Abstract
We report the results of a study of the prevalences of three clinically relevant Borrelia burgdorferi sensu lato genospecies (Borrelia burgdorferi sensu stricto, Borrelia afzelii, and Borrelia garinii) in 1,040 questing Ixodes ticks from all regions of Latvia, where Lyme borreliosis is endemic. The prevalences of Borrelia in Ixodes ricinus and Ixodes persulcatus were 22.6 and 27.9%, respectively. Molecular typing of B. burgdorferi from infected ticks was performed by restriction fragment length polymorphism (RFLP) analysis of PCR-amplified fragments of the 16S-23S (rrs-rrlA) rRNA intergenic spacer by using species-specific primers and subsequent sequencing. The dominant Borrelia species in both Ixodes species was B. afzelii. In addition, different restriction patterns of B. garinii and B. afzelii were also identified. This study demonstrates that the 16S-23S rRNA PCR-RFLP typing method is simple, sensitive, and fast and that it allows one to differentiate among B. burgdorferi species and subspecies with various degrees of pathogenic potential directly in ticks. These features are important in monitoring Lyme disease.
Lyme disease (Lyme borreliosis [LB]), a disorder that can affect multiple organ systems, results from an infection with the spirochete Borrelia burgdorferi sensu lato and is the most common tick-borne human disease in Europe and the United States. Since B. burgdorferi, the spirochete responsible for this zoonotic infection, is phenotypically and genotypically heterogeneous, it causes variability in the clinical aspects of the disease. Arthritis and carditis are preferentially associated with B. burgdorferi sensu stricto, the degenerative skin disorder acrodermatitis chronica et atrophicans is primarily associated with Borrelia afzelii, and neuroborreliosis is primarily associated with Borrelia garinii (13, 18). However, B. burgdorferi sensu stricto is also responsible for human neuroborreliosis in the United States (31).
B. burgdorferi sensu lato spirochetes are maintained in enzootic spirochete-tick vector-vertebrate cycles. Incidental human exposure occurs when enzootic ticks from such maintenance cycles bite humans. The principal vectors of B. burgdorferi sensu lato are ticks of the Ixodes ricinus complex (2). Two tick species, I. ricinus and Ixodes persulcatus, are present in Latvia, and vector competence for these two species has been experimentally confirmed (3, 5, 8, 10, 24, 28). Identification of the pathogenic B. burgdorferi species in ticks has epidemiological importance, since they are prime factors in determining the risk of acquiring LB and its clinical presentation. However, in Latvia, only I. ricinus ticks collected in the vicinity of Riga have been examined for the presence of Borrelia species in previous studies (14).
The first cases of LB in Latvia were registered in 1986. There were 583 cases of LB in Latvia in 1998, 281 cases in 1999 (an incidence of 11.52 cases per 100,000 inhabitants), 472 cases in 2000 (19.35 cases per 100,000 inhabitants), and 379 cases in 2001 (16.02 cases per 100,000 inhabitants). Cases of LB were registered in all four regions of Latvia (Kurzeme, Vidzeme, Zemgale, and Latgale). In spite of the relatively small territories occupied by each of these regions, some of their differences could influence the prevalence of LB. First, only I. ricinus is found in the western part of Latvia (Kurzeme and Zemgale), whereas both tick species, I. ricinus and I. persulcatus, are found in the central and eastern parts of Latvia (Vidzeme and Latgale). Secondly, many colonial seabirds that may be reservoirs for LB, especially for B. garinii (9, 26), migrate primarily along the coast of western and central Latvia (Kurzeme and Vidzeme). Most patients get infected in the vicinity of their homes, and erythema migrans appears in the early stages of B. burgdorferi sensu lato infections in most Latvian LB patients (I. Lucenko and A. Bormane, Abstr. 5th Baltic-Nordic Conf. Tick-Borne Zoonosis, abstr. 4, 1998). Neuroborreliosis is a frequent manifestation of disseminated infection in Latvia, as confirmed by serologic testing in all of the cases (I. Logina and I. Supe, Abstr. 5th Baltic-Nordic Conf. Tick-Borne Zoonosis, abstr. 6, 1998). However, there are no data about the Borrelia genospecies causing neuroborreliosis in Latvia.
In order to establish an epidemiological context for the prevalence of LB in Latvia, we surveyed the prevalence of clinically relevant B. burgdorferi sensu lato species in I. ricinus and I. persulcatus ticks collected during 4 years from all four regions of Latvia.
MATERIALS AND METHODS
Collection of ticks.
Questing ticks were collected by flagging in the four regions of Latvia (Kurzeme, Vidzeme, Zemgale, and Latgale) in 1998, 1999, 2000, and 2001. Ticks attached to the flag were removed with tweezers, placed singly in a 1.5-ml Eppendorf tube, and kept frozen at −20°C until they were used. An average of 400 ticks was collected in each region of Latvia each year, altogether >6,000. By random choice, 1,040 ticks were selected (260 ticks from each year) for the studies of Borrelia prevalence.
