Abstract
Transition metals occupy an essential niche in biological systems. Their electrostatic properties stabilize substrates or reaction intermediates in the active sites of enzymes, while their heightened reactivity is harnessed for catalysis. However, the latter property renders transition metals toxic at high concentrations. Bacteria, like all living organisms, must regulate the levels of these elements to satisfy their physiological needs while avoiding harm. It is therefore not surprising that the host capitalizes on both the essentiality and toxicity of transition metals to defend against bacterial invaders. This review will discuss established and emerging paradigms in nutrient metal homeostasis at the pathogen-host interface.
Transition metals are involved in many crucial biological processes and are therefore necessary for the survival of all living organisms. These metals are frequently incorporated into metalloproteins including metalloenzymes, storage proteins and transcription factors. The functional roles of transition metals in biological systems can be broken down broadly into non-catalytic functions, redox catalysis and non-redox catalysis. Of the redox-active metals, Fe is most commonly used, followed by Cu and Mo1. In both eukaryotes and prokaryotes, approximately 50% of non-haem Fe and Cu proteins are oxidoreductases or other electron transfer proteins 1. In addition, haem Fe is an important cofactor for respiration, as well as various biosynthetic and metabolic processes. Although Mg is the most prevalent non-redox metal found in enzymes, Zn is the most common transition metal 1. Zn can serve structural as well as catalytic roles in proteins. Interestingly, the distribution of Zn-binding proteins differs significantly within bacteria, archaea and eukaryotes. Enzymes constitute approximately 80% of the zinc-containing proteins of archaea and bacteria but less than 50% in eukaryotes, however Zn-dependent transcription factors make up 44% of the zinc-containing proteins in eukaryotes, demonstrating that Zn plays an important role in gene regulation in these organisms 2. Consequently, Zn-binding proteins make up a larger proportion of the total proteome in eukaryotes as compared to bacteria and archaea3.
All living organisms require transition metals to survive; yet the catalytic activity of these metals also potentiates their toxicity and the levels of transition metals must therefore be carefully controlled. Moreover, the mechanisms used to limit the availability of free transition metals also serve as a countermeasure against invading bacteria. The human body is a rich reservoir of essential nutrients for those bacteria that have evolved to exploit this resource. To prevent infection with pathogenic organisms, humans, like other mammals, restrict access to essential metals in a process termed “nutritional immunity”. Originally coined to refer to restriction of iron availability by the host, the term “nutritional immunity” can also be applied to mechanisms for withholding other essential transition metals or directing the toxicity of these metals against microbial invaders. This review will focus on four of these metals, namely Fe, Mn, Zn and Cu, and will discuss the roles for these metals at the pathogen-host interface. In addition, emerging paradigms in nutritional immunity will be reviewed, including host strategies for metal intoxication, the interplay between host genetics and the outcome of bacterial infections, and the extension of nutritional immunity to include non-metals.
Fe limitation: a universal strategy in innate defence
Fe is the fourth most abundant element in the Earth's crust and the most abundant transition metal in the human body. In bacteria, Fe is a co-factor of many enzymes and as such plays a crucial role in diverse physiological processes such as DNA replication, transcription and central metabolism1. Furthermore, the Fe-containing protoporphyrin haem is incorporated into cytochromes, thus participating in energy generation through respiration. Fe is required by virtually all bacterial pathogens and vertebrates therefore limit access to Fe to exploit this requirement as a potent defence against infection4, 5. As a result, bacteria must elaborate Fe-acquisition systems in order to successfully colonize host tissues. Recent reviews have focused on bacterial systems for acquiring Fe and the mechanisms utilized by vertebrate hosts to withhold Fe from invading bacteria 6-10. These mechanisms are discussed briefly below to allow comparison with other systems.
