20]. These diseases are thought to arise secondary to an exaggerated immune response upon the constant stimulus of the gastrointestinal microbiota, [21] where abnormal endogenous control mechanisms such as the TGF-beta pathway may be involved. In accordance with this possibility, we observed an ulcerative colitis-like clinical picture and mild chronic inflammation in the colonic mucosa of two unrelated patients with SMAD4 mutation-associated JP/HHT.
The cases demonstrate that dominant SMAD4 mutations can associate with colitis in combination with JP/HHT in select individuals. Our findings in Smad4+/− mice support the clinical observations by showing that Smad4 haploinsufficiency can augment acute colitis susceptibility in mammals with variable penetrance, which may be transmitted by genetic and/or non-genetic (such as epigenetic [18, 22]) modifiers [23] in association with the intestinal microbiota [24]. More specifically, the Enterococcus genus and Enterococcus faecalis in particular was increased over 5 -fold on average (Table 1) in the colonic mucosa of the colitis-prone haploinsufficient animals. Although statistically significant by uncorrected parametric comparisons, these differences would not be significant after correction for multiple testing (which correction is feasible to apply when assessing for significance in case samples with massive data-sets are compared) because of the relatively few samples studied. Therefore, these results thus far only represent a trend with respect to bacterial dysbiosis in colitis prone Smad4+/− mice. Moreover, we can not rule out that the observed dysbiosis in the colitis prone happloinsufficient animals is not the cause, but a non-specific and sustained result of the more severe colinic inflammation in those. Nevertheless, our findings are consistent with reports of Enterococcus faecalis being associated with greater severity of colitis in murine models of IBD [25, 26]. Even more interestingly, Enterococcus was increased in the feces of colorectal cancer patients [27]. Importantly, JP/HHT patients have been observed to develop early onset colorectal cancer (mean age 28 year) in 30% of the cases [28]. Based on our results and the literature, it is possible that those patients with JP/HHT who harbor increased amounts of Enterococci may be more prone to develop intestinal inflammation and colorectal cancer than others with this disorder. Therefore, our findings may be relevant not only in respect to SMAD4 haploinsufficiency-associated colitis, but colorectal cancer as well. Further studies towards the connections between the intestinal microbiota, and colorectal inflammation and/or malignancy in the face of genetic predisposition are warranted.
Acknowledgements
The authors wish to thank Dr. Elizabeth Robertson (University of Oxford) for the Smad4 mutant line and Alfred Balasa for technical support.
These studies were supported by National Institutes of Health grant CA138628 (to S.A.P.) and a Burroughs Wellcome Career Award in the Biomedical Sciences (to S.A.P.). R.K. was supported in part by the Broad Medical Research Program, the Broad Foundation (IBD-0252); the Child Health Research Career Development Agency of the Baylor College of Medicine (NIH # 5K12 HD041648); and a Public Health Service grant DK56338, funding the Texas Medical Center Digestive Diseases Center
Footnotes
Author contributions: R.S.: conception, design, data collection, figures, manuscript writing; S.A.P.: design, data collection, manuscript writing; D.N.: conduct of experiment, manuscript writing; S.E.D.: data interpretation, figures, manuscript writing; R.J.S.: data collection, manuscript writing; A.P.O.: data collection, manuscript writing; E.J.P.: data collection; M.J.F.: data collection, manuscript writing; R.K.: conception, design, conduct of experiment, data collection, figures, manuscript writing.