Extraction of DNA from ticks.
One hundred microliters of Tris-EDTA buffer (10 mM Tris-HCl, 1 mM EDTA, pH 7.6) was added to each sample tube, and each tick was crushed with a sterile plastic rod. Buffer containing the tick triturate was extracted with 100 μl of phenol-chloroform-isoamyl alcohol mixture (25:24:1; pH 8.0) by vortexing and subsequent centrifugation (11,000 rpm for 2 min; Eppendorf centrifuge 5417C; rotor FA 45-24-11) to separate the DNA from proteins. After centrifugation, the upper layer was transferred to a new tube and the DNA was reextracted with 100 μl of chloroform by vortexing and centrifugation (11,000 rpm for 2 min). The upper layer was again transferred to a new tube and heated for 15 min (90°C). Aliquots were frozen at −20°C until they were used.
Reference DNA.
DNA samples isolated from six reference strains (B. burgdorferi sensu stricto B31, B. burgdorferi sensu stricto N40, B. afzelii F1, B. afzelii ACA I, B. afzelii VS461, and B. garinii IP90) kindly donated by S. Bergstrom, Umeå, Sweden, were used as positive controls in all PCR-based methods.
B. burgdorferi detection by PCR amplification.
Nested PCR was carried out by the method described by Priem et al. (27). We used a primer set targeting the B. burgdorferi-specific segment of the ospA gene, located in linear plasmid lp54. A 3-μl aliquot of isolated DNA was employed for the PCR. The 50-μl reaction mixture contained 5 μl of 10-fold PCR buffer with (NH4)2SO4 (MBI Fermentas, Vilnius, Lithuania) [final concentrations, 75 mM Tris-HCl, 20 mM (NH4)2SO4, 1.5 mM MgCl2, 0.01% Tween 20], 2.5 μl of bovine serum albumin (0.1 mg · ml−1; MBI Fermentas), 100 mM each deoxynucleoside triphosphate, 1.5 U of Taq DNA polymerase (MBI Fermentas), and 30 pmol of each primer. First-round amplification employed PrZS7 (5′-GGGAATAGGTCTAATATTAGCC-3′; positions 18 to 39 of the ospA gene) as the forward primer and Osp5 (5′-CACTAATTGTTAAAGTGGAAGT-3′; positions 660 to 682 of the ospA gene) as the reverse primer. The amplification profile for the first-round PCR consisted of 35 cycles of denaturation at 95°C for 15 s, annealing at 50°C for 20 s, and extension at 72°C for 60 s. Three microliters of the first-round PCR product was employed as a template in a second-round PCR with Osp6 (5′-GCAAAATGTTAGCAGCCTTGAT-3′; positions 54 to 75 of the ospA gene) as the forward primer and Osp8 (5′-CGTTGTATTCAAGTCTGGTTCC-3′; positions 423 to 444 of the ospA gene) as the reverse primer. The amplification profile for the second-round PCR consisted of 25 cycles of denaturation at 95°C for 15 s, annealing at 55°C for 20 s, and extension at 72°C for 30 s. PCR amplification resulted in a 391-bp product.
Amplicons were visualized on a 1.5% agarose gel stained with ethidium bromide.
To monitor the occurrence of false-positive PCR results, negative controls were included during extraction of the tick samples (1 control sample for every 20 tick samples). Each time the PCR was performed, negative and positive control samples were included. False-negative results due to inhibition of the PCR were excluded by the use of internal spike controls.
To avoid cross-contamination and sample carryover, pre- and post-PCR sample processing and PCR amplification were performed in separate rooms.
B. burgdorferi typing by PCR-restriction fragment length polymorphism (RFLP).
ospA gene-positive DNA samples were used for further analysis.
(i) PCR.
A region of the B. burgdorferi 16S-23S (rrs-rrlA) ribosomal DNA (rDNA) spacer region was amplified by nested PCR with primers originally described by Liveris at al. (17). The use of the nested-PCR procedure resulted in an increased yield of product, allowing the use of this method directly on DNA extracted from ticks, thus obviating the necessity of culture. Six microliters of isolated DNA were employed for the PCR. The 50-μl reaction mixture contained 5 μl of 10-fold PCR buffer with (NH4)2SO4 [final concentrations, 75 mM Tris-HCl, 20 mM (NH4)2SO4, 1.5 mM MgCl2, 0.01% Tween 20; MBI Fermentas], 2.5 μl of bovine serum albumin (final concentration, 0.1 mg · ml−1; MBI Fermentas), 100 mM each deoxynucleoside triphosphate, 1.5 U of Taq DNA polymerase (MBI Fermentas), and 30 pmol of each primer. First-round amplification employed Pa (5′-GGTATGTTTAGTGAGGG-3′) as the forward primer and P95 (5′-GGTTAGAGCGCAGGTCTG-3′) as the reverse primer. The amplification profile for the first-round PCR consisted of 35 cycles of denaturation at 95°C for 15 s, annealing at 50°C for 20 s, and extension at 72°C for 60 s. Three microliters of the first-round PCR product was used as a template in a second-round PCR with Pb (5′-CGTACTGGAAAGTGCGGCTG-3′) as the forward primer and P97 (5′-GATGTTCAACTCATCCTGGTCCC-3′) as the reverse primer. The amplification profile for the second-round PCR consisted of 25 cycles of denaturation at 95°C for 15s, annealing at 55°C for 20 s, and extension at 72°C for 30 s. A final extension step at 72°C for 5 min was carried out after both PCRs. Amplicons were visualized on a 1.5% agarose gel stained with ethidium bromide.