Host mechanisms for withholding iron
To prevent access to Fe, vertebrates use a number of proteins that render this valuable nutrient largely inaccessible to bacteria that lack sophisticated Fe-capturing systems (Figure 1a). In vertebrates, the majority of Fe is complexed to haem, a tetrapyrrole ring encircling a singular Fe atom and the cofactor of the oxygen transport protein haemoglobin. Haemoglobin is further contained within circulating erythrocytes, representing an additional barrier to access by pathogens. If free haemoglobin or haem is released from erythrocytes, these molecules are rapidly bound by haptoglobin and haemopexin, respectively. Therefore, for bacterial pathogens to access this rich pool of Fe, they must lyse erythrocytes, remove haem from haemoglobin or haemopexin, then liberate the Fe from the macrocyclic conjunction of haem.
Figure 1. Fe limitation and Fe acquisition during bacterial infections.
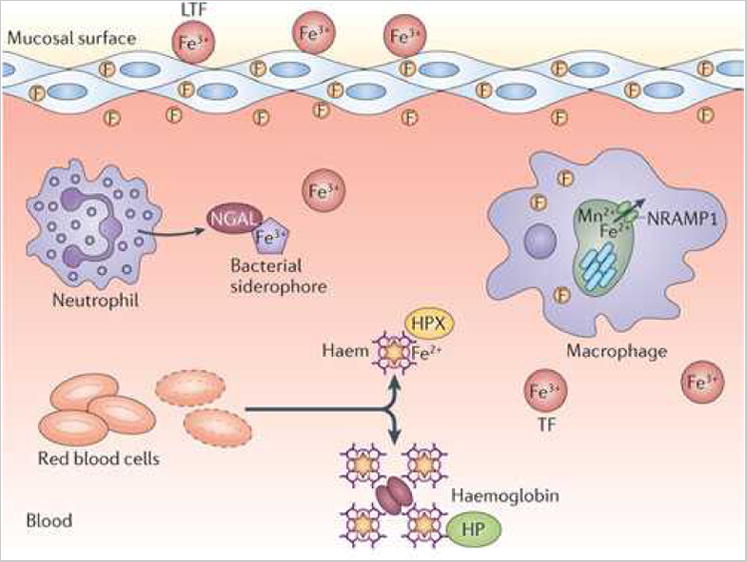
a. Overview of Fe limitation strategies in the vertebrate host. Fe3+ is stored intracellularly in complex with ferritin (F), bound by serum transferrin (TF) or bound by lactoferrin (LTF) at mucosal surfaces. In the blood, Fe2+ is complexed with haem, which is bound by haemoglobin within red blood cells (RBCs). Upon red cell lysis, haemoglobin is bound by haptoglobin (Hpt) and free haem is scavenged by haemopexin (Hpx). In addition to these haem-binding proteins, neutrophil gelatinase-associated lipocalin (NGAL) binds and sequesters bacterial siderophores. b. Representative Fe acquisition systems expressed by Gram-negative and Gram-positive pathogens. Both Gram-negative and Gram-positive pathogens possess systems to acquire Fe3+-siderophores, Fe2+-haem, Fe3+ from transferrin and/or free Fe2+. Not all systems are expressed by the same organism. TF, transferrin; SIP, siderophore interacting protein; HO, haem oxygenase; Hpt, haptoglobin; OM, outer membrane; P, periplasm; IM, inner membrane; CW, cell wall; CM, cytoplasmic membrane.
In addition to haem, Fe is stored intracellularly in the Fe storage protein ferritin and is therefore only accessible to intracellular pathogens following host cell lysis. Moreover, natural resistance-associated macrophage protein 1 (NRAMP1) localizes to the phagosomal membrane where it pumps Fe and Mn out of the phagosomal compartment, further reducing access to these metals by intracellular pathogens that reside within a phagosome 11. At physiological pH, extracellular Fe2+ is oxidized to the insoluble Fe3+ and mobilized by the serum protein transferrin, which binds Fe3+ with exceptionally high affinity. Free Fe3+ is also bound by lactoferrin, a globular glycoprotein of the transferrin family that is present in secretions such as breast milk, tears, and saliva. Notably, lactoferrin is present within the granules of polymorphonuclear leukocytes and is therefore a crucial component of the mucosal innate response to infection 12.
