Abstract
Successful preemptive cytomegalovirus (CMV) therapy in transplant patients depends on the availability of sensitive, specific, and timely diagnostic tests for CMV infections. The pp65 antigenemia assay has been used for this purpose with considerable success. Quantification of CMV DNA is currently regarded to be an alternative diagnostic approach. The precise relationship between these two methods has still to be defined, but is essential to compare diagnostic results. This study compared the results of both assays with a large series of transplant recipients in different categories. An internally controlled quantitative real-time CMV DNA PCR was used to test 409 plasma samples from solid organ transplant (SOT) and stem cell transplant (SCT) patients. Levels of CMV DNA in plasma correlated well with classified outcomes of the pp65 antigenemia test. Despite this correlation, the quantitative CMV PCR values in a class of antigen test results were within a wide range, and the definition of an optimal cutoff value for initiating treatment required further analysis by a receiver-operating characteristic curve analysis. This is essential for reactivating infections in particular. For the SCT patients the optimal cutoff value of CMV DNA load defining relevant viral reactivation (in this assay, 10,000 copies/ml) was slightly higher than that for the SOT patients (6,300 copies/ml). Based on a comparison with the established pp65 antigenemia assay, quantification of CMV DNA in plasma appeared to be capable of guiding the clinical management of transplant recipients. This approach may have important advantages, which include a superior reproducibility and sensitivity, allowing the inclusion of kinetic criteria in clinical guidelines.
Human cytomegalovirus (CMV) is a ubiquitous member of the human herpesvirus family of viruses. Up to 80% of healthy adults in western countries are seropositive, indicating previous exposure and established latency with the capability of viral reactivation. The mechanism of reactivation is largely unknown but appears to be strongly related to impaired immune control of the virus. For this reason, CMV is one of the most common opportunistic pathogens complicating the care of transplant recipients; potentially it is a major cause of morbidity and mortality. Treatment of CMV disease with specific antiviral drugs such as ganciclovir and foscarnet reduces disease severity and mortality in these patients. Prophylactic and preemptive antiviral strategies have been developed and aim at avoiding aggressive treatment of established end-organ disease. Prophylactic treatment involves the administration of antiviral drugs to all patients at risk for an extended period. Preemptive therapy is specifically directed towards patients identified as having a high risk for CMV disease, thus sparing many from the toxicity of universally applied antiviral prophylaxis. The success of preemptive therapy is dependent upon the availability of appropriate diagnostic tests for early stages of CMV infections. The pp65 antigenemia assay has been used for this purpose with considerable success (2, 10, 14). However, this assay is labor-intensive and requires samples to be processed within a few hours. In addition, its reading is subjective and therefore requires skilled interpretation. Finally, the assay can be seriously complicated by leukopenia in stem cell transplant (SCT) recipients before engraftment.
Qualitative PCR detection of CMV DNA in leukocytes or plasma appeared to be a sensitive method for detection of CMV in blood but lacked specificity for the diagnosis of CMV disease (2). Quantification of CMV DNA should be able to define more specifically the levels associated with disease (2).
Real-time PCR-based assays (8) are able to quantify viral DNA accurately over a broad range of input target copies without the necessity for post-PCR handling. As such, these assays provide fast results with less risk of contamination. Recent studies have reported on the application of real-time PCR for the quantification of CMV DNA (13, 15, 18, 19, 21, 23, 25).
The clinical use of these methods could be evaluated in comparison to the currently widely employed pp65 antigenemia assay, with respect to the diagnosis of CMV infection but also in monitoring of individual transplant recipients during active infection. It is obvious that patients undergoing solid organ transplantation (SOT) or SCT nowadays will always be protected from clinical disease by a monitoring strategy or a preventive regimen, excluding an evaluation solely based on clinical outcome. The establishment of the precise relationship between the two methods is essential to compare diagnostic results, particularly when laboratories consider replacement of the antigen assay.
In this study, an internally controlled quantitative real-time PCR assay has been used to determine the CMV DNA load in plasma. The assay also monitors the efficacy of nucleic acid extraction from the clinical sample and the presence of inhibitors in the PCR. The correlation of this optimized assay to the classical pp65 antigenemia assay has been evaluated with SOT recipients as well as SCT recipients. The aim of this study was to validate the clinical application of real-time measurement of the CMV DNA load in plasma specimens from SOT and SCT recipients and to define criteria for treatment in these groups of patients.
MATERIALS AND METHODS
Patients and samples.