(ii) RFLP analysis.
Ten-microliter aliquots of the nested-PCR amplification products were subjected to RFLP analysis by digestion with 2 U of either HinfI or TruI (MBI Fermentas). HinfI-digested fragments were analyzed by electrophoresis in a 1.5% agarose gel stained with ethidium bromide. TruI-digested fragments were analyzed by electrophoresis in 18% polyacrylamide gel stained with ethidium bromide or silver.
DNA sequencing.
For questionable samples, DNA sequencing was used. A DNA extraction kit (MBI Fermentas) was used to purify PCR products from agarose gels according to the manufacturer's instructions.
If only one PCR product was obtained (as a confirmed gel analysis), the DNA product was purified directly from the reaction mixture. Purification of the PCR products was carried out in 26-μl reaction volumes. Each reaction volume contained 25 μl of nested PCR product, 1 U of exonuclease I (U.S. Biochemicals, Cleveland, Ohio), and 0.9 U of shrimp alkaline phosphatase (U.S. Biochemicals). The reaction profile was 20 min at 37°C, 15 min at 80°C, and 5 min at 4°C. Four or 5 ml of the PCR product, depending on the amount of DNA, was used for DNA-sequencing reactions. For these reactions, fluorescence-labeled dideoxynucleotide technology was used (Applied Biosystems, Inc., Foster City, Calif.). The sequenced fragments were separated, and the data were registered on a PRISM 3100 automated DNA sequencer (Applied Biosystems, Inc.).
Species-specific PCR.
Species-specific PCR that targeted the 16S rRNA gene for the typing of B. garinii and B. burgdorferi sensu stricto was performed as described elsewhere (15).
Statistical methods.
Statistical significance was calculated using the χ2 test.
Nucleotide sequence accession numbers.
The rrs-rrlA intergenic spacer sequences of B. burgdorferi found in this study are available in the GenBank database under accession numbers AY121817, AY121816, AY163781, AY163782, and AY163783.
RESULTS
Prevalence of Borrelia in ticks.
In this study, 517 I. ricinus and 523 I. persulcatus ticks were analyzed by nested PCR for the presence of B. burgdorferi sensu lato. The results showed that 117 (22.6%) of the I. ricinus and 146 (27.9%) of the I. persulcatus ticks were PCR positive. In summary, 263 (25.3%) of the 1,040 ticks collected in four regions of Latvia in different years were infected with B. burgdorferi sensu lato species.
Tests of the sensitivity of the method showed that the smallest amount of DNA that gave a positive result represented three spirochetes in each PCR mixture (data not shown).
Typing of B. burgdorferi directly in ticks by RFLP analysis.
By random choice, a total of 100 DNA samples prepared from field-collected ticks that were positive for the presence of Borrelia and six reference strains (B. burgdorferi sensu stricto strain B31, B. burgdorferi sensu stricto strain N40, B. afzelii strains F1, ACAI, and VS461, and B. garinii strain IP90) were analyzed by PCR-RFLP using the two restriction enzymes HinfI and TruI. The smallest amount of DNA that gave a positive result represented six spirochetes in each PCR mixture (data not shown). A PCR amplicon was obtained from each of 71 DNA samples (71%). The PCR amplicon sizes differed and were species specific (Fig. 1). Five different HinfI (H1 to H5) and six different TruI (A, B, C, D, E, and F) restriction patterns were obtained from isolated tick DNAs. A typical result of HinfI RFLP analysis is shown in Fig. 2, and the results of all the analyses are summarized in Table 1.
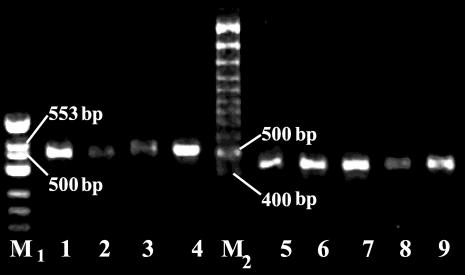
FIG. 1.