References
- 1.Waite KA, Eng C. From developmental disorder to heritable cancer: it’s all in the BMP/TGF-beta family. Nat Rev Genet. 2003;4:763–73. doi: 10.1038/nrg1178. [DOI] [PubMed] [Google Scholar]
- 2.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–21. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 3.Gallione CJ, Repetto GM, Legius E, Rustgi AK, Schelley SL, Tejpar S, Mitchell G, Drouin E, Westermann CJ, Marchuk DA. A combined syndrome of juvenile polyposis and hereditary haemorrhagic telangiectasia associated with mutations in MADH4 (SMAD4) Lancet. 2004;363:852–9. doi: 10.1016/S0140-6736(04)15732-2. [DOI] [PubMed] [Google Scholar]
- 4.Fiocchi C. TGF-beta/Smad signaling defects in inflammatory bowel disease: mechanisms and possible novel therapies for chronic inflammation. J Clin Invest. 2001;108:523–6. doi: 10.1172/JCI13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monteleone G, Pallone F, MacDonald TT. Smad7 in TGF-beta-mediated negative regulation of gut inflammation. Trends Immunol. 2004;25:513–7. doi: 10.1016/j.it.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 6.Monteleone G, Kumberova A, Croft NM, McKenzie C, Steer HW, MacDonald TT. Blocking Smad7 restores TGF-beta1 signaling in chronic inflammatory bowel disease. J Clin Invest. 2001;108:601–9. doi: 10.1172/JCI12821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallione C, Aylsworth AS, Beis J, Berk T, Bernhardt B, Clark RD, Clericuzio C, Danesino C, Drautz J, Fahl J, Fan Z, Faughnan ME, Ganguly A, Garvie J, Henderson K, Kini U, Leedom T, Ludman M, Lux A, Maisenbacher M, Mazzucco S, Olivieri C, Ploos van Amstel JK, Prigoda-Lee N, Pyeritz RE, Reardon W, Vandezande K, Waldman JD, White RI, Jr, Williams CA, Marchuk DA. Overlapping spectra of SMAD4 mutations in juvenile polyposis (JP) and JP-HHT syndrome. Am J Med Genet A. 2010;152A:333–9. doi: 10.1002/ajmg.a.33206. [DOI] [PubMed] [Google Scholar]
- 8.Calva-Cerqueira D, Chinnathambi S, Pechman B, Bair J, Larsen-Haidle J, Howe JR. The rate of germline mutations and large deletions of SMAD4 and BMPR1A in juvenile polyposis. Clin Genet. 2009;75:79–85. doi: 10.1111/j.1399-0004.2008.01091.x. [DOI] [PubMed] [Google Scholar]
- 9.Chu GC, Dunn NR, Anderson DC, Oxburgh L, Robertson EJ. Differential requirements for Smad4 in TGFbeta-dependent patterning of the early mouse embryo. Development. 2004;131:3501–12. doi: 10.1242/dev.01248. [DOI] [PubMed] [Google Scholar]
- 10.Liu ES, Ye YN, Shin VY, Yuen ST, Leung SY, Wong BC, Cho CH. Cigarette smoke exposure increases ulcerative colitis-associated colonic adenoma formation in mice. Carcinogenesis. 2003;24:1407–13. doi: 10.1093/carcin/bgg094. [DOI] [PubMed] [Google Scholar]
- 11.Kitajima S, Takuma S, Morimoto M. Histological analysis of murine colitis induced by dextran sulfate sodium of different molecular weights. Exp Anim. 2000;49:9–15. doi: 10.1538/expanim.49.9. [DOI] [PubMed] [Google Scholar]
- 12.Perret V, Lev R, Pigman W. Simple method for the preparation of single cell suspensions from normal and tumorous rat colonic mucosa. Gut. 1977;18:382–5. doi: 10.1136/gut.18.5.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kellermayer R, Balasa A, Zhang W, Lee S, Mirza S, Chakravarty A, Szigeti R, Laritsky E, Tatevian N, Smith CW, Shen L, Waterland RA. Epigenetic maturation in colonic mucosa continues beyond infancy in mice. Hum Mol Genet. 2010;19:2168–76. doi: 10.1093/hmg/ddq095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bailey MT, Walton JC, Dowd SE, Weil ZM, Nelson RJ. Photoperiod modulates gut bacteria composition in male Siberian hamsters (Phodopus sungorus) Brain Behav Immun. 