From August 2001 to June 2002, 3,100 EDTA-plasma and whole-blood specimens from SOT (kidney, kidney-pancreas, and liver) and SCT (pediatric and adult) recipients admitted to the Leiden University Medical Center were prospectively collected. From this group, 409 plasma samples from 128 SOT (36 liver and 51 kidney or kidney-pancreas) and SCT (41 adult and pediatric) patients were randomly selected for analysis, irrespective of the CMV serostatus of the donors and recipients. These 409 plasma samples were classified into five groups according to the results of the pp65 antigenemia assay. Group I (n = 195) corresponded to CMV antigenemia-negative samples. Group II (n = 79) corresponded to samples with low CMV antigenemia values (1 to 3 positive cells), and groups III (n = 57), IV (n = 50), and V (n = 28) corresponded to samples with moderate (4 to 20 positive cells), high (21 to 100 positive cells), and very high (>100 positive cells) CMV antigenemia values, respectively.
Also, 295 corresponding whole-blood samples were selected to address the correlation between CMV DNA loads in plasma and whole blood. During the study period, antigenemia assays were used for patient management, while real-time quantitative CMV PCR was performed retrospectively on the EDTA-plasma and whole blood samples frozen at −80°C.
Additionally, 10 CMV-seronegative kidney or kidney-pancreas transplant patients identified as undergoing a primary CMV infection (donor positive/recipient negative [D+/R−] combinations) were analyzed longitudinally, with a mean follow-up time of 82 days (range, 72 to 180 days) posttransplantation.
Viral standards and controls.
A sucrose gradient-purified and electron microscopy-counted HCMV AD169 strain (5.28 × 1010 virus particles/ml; ABI, Columbia, Md.) was used as a standard for quantification. The strain was diluted to a concentration of 108 particles/ml and subsequently serially diluted to determine a standard curve. The agreement of particle numbers and DNA copies was confirmed by using quantified plasmid DNA (IQ Products, Groningen, The Netherlands) containing the CMV PCR fragment (data not shown). A phocine herpes virus (PhHV) strain, propagated in cell culture, was used as internal control in the real time PCR. For specificity testing, patient samples positive for herpes simplex virus types 1 and 2, varicella-zoster virus, Epstein-Barr virus, human herpesvirus 6, adenovirus, parvovirus B19, and hepatitis B virus were used.
CMV antigenemia assay.
The CMV antigenemia assay was performed with the CMV Brite Turbo kit (IQ Corporation BV, Groningen, The Netherlands), according to the manufacturer's instructions. Briefly, 2.0 × 106 leukocytes were applied to a glass slide by cytospin, fixed, and permeabilized to allow subsequent detection of CMV pp65 antigen. The presence of pp65 antigen was detected by the C10/C11 antibody cocktail and visualized by means of a specific secondary fluorescein isothiocyanate-labeled antibody. The number of CMV antigen-positive cells per duplicate stain was counted.
CMV serology.
CMV-specific immunoglobulin M (IgM) and IgG antibodies in sera from patients were determined by using the Vironostika CMV-IgM assay (BioMerieux/Organon Teknika, Boxtel, The Netherlands) and the AxSYM CMV-IgG assay (Abbott Laboratories, North Chicago, Ill.) according to manufacturers' instructions.
Extraction of CMV DNA.
Nucleic acids were extracted from 0.2-ml plasma and whole blood samples by using the MagnaPure LC total nucleic acid isolation kit (Roche Molecular Systems, Almere, The Netherlands). During this fully automated purification procedure, lysis-binding buffer is added to the samples, resulting in complete cell lysis and protein denaturation. Subsequently, protein K is added to the samples and cellular proteins are digested. DNA binds to the silica surface of added magnetic glass particles due to the chaotropic salt conditions and the high ionic strength of the lysis-binding buffer. In the next step, wash buffer I removes unbound substances such as proteins, cell membranes, and PCR inhibitors such as heparin and hemoglobin. Wash buffer II further removes impurities and reduces the chaotropic salt concentration. Eventually, purified DNA is eluted in buffer at an elevated temperature.
Quantitative real-time PCR.
The CMV-specific PCR primers (Table 1) were derived from those previously described (3), and a specific TaqMan probe was developed. The primers (Eurogentec, Seraing, Belgium) amplified a 126-bp fragment from the CMV immediate-early antigen region. The TaqMan probe was labeled at the 5′ end with 6-carboxyfluorescein (FAM) and at the 3′ end with the fluorescent quencher 6-carboxytetramethylrhodamine. The 3′ end was phosphorylated to prevent probe extension during amplification.
TABLE 1.