PCR products of the 16S-23S rDNA spacer from B. burgdorferi sensu lato tick isolates. The species assignments of the strains are given in Table 1. Amplification was carried out using the Pa, Pb, P95, and P97 primer set. DNAs were electrophoresed on a 1.5% agarose gel in TAE buffer (0.04 M Tris-acetate, 0.001 M EDTA), stained with ethidium bromide, and visualized by UV illumination. A φX174 DNA/HinfI marker (M1; Fermentas) and a 100-bp Gene Ruler (M2; Fermentas) were used as DNA molecular size markers. Lane 1, tick Nr77; lane 2, tick Nr22; lane 3, tick Nr65; lane 4, B. garinii strain IP90; lane 5, B. afzelii strain ACAI; lane 6, tick Nr27; lane 7, tick Nr26; lane 8; tick Nr25; lane 9, tick Nr21.
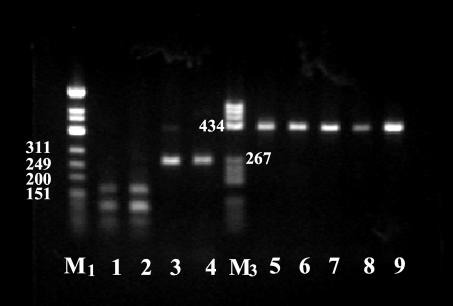
FIG. 2.
HinfI restriction patterns of the amplified 16S-23S rDNA spacer from B. burgdorferi tick isolates. The species assignments of isolates are indicated in Table 1. DNAs were electrophoresed on a 1.5% agarose gel in TAE buffer (0.04 M Tris-acetate, 0.001 M EDTA), stained with ethidium bromide, and visualized by UV illumination. A φX174 DNA/HinfI marker (M1; Fermentas) and a pBR322 DNA/BsuRI marker, (M3; Fermentas) were used as DNA molecular size markers. Lane 1, tick Nr77; lane 2, tick Nr22; lane 3, tick Nr65; lane 4, B. garinii strain IP90; lane 5, B. afzellii strain ACAI; lane 6, tick Nr27; lane 7, tick Nr26; lane 8, tick Nr25; lane 9, tick Nr21.
TABLE 1.
PCR-RFLP typing of B. burgdorferi isolates from Ixodes ticks
| Digestion patterna
|
No. (%) of DNA samples
|
Genospecies of B. burgdorferi sensu lato | |||
|---|---|---|---|---|---|
| HinfI | TruI | Total | I. ricinus | I. persulcatus | |
| H1 | A | 39 (54.9) | 18 | 21 | B. afzelii |
| H1 | B | 7 (9.9) | 0 | 7 | B. afzelii |
| H2 | C | 9 (12.7) | 8 | 1 | B. garinii |
| H3 | D | 9 (12.7) | 1 | 8 | B. garinii |
| H4 | E | 2 (2.8) | 2 | 0 | B. burgdorferi sensu stricto |
| H5 | F | 2 (2.8) | 0 | 2 | Untyped |
| H1 and H2 | A and C | 3 (4.2) | 2 | 1 | B. afzelii + B. garinii |
Based on digestion pattern with either HinfI or TruI as defined in the text.
PCR-RFLP patterns obtained from tick samples were compared with those obtained from reference strains. Interestingly, only the two B. burgdorferi sensu stricto reference strains used and two of the tick samples gave the expected 940-bp amplicon with the Pa, Pb, P95, and P97 primers. After digestion with HinfI, three different PCR-RFLP patterns were observed; both tick samples gave identical patterns (pattern H4E [Fig. 3, lanes 5 and 6]). The B. burgdorferi sensu stricto B31 PCR-RFLP pattern with digestion enzyme HinfI corresponded to its sequence in GenBank (372, 306, and 262 bp).
FIG. 3.
PCR products (lines 1 and 2) and HinfI restriction patterns (lines 3 to 6) of the 16S-23S rDNA spacer from B. burgdorferi sensu stricto tick isolates. The species assignments of strains are given in Table 1. DNAs were electrophoresed on a 1.5% agarose gel in TAE buffer (0.04 M Tris-acetate, 0.001 M EDTA), stained with ethidium bromide, and visualized by UV illumination. A 100-bp Gene Ruler (M2; Fermentas) was used as a DNA molecular size marker. Lane 1, B. burgdorferi sensu stricto strain B31; lane 2, tick Nr201; lane 3, B. burgdorferi sensu stricto strain N40; lane 4, B. burgdorferi sensu stricto strain B31; lane 5, tick Nr201; lane 6, tick Nr211.