2010;24:577–84. doi: 10.1016/j.bbi.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 15.Dowd SE, Zaragoza J, Rodriguez JR, Oliver MJ, Payton PR. Windows .NET Network Distributed Basic Local Alignment Search Toolkit (W.ND-BLAST) BMC Bioinformatics. 2005;6:93. doi: 10.1186/1471-2105-6-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–5. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, Weber J, Hoffmann U, Schreiber S, Dietel M, Lochs H. Mucosal flora in inflammatory bowel disease. Gastroenterology. 2002;122:44–54. doi: 10.1053/gast.2002.30294. [DOI] [PubMed] [Google Scholar]
- 18.Kellermayer R, Dowd SE, Harris RA, Balasa A, Schaible TD, Wolcott RD, Tatevian N, Szigeti R, Li Z, Versalovic J, Smith CW. Colonic mucosal DNA methylation, immune response, and microbiome patterns in Toll-like receptor 2-knockout mice. FASEB J. 2011;25:1449–60. doi: 10.1096/fj.10-172205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schaible TD, Harris RA, Dowd SE, Smith CW, Kellermayer R. Maternal methyl-donor supplementation induces prolonged murine offspring colitis susceptibility in association with mucosal epigenetic and microbiomic changes. Hum Mol Genet. 2011;20:1687–96. doi: 10.1093/hmg/ddr044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monteleone G, Boirivant M, Pallone F, MacDonald TT. TGF-beta1 and Smad7 in the regulation of IBD. Mucosal Immunol. 2008;1(Suppl 1):S50–3. doi: 10.1038/mi.2008.55. [DOI] [PubMed] [Google Scholar]
- 21.Sartor RB. Key questions to guide a better understanding of host-commensal microbiota interactions in intestinal inflammation. Mucosal Immunol. 2011;4:127–32. doi: 10.1038/mi.2010.87. [DOI] [PubMed] [Google Scholar]
- 22.Harris RA, Nagy-Szakal D, Pedersen N, Opekun A, Bronsky J, Munkholm P, Jespersgaard C, Andersen P, Melegh B, Ferry G, Jess T, Kellermayer R. Genome-wide peripheral blood leukocyte DNA methylation microarrays identified a single association with inflammatory bowel diseases. Inflamm Bowel Dis. 2012 doi: 10.1002/ibd.22956. doi: 10.1002/ibd.22956. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kellermayer R. Genetic drift. “Omics”as the filtering gateway between environment and phenotype: The inflammatory bowel diseases example. Am J Med Genet A. 2010;152A:3022–5. doi: 10.1002/ajmg.a.33726. [DOI] [PubMed] [Google Scholar]
- 24.Nagy-Szakal D, Kellermayer R. The remarkable capacity for gut microbial and host interactions. Gut Microbes. 2011;2:178–82. doi: 10.4161/gmic.2.3.16107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann M, Messlik A, Kim SC, Sartor RB, Haller D. Impact of a probiotic Enterococcus faecalis in a gnotobiotic mouse model of experimental colitis. Mol Nutr Food Res. 2011;55:703–13. doi: 10.1002/mnfr.201000361. [DOI] [PubMed] [Google Scholar]
- 26.Steck N, Hoffmann M, Sava IG, Kim SC, Hahne H, Tonkonogy SL, Mair K, Krueger D, Pruteanu M, Shanahan F, Vogelmann R, Schemann M, Kuster B, Sartor RB, Haller D. Enterococcus faecalis metalloprotease compromises epithelial barrier and contributes to intestinal inflammation. Gastroenterology. 2011;141:959–71. doi: 10.1053/j.gastro.2011.05.035. [DOI] [PubMed] [Google Scholar]
- 27.Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Jia W, Cai S, Zhao L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012;6:320–9. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schwenter F, Faughnan ME, Gradinger AB, Berk T, Gryfe R, Pollett A, Cohen Z, Gallinger S, Durno C. Juvenile polyposis, hereditary hemorrhagic telangiectasia, and early onset colorectal cancer in patients with SMAD4 mutation. J Gastroenterol. 2012;47:795–804. doi: 10.1007/s00535-012-0545-8. [DOI] [PubMed] [Google Scholar]