CMV and PhHV primers and probes
| Primer or probe | Sequence |
|---|---|
| Forward CMV primer | 5′-CAAGCGGCCTCTGATAACCA-3′ |
| Reverse CMV primer | 5′-ACTAGGAGAGCAGACTCTCAGAGGAT-3′ |
| TaqMan CMV probe | FAM-TGCATGAAGGTCTTTGCCCAGTACATTCT-TAMRA |
| Forward PhHV primer | 5′-GGG CGA ATC ACA GAT TGA ATC-3′ |
| Reverse PhHV primer | 5′-GCG GTT CCA AAC GTA CCA A-3′ |
| TaqMan PhHV probe | Cy5-GTC GCC CCT GGT TTT TAT CGT ACG GGA ACA GGC GAC-BHQ2 |
The PCR was carried out by using the HotStar Taq master mix (Qiagen, Hilden, Germany) in an I-Cycler IQ DNA detection system (Bio-Rad, Veenendaal, The Netherlands). Briefly, 10 μl of either the standard-curve DNA or DNA extracted from the samples was added to 40 μl of PCR mixture containing a 400 μM concentration of each deoxynucleoside triphosphate, a 0.25 μM concentration of each primer, a 0.625 μM concentration of the fluorogenic probe, 4.5 mM MgCl2, and HotStar Taq DNA polymerase in HotStar PCR buffer. Template denaturation and activation of HotStar Taq DNA polymerase for 15 min at 95°C were followed by 50 cycles of denaturation at 95°C for 20 s, annealing at 63°C for 20 s, and extension at 72°C for 1 min.
To monitor the efficiency of the DNA extraction and PCR inhibition, all clinical samples were spiked with a fixed amount of PhHV virus particles prior to DNA extraction. PhHV DNA was amplified by using a PhHV-specific PCR assay as described previously (17). Primers used for the PhHV assay amplified a 89-bp fragment of the glycoprotein B gene. The probe was labeled with Cy5 and BHQ2 (Biolegio, Malden, The Netherlands). Primer and probe sequences are shown in Table 1. The CMV and PhHV assays were performed as a duplex PCR in a single tube. The PCR was performed under the same conditions as the CMV assay. During amplification the CMV and PhHV targets generated different reporter fluorescence signals (FAM and indodicarbocyanine, respectively).
ROC curve analysis.
Receiver-operating characteristic (ROC) curves represent the joined values of the true-positive ratio (sensitivity) and false-positive ratio (1 − specificity) for each value of the diagnostic variable (1, 26). In this study, ROC plot analysis was performed to determine a threshold value of the CMV DNA load in plasma for initiating treatment. Current clinical practice is based on the pp65 antigenemia assay, and therefore this assay was chosen to determine the optimal cutoff value for the DNA-based assay. In order to avoid the risk of nonspecific results of the lowest antigen level of one and two positive cells, the outcome for more than three positive cells in the pp65 antigenemia test was taken to be the lower predictive threshold for CMV disease in R+ transplant recipients. The level of three positive cells was deliberately chosen as the lowest convincing positive result. All database entry and statistical analysis were performed with SPSS version 10.0.7.
RESULTS
CMV DNA levels.
With the selected primers and probes, efficient amplification of dilution series of CMV DNA was obtained. When the Ct values were plotted, a standard line with a slope of 3.33 could be generated, indicating a PCR efficiency of 99.7%. Based on the dilution series of the CMV AD169 strain, the sensitivity of the assay was found to be approximately 100 to 250 copies/ml, which is 2 to 5 copies of CMV DNA in the reaction.
The specificity was tested with DNAs from a range of other viruses. No amplification was observed with herpes simplex virus types 1 and 2, varicella-zoster virus, Epstein-Barr virus, human herpesvirus 6, adenovirus, parvovirus B19, and hepatitis B virus.
The intra-assay variation was determined by using three CMV standard-curve DNA dilutions with low (103 copies/well), medium (105 copies/well), and high (108 copies/well) concentrations. The Ct values obtained for the low, medium, and high concentrations of standard CMV DNA in this test of intra-assay variation were 41.9 ± 1.0, 34.5 ± 0.3, and 24.7 ± 0.4, respectively (values are means ± standard deviations).
To determine the interassay variation, CMV standard DNA dilutions with low, medium, and high concentrations were subjected to the real-time PCR in 15 distinct experiments. These 15 distinct experiments also included 15 separate DNA extractions. The mean Ct values were 39.8 ± 1.4, 33.5 ± 1.1, and 23.5 ± 0.9 (means ± standard deviations), respectively.
Monitoring of DNA extraction and detection of PCR inhibition.