All three B. afzelii reference strains (F1, ACAI, and VS461), B. garinii reference strain IP90, and most samples from ticks (except those with the H4E and H5F patterns [four samples]) gave much shorter amplicons (532 to 430 bp). The H1A and the H1B RFLP types, obtained from 39 (54.9%) and 6 (8.5%) of the Borrelia-infected ticks, respectively, were sequenced. These patterns showed 100 and 98% similarity to the B. afzelii reference strains ACAI and VS461 (the two reference strains showed identical PCR-RFLP patterns). The H2C RFLP pattern, obtained from nine (12.7%) samples, was identical to the restriction pattern obtained from the B. garinii reference strain IP90. The PCR amplicon obtained from the B. afzelii reference strain F1 was ∼500 bp long. However, similar PCR-RFLP patterns were not obtained from any of the tick samples (data not shown).
The H3D and the H4E RFLP patterns were obtained from 10 (14.1%) and 2 (2.8%) of the tick DNA samples, respectively. These patterns differed from the patterns given by the reference strains used in this study, and the strains were classified as B. garinii (H3D samples) and B. burgdorferi sensu stricto (H4E samples) based on the results of the species-specific PCR analysis.
Two of the DNA samples (2.8%) had an identical and definitely atypical H5F RFLP pattern that was not classified in this study.
Three tick samples (4.2%) had mixed RFLP patterns (both H1A and H2C), suggesting a double infection with both B. afzelii and B. garinii in these ticks. Both of the B. burgdorferi sensu stricto-infected ticks also showed evidence of a mixed infection, one with the B. afzelii H1A RFLP type and the second with the B. garinii H2C RFLP type.
The H1A RFLP pattern of B. afzelii was dominant in both Ixodes species. Interestingly, the H1B RFLP pattern was observed only in I. persulcatus ticks. The H2C RFLP pattern was observed mainly in I. ricinus ticks (eight ticks versus one tick; P = 0.003), whereas the H3D pattern predominated in I. persulcatus ticks (eight ticks versus one tick; P = 0.035).
The observed RFLP patterns from the DNA samples were completely consistent with the classification of B. burgdorferi into three distinct species (B. afzelii, B. garinii, and B. burgdorferi sensu stricto). The data presented in Table 1 demonstrate that all three clinically relevant B. burgdorferi sensu lato genotypes are found in Latvia.
The present study established that 16S-23S rDNA spacer analysis is both species and subspecies specific and can serve to identify B. burgdorferi sensu lato genotypes.
DISCUSSION
In the present study, for the first time in Latvia, Ixodes ticks collected from ecologically different regions of the country were examined and the prevalences of B. burgdorferi species in Ixodes persulcatus ticks were determined. Also in this study, the 16S-23S rDNA nested-PCR-RFLP typing method, originally used for the typing of B. burgdorferi in clinical specimens from early LB patients (12, 17), was extended to include the typing of B. garinii and B. afzelii species directly in field-collected ticks. Only a brief mention of such an approach was found in the literature (17).
In our study, the prevalence of Borrelia in field-collected ticks was 25.3%. This is in general agreement with the results obtained in other European countries, where the reported mean rates for adult unfed I. ricinus ticks infected with B. burgdorferi vary from 3 to 58% (11). An extremely high prevalence of B. burgdorferi-infected ticks was found in Portugal (75%), where the majority belonged to the Borrelia lusitanie type (4). In the Baltic St. Petersburg and Kaliningrad regions of Russia, infection rates of 26.3% for I. persulcatus and 11.5% for I. ricinus have been reported, whereas in the Western Siberian region of Russia, 38% of the I. persulcatus ticks were infected (1, 22). The infection rate of I. persulcatus ticks in our study (27.9%) is similar to that obtained in the neighboring Baltic regions of Russia, but the prevalence of Borrelia in I. ricinus in Latvia is higher (22.6%). There was no significant difference between rates of infection of I. ricinus and I. persulcatus with B. burgdorferi in Latvia. However, an infection rate of 31.3% was reported in I. ricinus ticks from the Riga area of Latvia in a previous study (14). These differences can be explained by differences in the strategy used in collecting the ticks. A recent study by Jenkins et al. (13) shows that samples taken at different times and from different locations may differ greatly in the prevalence of Borrelia. Therefore, in order to minimize these differences, we examined ticks randomly chosen from those collected in all four regions of Latvia throughout the 4-year period studied.