Bacterial iron acquisition
All bacterial pathogens must have mechanisms to circumvent nutritional immunity. In the case of Fe, these strategies are numerous and varied. Perhaps the most elegant strategy to circumvent host-mediated Fe sequestration is that of Borrelia burgdorferi, the causative agent of Lyme disease: by substituting Mn in place of Fe within its Fe-requiring enzymes, this organism does not require Fe to infect its host 13. However, as discussed in subsequent sections, the host encodes additional mechanisms to restrict Mn availability.
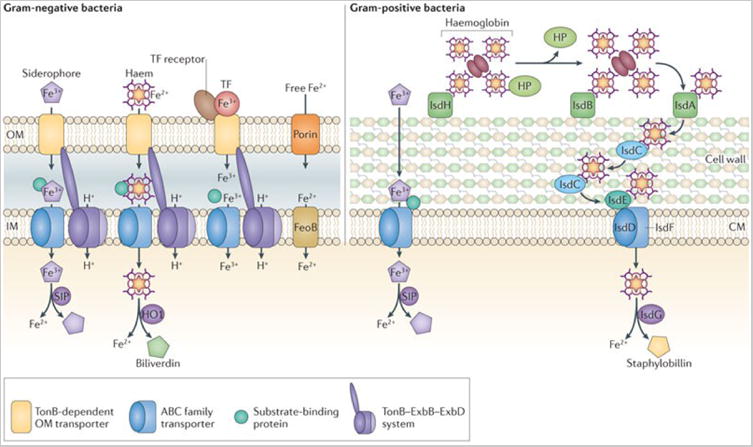
Strategies for Fe acquisition can be generally divided into siderophore, haem and free Fe2+ acquisition systems (Figure 1b). Siderophores are low molecular weight Fe chelators that are secreted by bacteria and bind Fe3+ with an affinity that surpasses that of transferrin and lactoferrin (1023 M−1) 14. As these molecules are too large to diffuse through non-selective porins in the outer membranes of Gram-negative bacteria, energy-dependent transport of siderophores is mediated through TonB-dependent receptors. The periplasm of Gram-negative bacteria lacks ATP or ionic gradients that can drive transport across the outer membrane. Therefore, energy from the proton motive force generated at the inner membrane is harnessed by the TonB-ExbB-ExbD system to mediate outer membrane transport. In the periplasm, substrate binding proteins (SBPs), members of the ATP-binding cassette (ABC) transporter family, recognize the siderophore-Fe complex and ultimately shuttle this complex to the cognate transporter. Gram-positive bacteria also express SBPs, however these proteins are tethered to the cytoplasmic membrane. In both Gram-positive and Gram-negative bacteria, once siderophores are in the cytoplasm, the Fe3+ is released through reduction to Fe2+ or through enzymatic degradation of the siderophore. The end result is the release of Fe2+ for use as a nutrient source. To combat siderophore-mediated Fe3+ acquisition, vertebrates produce neutrophil gelatinase-associated lipocalin (NGAL; also known as lipocalin 2 or siderocalin), which binds and sequesters certain siderophores (Fig. 1a) 15. However, some bacteria produce ‘stealth’ siderophores that evade siderocalin by chemical modification 16, 17.
Haem acquisition systems typically involve a cell surface receptor for either haem or haemoproteins, which pass haem through a membrane transport system into the cytoplasm. There are several well-characterized systems in Gram-negative bacteria 10, 18, 19. In Gram-positive bacteria, the major systems described to date include the Fe-regulated surface determinant (Isd) system found in many Firmicutes as well as the Shr, Shp and HtsABC proteins found in the streptococci 6-8, 20. The first step in haem transport involves binding of haem, haemoglobin, or haemoglobin-haptoglobin complexes by cell wall-anchored receptors (Gram-positive) or TonB-dependent receptors (Gram-negative). Haem is then extracted from haemoglobin and relayed to a SBP associated with a haem-specific ABC family transporter that mediates translocation into the cytoplasm. In addition to surface-bound receptors, some bacteria produce secreted proteins that complex haem, which are known as hemophores and are functionally analogous to siderophores 21, 22. Once bound to haem, haemophores are recognized by haemophore receptors and the haem is internalized.