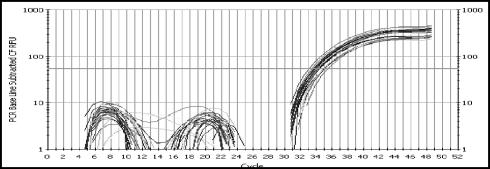
Reliable implementation of quantitative assays requires internal controls to avoid false-negative results or underestimation of values. Here an internal control reaction was used to monitor the nucleic acid extraction procedure and the presence of PCR inhibitors. The amount of internal control spike was arbitrarily set at a concentration which resulted in a Ct value of 34 ± 2 (Fig. 1). It was arbitrarily chosen that Ct values of the internal control that differed by more than two cycles from the value in the negative control sample were regarded as inhibitory.
FIG. 1.
Amplification plots obtained with the internal control (PhHV) DNA in a group of plasma samples. No inhibitory samples are detected, as the Ct value of the PhHV control was 34 ± 2.
To analyze possible competition between the CMV and PhHV DNA amplifications, serial dilutions from 108 to 103 of the electron microscopy-counted CMV AD169 strain were spiked with high and low concentrations of PhHV DNA and subjected to the PCR run. In addition, a serial dilution of PhHV DNA was spiked with low (103) and high (108) concentrations of CMV AD169 DNA. The results showed no significant difference (data not shown).
Correlation between CMV DNA loads in plasma and whole blood.
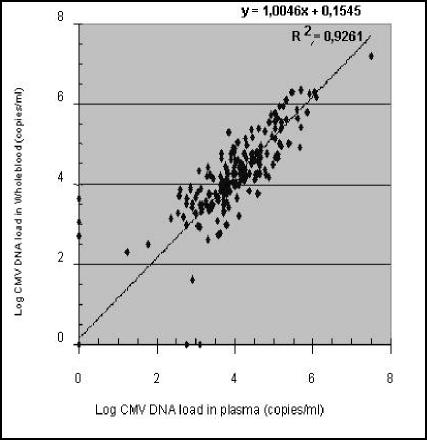
The CMV DNA loads in corresponding whole blood samples from 295 out of the 409 selected plasma samples were determined. The CMV DNA load in plasma was plotted against the CMV DNA load in whole blood (Fig. 2), and the correlation coefficient (r) of 0.962 indicated a high correlation between CMV DNA loads in plasma and whole blood. As can be derived from Fig. 2, the CMV DNA load in whole blood tends to be slightly but not significantly higher than that in plasma. An overall difference of 0.15 log unit (1.4 times) was found (Fig. 2).
FIG. 2.
Comparison of the CMV DNA levels in plasma and whole blood from the same patients.
Correlation between pp65 antigenemia and quantitative real-time CMV PCR.
The CMV real-time PCR assay was evaluated with 409 plasma samples from 128 SOT and SCT recipients. The corresponding pp65 antigenemia results were used to group the samples into five clinical categories. The median CMV DNA copy numbers in plasma were 0.00 copies/ml (mean, 2,82 copies/ml) for samples in group I and 4.47 × 103 copies/ml (mean, 2.14 × 103 copies/ml), 1.45 × 104 copies/ml (mean, 2.66 × 104 copies/ml), 5.50 × 104 copies/ml (mean, 6.92 × 104 copies/ml), and 1.91 × 105 copies/ml (mean, 2.82 × 105 copies/ml) for samples in groups II, III, IV, and V, respectively. As shown in Fig. 3 the CMV DNA copy numbers in plasma and the CMV antigenemia values seem to correlate well. Despite the correlation, the values for the CMV DNA load within the distinct pp65 groups showed a wide range (Fig. 3).
FIG. 3.
Comparison of CMV DNA loads in plasma specimens from SOT and SCT recipients by the pp65 antigenemia assay. The CMV DNA load was plotted to five pp65 antigenemia groups. For each group, the median load, the interquartile 50% range, and the range of values are represented. Open circles indicate the outliers (values between 1.5 and 3 box lengths from the upper or lower edge of the box). Asterisks represent the extreme values (values more than three box lengths).
Discrepancy analysis.
The quantitative real-time CMV PCR viral load values generally correlated well with the groups of antigenemia values (Fig. 3). However, some discrepancies between the pp65 antigenemia test and the CMV viral load were observed. Of the 195 antigenemia-negative samples in group I, 24 samples (from 16 patients) had a detectable CMV DNA load (mean CMV DNA load, 4.37 × 103 copies/ml; median, 3.09 × 103 copies/ml). In the CMV serostatus analysis, 15 of these 16 patients were either CMV IgG positive or had seroconverted (IgM positive), indicating that a positive CMV DNA load can be expected. For one patient, no serostatus or other follow-up data were available.