Various methods have been used to type B. burgdorferi sensu lato in culture isolates, but only a few of them are useful for typing Borrelia directly in ticks. The most popular are the PCR-based methods, such as species-specific PCR, in which the conserved 16S rRNA gene (15) or species-specific plasmid gene locus (20, 21) is targeted, the 23S-5S rRNA spacer region PCR hybridization method (29), and the rDNA PCR-RFLP analysis of the rrs-rrlA intergenic spacer (17). The 16S-23S rDNA nested-PCR-RFLP method used in our study was chosen because it is relatively inexpensive and could be applied without isolating Borrelia from culture. The rDNA cluster of most B. burgdorferi sensu lato strains is located in the center of the linear chromosome and is arranged in the following order: rrs-rrlA-rrfA-rrlB-rrfB (6, 7, 25, 30). One of the unusual properties of the rRNA gene cluster is the presence of a large (>3-kb) and highly variable intergenic spacer between the 16S (rrs) and 23S (rrlA) rRNA genes (16). To our knowledge, this method had not been used previously for typing B. garinii and B. afzelii directly in ticks from Europe. The amplification of the partial rrs-rrlA spacer, followed by digestion with HinfI and TruI, gave both species- and subspecies-specific RFLP patterns. In agreement with the original reference describing this method (17), the amplicon size of B. burgdorferi sensu stricto B31, B. burgdorferi sensu stricto N40, and two of our tick samples, classified as B. burgdorferi sensu stricto, was 940 bp. However, the amplicon sizes of the three B. afzelii reference strains, the B. garinii reference strain, and most of our tick samples belonging to these strains were much smaller, 532 to 430 bp. This finding does not agree with the results of Liveris et al. (16), who reported that both B. afzelii and B. garinii have larger 16S-23S rDNA spacers than B. burgdorferi sensu stricto. However, more detailed information about the 16S-23S rDNA intergenic spacer sequences in strains other than those of B. burgdorferi sensu stricto is lacking. To address this requires the sequencing of the whole 16S-23S rRNA spacer in B. burgdorferi genotypes other than B. burgdorferi sensu stricto. One of the objectives of our study was to partially sequence this spacer in the B. garinii and B. afzelii genotypes and subtypes by using PCR amplicons. The sequences obtained were submitted to GenBank.
Five different HinfI (H1 to H5) and six different TruI (A, B, C, D, E, and F) restriction patterns were obtained from the isolated tick DNA. Two different B. afzelii subspecies were identified in Latvia: 54.9% of all infected Ixodes ticks carried the H1A RFLP type, and 9.9% had the H1B RFLP type. Interestingly, the B. afzelii H1B RFLP type was detected only in I. persulcatus ticks, common in far eastern Europe and Asia (P = 0.0142). There are also significant differences in the numbers of tick species carrying different B. garinii RFLP types: the H2C RFLP type was found predominantly in I. ricinus (eight ticks versus one tick; P = 0.003), whereas the H3D RFLP type was more common in I. persulcatus (eight ticks versus one tick; P = 0.035). This finding suggests that the infection is vector specific. To determine to what extent an association between a pathogen and a tick species exists, additional studies are required.
The B. burgdorferi sensu stricto strain detected only in two I. ricinus ticks from Latvia differed from the reference strains B31 and N40. The presence of different subtypes in Latvia is not surprising, since many previous studies have identified strain heterogeneity within these genomic groups (19, 32). To determine if genetic heterogeneity may be responsible for the wide variation in the symptomatology and severity of LB (23), there is a need for further studies of the pathogenicities of different subtypes of Borrelia.
Some mixed infections were also detected in our study. Three ticks were infected with B. garinii and B. afzelii, one with B. burgdorferi sensu stricto and B. garinii, and one with B. burgdorferi sensu stricto and B. afzelii spirochetes. Two I. persulcatus tick isolates with identical but atypical RFLP patterns that could not be identified require further investigation.
The seroepidemiological studies of the population living in LB areas of endemicity in Latvia showed that 2.3% had positive (immunoglobulin G) serum samples, indicating the existence of abortive, subclinical, and/or unnoticed clinical forms of LB. In contrast, 21.2% of neurological patients whose exposure to an infected tick in the area of endemicity was suspected were seropositive (Logina and Supe, Abstr. 5th Baltic-Nordic Conf. Tick-Borne Zoonosis). The high incidence of Borrelia-infected ticks and the prevalence of clinically relevant B. burgdorferi sensu lato genospecies in these ticks, together with the existence of different clinical forms of LB in Latvia, indicates that there may be a correlation between the clinical form and the genospecies of the causative agent.
This study showed that a relatively large human population is at considerable risk of contracting a Borrelia infection in Latvia. All three clinically relevant B. burgdorferi species are distributed in Latvia, with the dominant genospecies being B. afzelii. The 16S-23S rDNA nested-PCR-RFLP typing method was able to distinguish differences within all three genomic groups investigated and demonstrated that significant genetic heterogeneity exists among the B. burgdorferi subtypes in I. ricinus and I. persulcatus ticks. This study confirms that simple RFLP analysis can be a fast and useful tool for typing of B. burgdorferi isolates without culturing.
Acknowledgments
This work was supported by a Council of Sciences of Latvia grant (no. 02.0924).
We thank Inta Jansone for her continuous support and help in this project and Guntis Brumelis and Vitauts Kalnins for correcting the manuscript. We also acknowledge the donation of reference strains by S. Bergstrom (Umeå, Sweden).