In nine patients the CMV DNA load was positive at the moment that CMV IgG was negative. Follow-up of these nine patients revealed that all of them seroconverted subsequently, indicating that a persisting seronegative status with a detectable load did not occur.
Finally, one pp65-positive sample (three cells) showed an undetectable CMV DNA load. The negative CMV DNA PCR was caused by either PCR inhibition or inefficient DNA isolation, since the PhHV internal control PCR remained negative during 50 amplification cycles.
Determination of CMV DNA load threshold values.
As the pp65 assay has been used for guiding CMV therapy in current clinical practice, it is important to establish the corresponding threshold values for the CMV DNA assay. In order to define the optimal cutoff value of CMV DNA load for initiating treatment in transplant patients at risk for CMV disease, an ROC curve analysis was performed, using existing treatment criteria based on pp65 test results. The outcome of more than three positive cells in the pp65 antigenemia test was taken to be the lower predictive threshold for CMV disease in R+ transplant patients. If subsequently a CMV DNA level of 100 copies/ml was used as a threshold for predicting CMV disease, the sensitivity and specificity were 99 and 66%, respectively (Table 2). The sensitivity decreased to 81% and the specificity increased to 90% when the CMV DNA level was increased to 10,000 copies/ml. When the ROC curve analysis was performed for SOT and SCT patients separately, the sensitivity and specificity for the above-mentioned CMV DNA levels were essentially the same (Table 2). For the SCT patients the optimal cutoff value of CMV DNA load was 104 copies/ml (sensitivity and specificity of 83 and 82%, respectively), and for the SOT recipients the optimal cutoff value was 5.37 × 103 copies/ml (sensitivity and specificity of 87 and 90%, respectively). The results are shown in Table 2.
TABLE 2.
Sensitivities and specificities of different threshold levels of CMV DNA load in plasmaa
| Group | Threshold CMV DNA level (copies/ml) in plasma as reference | Sensitivity (%) | Specificity (%) | Optimal CMV DNA levelb (% sensitivity, % specificity) |
|---|---|---|---|---|
| SCT recipients | 102 | 98 | 48 | 1.00 × 104 (83, 82) |
| 103 | 98 | 56 | ||
| 104 | 83 | 82 | ||
| SOT recipients | 102 | 99 | 72 | 5.37 × 103 (87, 90) |
| 103 | 98 | 78 | ||
| 104 | 80 | 93 | ||
| SCT and SOT recipients | 102 | 99 | 66 | 1.00 × 104 (81, 90) |
| 103 | 98 | 72 | ||
| 104 | 81 | 90 |
ROC analysis was performed by using the outcome of more than three positive cells in the pp65 antigenemia test as the value indicating which category should be considered positive. SOT and SCT recipients were considered as one group as well as separately.
Values are numbers of copies per milliliter.
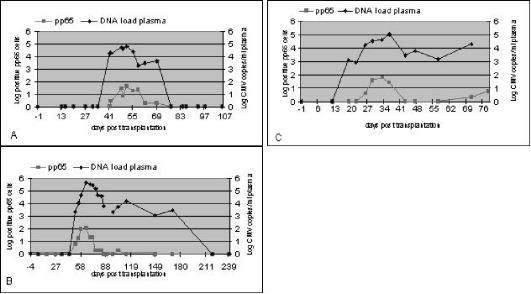
To assess the relationship between the CMV DNA level and the number of pp65-positive cells in transplant patients who are at risk for a primary CMV infection (D+/R−), 10 kidney or kidney-pancreas transplant recipients with a mean follow-up time of 82 days (range, 72 to 180 days) posttransplantation were longitudinally analyzed. In three (30%) of the patients, the CMV pp65 assay remained negative during the follow-up period. In the other seven patients (70%), positive pp65 results were observed (representative cases are shown in Fig. 4). The mean (median) numbers of days to the first positive test were 75 (41) and 78 (42) for CMV DNA load and antigenemia, respectively. In two patients from whom samples were obtained more frequently, a positive DNA load was detected prior to antigenemia positivity (18 versus 25 days in one patient and 19 versus 26 days in the other patient). CMV DNA never became positive later than pp65.
FIG. 4.
Three observed patterns in pp65 antigenemia and CMV DNA load follow-up in 7 of the 10 D+/R− kidney or kidney-pancreas transplant recipients studied. Three patients remained negative in both assays during follow-up. (A) The assays demonstrated that the patient responded well to antiviral treatment. This pattern was seen in four patients. (B). After antiviral treatment, the CMV DNA load persisted for some weeks at a level below the cutoff value of 104 copies/ml before it decreased to an undetectable level (one patient). (C) The pp65 assay and CMV DNA load measurement showed two episodes of CMV activation. Between these episodes, pp65 are negative, whereas the CMV DNA load persisted at levels below the defined cutoff value (two patients).
DISCUSSION
This study describes the application of an internally controlled real-time quantitative CMV PCR to plasma and whole-blood samples of SOT and SCT recipients. When these quantitative PCR results were compared with results of the pp65 antigenemia assay, the median CMV DNA copy numbers in plasma increased proportionally with the CMV antigenemia values, confirming results obtained in earlier studies (5, 6, 11, 12, 13, 16, 20, 23, 24). Nevertheless, some discrepancies were observed in this analysis of 409 plasma samples. These discrepancies could largely be explained by the increased sensitivity of the PCR compared to the pp65 test (7, 9). CMV DNA in plasma thus can be detected earlier than pp65 antigen in leukocytes. In addition, it should be noted that CMV DNA in plasma tended to persist longer during or after therapy than pp65 antigens; the rates of decline in the values of the two assays may well be different. Therefore, it is likely that these factors contribute to the finding that CMV DNA can be detected in antigenemia-negative samples. Since anti-CMV IgG antibodies were detectable in all of these pp65 and CMV DNA discrepant samples, lack of specificity of the CMV PCR is unlikely to be an issue. It was also shown that persisting seronegative status with a detectable DNA load did not occur.
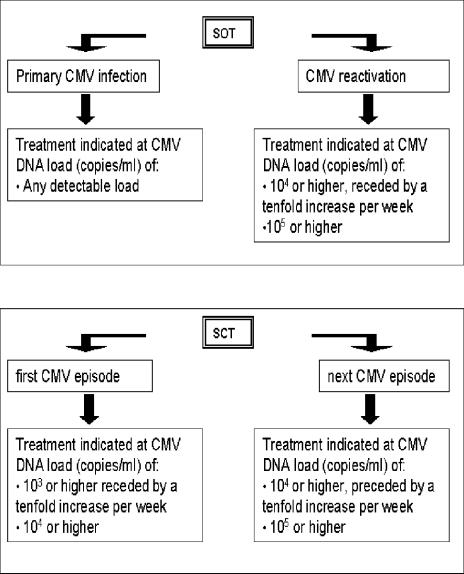
The analytical results of the two assays correlated well, and subsequently the clinical application of the results was assessed. This implies the development of criteria to initiate treatment based on CMV DNA values. For this purpose, patients at risk for a primary CMV infection and patients at risk for a CMV reactivation must be considered separately. Since patients at risk for a primary infection have never encountered CMV, any level of CMV DNA load will be predictive for the development of CMV infection. Treatment can be initiated on the basis of any positive result. However, patients at risk for a CMV reactivation may have a background CMV DNA level in plasma which is not necessarily correlated with disease. Determination of an optimal cutoff value for the CMV DNA level in plasma that is predictive for CMV disease is essential for the management of these patients. ROC analysis indicated optimal cutoff values of 10,000 copies of CMV DNA/ml in SCT patients and 5,370 copies/ml in SOT patients, with a sensitivity and specificity of more then 80% each. If, during monitoring, viral DNA loads exceed these threshold levels, antiviral therapy could be initiated to prevent disease. However, it has been shown previously that in addition to the viral DNA load, the kinetics of the DNA load should be taken into account for treatment decisions (4, 22). The present study results might enable the replacement of pp65 antigenemia tests by quantitative real-time CMV PCR. Figure 5 provides an illustration of possible practical guidelines which can be formulated for monitoring and management of CMV-related problems in transplant recipients based on the CMV DNA load in plasma. For further validation, the CMV DNA load measurement and pp65 antigenemia assays were performed simultaneously for all samples from SOT and SCT recipients during 2 months. A comparison of clinical decisions based on the guidelines as depicted in Fig. 5 with the clinical decisions based on the simultaneously obtained pp65 values demonstrated that the number of treatment periods was identical (results not shown). However, most of the CMV episodes were detected earlier with the CMV DNA load measurement.
FIG. 5.