REFERENCES
- 1.Alekseev, A. N., H. V. Dubinina, I. Van De Pol, and L. M. Schouls. 2001. Identification of Ehrlichia spp. and Borrelia burgdorferi in Ixodes ticks in the Baltic regions of Russia. J. Clin. Microbiol. 39:2237-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burgdorfer, W., J. F. Anderson, L. Gern, R. S. Lane, J. Piesman, and A. Spielman. 1991. Relationship of Borrelia burgdorferi to its arthropod vectors. Scand. J. Infect. Dis. Suppl. 77:33-40. [PubMed] [Google Scholar]
- 3.Burgdorfer, W., A. G. Barbour, S. F. Hayes, O. Peter, and A. Aeschlimann. 1983. Erythema chronicum migrans—a tickborne spirochetosis. Acta Trop. 40:79-83. [PubMed] [Google Scholar]
- 4.De Mishelis, S., H. S. Sewell, M. Collares-Pereira, M. Santos-Reis, L. M. Schouls, V. Benes, E. C. Holmes, and K. Kurtenbach. 2000. Genetic diversity of Borrelia burgdorferi sensu lato in ticks from mainland Portugal. J. Clin. Microbiol. 38:2128-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dolan, M. C., J. Piesman, M. L. Mbow, G. O. Maupin, O. Peter, M. Brossard, and W. T. Golde. 1998. Vector competence of Ixodes scapularis and Ixodes ricinus (Acari: Ixodidae) for three genospecies of Borrelia burgdorferi. J. Med. Entomol. 35:465-470. [DOI] [PubMed] [Google Scholar]
- 6.Fukunaga, M., Y., Yanagihara, and M. Sohnaka. 1992. The 23S/5S ribosomal RNA genes (rrl/rrf) are separate from the 16S ribosomal RNA gene (rrs) in Borrelia burgdorferi, the etiological agent of Lyme disease. J. Gen. Microbiol. 138:871-877. [DOI] [PubMed] [Google Scholar]
- 7.Gazumyan, A., J. J. Schwartz, D. Liveris, and I. Schwartz. 1994. Sequence analysis of the ribosomal RNA operon of the Lyme disease spirochete, Borrelia burgdorferi. Gene 146:57-65. [DOI] [PubMed] [Google Scholar]
- 8.Gern, L., and O. Rais. 1996. Efficient transmission of Borrelia burgdorferi between cofeeding Ixodes ricinus ticks (Acari: Ixodidae). J. Med. Entomol. 33:189-192. [DOI] [PubMed] [Google Scholar]
- 9.Gylfe, Å., B. Olsen, D. Straševièius, N. M. Ras, P. Weihe, L. Noppa, Y. Östberg, G. Baranton, and S. Bergstrom. 1999. Isolation of Lyme disease Borrelia from puffins (Fratercula arctica) and seabird ticks (Ixodes uriae) on the Faeroe Islands. J. Clin. Microbiol. 37:890-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu, C. M., B. Wilske, V. Fingerle, Y. Lobet, and L. Gern. 2001. Transmission of Borrelia garinii OspA serotype 4 to BALB/c mice by Ixodes ricinus ticks collected in the field. J. Clin. Microbiol. 39:1169-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hubalek, Z., and J. Halouzka. 1997. Distribution of Borrelia burgdorferi sensu lato genomic groups in Europe, a review. Eur. J. Epidemiol. 13:951-957. [DOI] [PubMed] [Google Scholar]
- 12.Iyer, R., D. Liveris, A. Adams, J. Nowakowski, D. McKenna, S. Bittker, D. Cooper, G. P. Wormser, and I. Schwartz. 2001. Characterization of Borrelia burgdorferi isolated from erythema migrans lesions: interrelationship of three molecular typing methods. J. Clin. Microbiol. 39:2954-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenkins, A., B. Kristiansen, A. Allum, R. Aakre, L. Strand, E. Kleveland, I. van de Pol, and L. Schouls. 2001. Borrelia burgdorferi sensu lato and Ehrlichia spp. in Ixodes ticks from southern Norway. J. Clin. Microbiol. 39:3666-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurtenbach, K., S. De Michelis, H. S. Sewell, S. Etti, S. M. Schafer, R. Hails, M. Collares-Pereira, M. Santos-Reis, K. Hanincova, M. Labuda, A. Bormane, and M. Donaghy. 2001. Distinct combinations of Borrelia burgdorferi sensu lato genospecies found in individual questing ticks from Europe. Appl. Environ. Microbiol. 67:4926-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liebisch, G., B. Sohns, and W. Bautsch. 1998. Detection and typing of Borrelia burgdorferi sensu lato in Ixodes ricinus ticks attached to human skin by PCR. J. Clin. Microbiol. 36:3355-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liveris, D., A. Gazumyan, and I. Schwartz. 1996. Molecular typing of Borrelia burgdorferi sensu lato by PCR-restriction fragment length polymorphism analysis. J. Clin. Microbiol. 33:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liveris, D., S. Varde, R. Iyer, S. Koenig, S. Bittker, D. Cooper, D. McKenna, J. Nowacowski, R. Nadelman, G. Wormser, and I. Schwartz. 