Illustration of possible guidelines for the interpretation of plasma CMV DNA values after transplantation, based on the observations described in this study. To formulate these guidelines, the threshold values as derived from the ROC analysis were rounded off to the nearest decimal. In these guidelines, some practical approaches are different for SOT and SCT recipients. In the case of SOT recipients, thresholds are defined only for recipients at risk for CMV reactivation. SOT recipients at risk for primary CMV infection (D+/R−) have never encountered CMV, and therefore any level of CMV DNA is considered to be evidence of imminent clinically relevant infection. Primary infections are more difficult to define in the case of SCT recipients, as the level of grafted donor immunity is highly variable with regard to CMV. Therefore, a distinction is made between the first CMV episode and any subsequent episodes after transplantation. During the first episode, the threshold is set more stringently than for the subsequent episodes.
It was shown that the CMV DNA loads in plasma and whole blood correlated very well. This finding demonstrated that both blood compartments are likely to be adequate for use in the diagnosis and monitoring of CMV disease in transplant recipients.
In summary, the quantitative CMV real-time PCR is a useful tool for monitoring the risk of development of CMV disease in transplant recipients. If the results obtained by this assay are compared with those obtained by the pp65 antigenemia assay, it is possible to define cutoff values for the CMV DNA load in plasma to be used in the management of transplant patients at risk for CMV reactivation.
Acknowledgments
We thank H. G. M. Niesters (Department of Virology, Erasmus Medical Center, Rotterdam, The Netherlands) for providing us with PhHV and primers for the PhHV PCR and R. Boom (Department of Medical Microbiology, Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands) for helpful discussions.
REFERENCES
- 1.Altman, D. G., and M. Bland. 1994. Diagnostic tests. 3. Receiver operating characteristic plots. Br. Med. J. 309:188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boeckh, M., and G. Boivin. 1998. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin. Microbiol. Rev. 11:533-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boom, R., C. J. Sol, T. Schuurman, A. Van Breda, J. F. Weel, M. Beld, I. J. Ten Berge, P. M. Wertheim-Van Dillen, and M. D. De Jong. 2002. Human cytomegalovirus DNA in plasma and serum specimens of renal transplant recipients is highly fragmented J. Clin. Microbiol. 40:4105-4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Emery, V. C, C. A. Sabin, A. V. Cope, D. Cor, A. F. Hasan-Walker, and P. D. Griffiths. 2000. Application of viral load kinetics to identify patients who develop cytomegalo disease after transplantation. Lancet 355:2032-2036. [DOI] [PubMed] [Google Scholar]
- 5.Gault, E., Y. Michel, A. Dehée, C. Belabani, J.-C. Nicolas, and A. Garbarg-Chenon. 2001. Quantification of human cytomegalovirus DNA by real-time PCR. J. Clin. Microbiol. 39:772-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guiver, M., A. J. Fox, K. Mutton, N. Mogulkoc, and J. Egan. 2001. Evaluation of CMV viral load using TaqMan CMV quantitative PCR and comparison with CMV antigenemia in heart and lung transplant recipients. Transplantation 71:1609-1615. [DOI] [PubMed] [Google Scholar]
- 7.Griscelli, F., M. Barrois, S. Chauvin, S. Lastere, D. Bellet, and J.-H. Bourhis. 2001. Quantification of human cytomegalovirus DNA in bone marrow transplant recipients by real-time PCR. J. Clin. Microbiol. 39:4362-4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real-time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 9.Humar, A., D. Gregson, A. M. Caliendo, A. McGeer, G. Malkan, M. Krajden, P. Corey, P. Greig, S. Walmsley, G. Levy, and T. Mazzulli. 1999. Clinical utility of quantitative cytomegalo viral load determination for predicting cytomegalovirus disease in liver transplant recipients. Transplantation 68:1305-1311. [DOI] [PubMed] [Google Scholar]
- 10.Kusne, S., P. Grossi, W. Irish, K. St. George, C. Rinaldo, J. Rakela, and J. Fung. 1999. Cytomegalovirus pp65 antigenemia monitoring as a guide for preemptive therapy: a cost effective strategy for prevention of cytomegalo disease in adult liver transplant recipients. Transplantation 68:1125-1131. [DOI] [PubMed] [Google Scholar]
- 11.Li, H., J. S. Dummer, W. R. Estes, S. Meng, P. F. Wright, and Y. W. Tang. 2003. Measurement of human cytomegalovirus loads by quantitative real-time PCR for monitoring clinical intervention in transplant recipients. J. Clin. Microbiol. 41:187-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Limaye, A. P., M. L. Huang, W. Leisenring, L. Stensland, L. Corey, and M. Boeckh. 2001. Cytomegalovirus (CMV) DNA load in plasma for the diagnosis of CMV disease before engraftment in hematopoietic stem-cell transplant recipients. J. Infect. Dis. 83:377-382. [DOI] [PubMed] [Google Scholar]
- 13.Machida, U., M. Kami, T. Fukui, Y. Kazuyama, M. Kinoshita, Y. Tanaka, Y. Kanda, S. Ogawa, H. Honda, S. Chiba, K. Mitani, Y. Muto, K. Osumi, S. Kimura, and H. Hirai. 2000. Real-time automated PCR for early diagnosis and monitoring of cytomegalovirus infection after bone marrow transplantation. J. Clin. Microbiol. 38:2536-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazzulli, T., R. H. Rubin, M. J. Ferraro, R. T. D'Aquila, S. A. Doveikis, B. R. Smith, T. H. The, and M. S. Hirsch. 1993. Cytomegalo antigenemia: clinical correlations in transplant recipients and in persons with AIDS. J. Clin. Microbiol. 31:2824-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Najioullah, F., D. Thouvenot, and B. Lina. 2001. Development of a real-time PCR procedure including an internal control for the measurement of HCMV viral load. J. Virol. Methods 92:55-64. [DOI] [PubMed] [Google Scholar]
- 16.Nazzari, C., A. Gaeta, M. Lazzarini, T. Delli Castelli, and C. Mancini. 2000. Multiplex polymerase chain reaction for the evaluation of cytomegalovirus DNA load in organ transplant recipients. J. Med. Virol. 61:251-258. [PubMed] [Google Scholar]
- 17.Niesters, H. G. M. 2001. Quantitation of viral loads using real-time amplification techniques. Methods 25:419-429. [DOI] [PubMed] [Google Scholar]
- 18.Nitsche, A., N. Steuer, C. A. Schmidt, O. Landt, H. Ellerbrok, G. Pauli, and W. Siegert. 2000. Detection of human cytomegalovirus DNA by real-time quantitative PCR. J. Clin. Microbiol. 38:2734-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nitsche, A., N. Steuer, C. A. Schmidt, O. Landt, and W. Siegert. 1999. Different real-time PCR formats compared for the quantitative detection of human cytomegalovirus DNA. Clin. Chem. 45:1932-1937. [PubMed] [Google Scholar]
- 20.Piiparinen, H., K. Hockerstedt, M. Lappalainen, J. Suni, and I. Lautenschlager. 2002. Monitoring of viral load by quantitative plasma PCR during active cytomegalovirus infection of individual liver transplant patients. J. Clin. Microbiol. 40:2945-2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Preiser, W., N. S. Brink, U. Ayliffe, K. S. Peggs, S. Mackinnon, R. S. Tedder, and J. A. Garson. 2003. Development and clinical application of a fully controlled quantitative PCR assay for cell-free cytomegalovirus in human plasma. J. Clin. Virol. 26:49-59. [DOI] [PubMed] [Google Scholar]
- 22.Schafer, P., W. Tenschert, L. Cremaschi, M. Schroter, B. Zollner, and R. Laufs. 2001. Area under the viraemia curve versus absolute viral load: utility for predicting symptomatic cytomegalovirus infections in kidney transplant patients. J. Med. Virol. 65:85-89. [PubMed] [Google Scholar]
- 23.Tanaka, N., H. Kimura, K. Iida, Y. Saito, I. Tsuge, A. Yoshimi, T. Matsuyama, and T. Morishima. 2000. Quantitative analysis of cytomegalovirus load using a real-time PCR assay. J. Med. Virol. 60:455-462. [DOI] [PubMed] [Google Scholar]
- 24.Yakushiji, K., H. Gondo, K. Kamezaki, K. Shigematsu, S. Hayashi, M. Kuroiwa, S. Taniguchi, Y. Ohno, K. Takase, A. Numata, K. Aoki, K. Kato, K. Nagafuji, K. Shimoda, T. Okamura, N. Kinukawa, N. Kasuga, M. Sata, and M. Harada. 2002. Monitoring of cytomegalovirus reactivation after allogeneic stem cell transplantation: comparison of an antigenemia assay and quantitative real-time polymerase chain reaction. Bone Marrow Transplant. 29:599-606. [DOI] [PubMed] [Google Scholar]
- 25.Yun, Z., I. Lewensohn-Fuchs, P. Ljungman, and A. Vahlne. 2000. Real-time monitoring of cytomegalovirus infections after stem cell transplantation using the TaqMan polymerase chain reaction assays. Transplantation 69:1733-1736. [DOI] [PubMed] [Google Scholar]
- 26.Zweig, M. H., and G. Campbell. 1993. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin. Chem. 39:561-577. [PubMed] [Google Scholar]