1999. Genetic diversity of Borrelia burgdorferi in Lyme disease patients as determined by culture versus direct PCR with clinical specimens. J. Clin. Microbiol. 37:565-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lünemann, J., S. Zarmas, S. Priem, J. Franz, R. Zschenderlein, E. Aberer, R. Klein, L. Schouls, G. Burmester, and A. Krause. 2001. Rapid typing of Borrelia burgdorferi sensu lato species in specimens from patients with different manifestations of Lyme borreliosis. J. Clin. Microbiol. 39:1130-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathiesen, D. A., J. H. Oliver, Jr., C. P. Kolobert, E. D. Tullson, G. L. Campbell, P. D. Mitchell, K. D. Reed, S. R. Telford III, J. F. Anderson, R. S. Lane, and D. H. Persing. 1997. Genetic heterogeneity of Borrelia burgdorferi in the United States. J. Infect. Dis. 175:98-107. [DOI] [PubMed] [Google Scholar]
- 20.Misonne, M. C., and P. P. Hoet. 1998. Species-specific plasmid sequences for PCR identification of the three species of Borrelia burgdorferi sensu lato involved in Lyme disease. J. Clin. Microbiol. 36:269-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Misonne, M. C., G. van Impe, and P. P. Hoet. 1998. Genetic heterogeneity of Borrelia burgdorferi sensu lato in Ixodes ricinus ticks collected in Belgium. J. Clin. Microbiol. 36:3352-3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morozova, O. V., A. K. Dobrotvorsky, N. N. Livanova, S. E. Tkachev, V. N. Bakhvalova, A. B. Beklemishev, and F. C. Cabello. 2002. PCR detection of Borrelia burgdorferi sensu lato, tick-borne encephalitis virus, and the human granulocytic ehrlichiosis agent in Ixodes persulcatus ticks from Western Siberia, Russia. J. Clin. Microbiol. 40:3802-3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nadelman, R. B., J. Nowakowski, G. Forseter, N. S. Goldberg, S. Bittker, D. Cooper, M. Aguero-Rosenfeld, and G. P. Wormser. 1996. The clinical spectrum of early Lyme borreliosis in patients with culture-confirmed erythema migrans. Am. J. Med. 100:502-508. [DOI] [PubMed] [Google Scholar]
- 24.Nakao, M., and Y. Sato. 1996. Refeeding activity of immature ticks of Ixodes persulcatus and transmission of Lyme disease spirochete by partially fed larvae. J. Parasitol. 82:669-672. [PubMed] [Google Scholar]
- 25.Ojami, C., B. E. Davidson, I. Saint Girons, and I. G. Old. 1994. Conservation of gene arrangement and unusual organization of rRNA genes in the linear chromosomes of the Lyme disease spirochaetes Borrelia burgdorferi, B. garinii and B. afzelii. Microbiology 140:2931-2940. [DOI] [PubMed] [Google Scholar]
- 26.Olsén, B., T. G. T. Jaenson, L. Noppa, J. Bunikis, and S. Bergstrom. 1993. A Lyme borreliosis cycle in seabirds and Ixodes uriae ticks. Nature 362:340-342. [DOI] [PubMed] [Google Scholar]
- 27.Priem, S., M. Rittig, T. Kamradt, G. Burmester, and A. Krause. 1997. An optimized PCR leads to rapid and highly sensitive detection of Borrelia burgdorferi in patients with Lyme borreliosis. J. Clin. Microbiol. 35:685-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sato, Y., and M. Nakao. 1997. Transmission of the Lyme disease spirochete, Borrelia garinii, between infected and uninfected immature Ixodes persulcatus during cofeeding on mice. J. Parasitol. 83:547-550. [PubMed] [Google Scholar]
- 29.Schouls, L. M., I. Van De Pol, S. G. T. Rijpkema, and C. S. Schot. 1999. Detection and identification of Ehrlichia, Borrelia burgdorferi sensu lato, and Bartonella species in Dutch Ixodes ricinus ticks. J. Clin. Microbiol. 37:2215-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwartz, J. J., A. Gazumyan, and I. Schwartz. 1992. rRNA gene organization in the Lyme disease spirochete, Borrelia burgdorferi. J. Bacteriol. 174:3757-3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, G., A. P. van Dam, I. Schwartz, and J. Dankert. 1999. Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological and clinical implications. J. Clin. Microbiol. Rev. 12:633-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, G., A. P. van Dam, L. Spanjaard, and J. Dankert. 1998. Molecular typing of Borrelia burgdorferi sensu lato by randomly amplified polymorphic DNA fingerprinting analysis. J. Clin. Microbiol. 36:768-776. [DOI] [PMC free article] [PubMed] [Google Scholar]