Upon translocation into the bacterial cytoplasm haem is degraded by haem catabolizing enzymes (Figure 1b). These haem oxygenases can be classified into three different enzyme families. The HO-1 family is evolutionary related to the eukaryotic haem degrading enzymes 23. HO-1 family members are present in both Gram-negative and Gram-positive bacteria and degrade haem to free Fe2+ and biliverdin 23. The IsdG-family haem oxygenases are found in both Gram-negative and Gram-positive bacteria, and these enzymes degrade haem to Fe2+ and staphylobilin, a chromophore24-27. More recently, a third family of haem oxygenases represented by the Campylobacteri jejuni ChuZ enzyme has been described 28, 29; the product of ChuZ-mediated haem degradation is not yet known.
Although Fe is predominantly transported in the chelated form, some bacteria also transport free Fe. In some cases, bacteria express receptors for host Fe-binding proteins such as the transferrin receptors found in Neisseria spp., Morexella catarrhalis and Haemophilus influenzae 30. These TonB-dependent receptors bind transferrin, then extract and transport Fe3+ across the outer membrane. Additionally, some bacteria transport free Fe2+ across the cytoplasmic membrane using the FeoB family of transporters 31-34 (Fig 1b). FeoB is a large membrane protein containing a GTP-binding domain that is similar to eukaryotic G proteins 32. GTPase activity is necessary for Fe2+ transport, and coupling of the G protein and membrane transporter domains within the same protein distinguishes FeoB from eukaryotic G-protein coupled receptors 32. FeoB is typically co-expressed with FeoA, a small SH3-domain protein that is probably found in the cytoplasm, and FeoC, which is thought to act as a Fe-S-dependent repressor 32. Overall, FeoB represents a unique family of bacterial transition metal transporters, members of which are important for virulence in numerous pathogens 31-34.
Nutritional immunity is not a defensive strategy that is exclusive to vertebrates. Mechanisms for Fe restriction exist in plants and invertebrates including the expression of ferritins and transferrins 35-37. In the entomopathogen, Photorhabdus luminescens, Fe availability is an important signal for the switch between symbiotic colonization and pathogenic infection. In these bacteria, certain iron acquisition systems are crucial for virulence but dispensable for symbiosis 38. Similarly, iron acquisition through siderophores serves as a virulence strategy for phytopathogens, while siderophores produced by symbiotic bacteria in the rhizosphere can be beneficial to plants (Box 1). Clearly, iron acquisition is important for both bacterial pathogenesis and symbiosis. Moreover, withholding of iron is a conserved innate immune strategy across multiple kingdoms of life.
Box 1. Roles for Fe in plant-microorganism interactions.
Plants use several strategies to restrict Fe including ferritins and transferrins. In addition, plants express at least four different members of the NRAMP family of Mn and Fe transporters. The role for NRAMP transporters in immunity was first appreciated in vertebrates, where phagosomal NRAMP1 reduces Fe and Mn availability to pathogens within this compartment 11. In Arabidopsis thaliana, AtNRAMP3 and AtNRAMP4 are up-regulated in the plant vacuole in response to Erwinia chrysamthemii infection and are important for Fe transport and host defence 146. It is not clear, however, whether Fe withholding from the bacterium is the primary mechanism of AtNRAMP3/4-mediated plant defences. AtNRAMP3/4 contribute to H2O2 accumulation in infected leaves suggesting that extrusion of Fe from the vacuole to the cytoplasm may be important for generating the oxidative burst 146. The role for nutritional immunity in plant defences is also evidenced by the fact that Fe acquisition systems are required for virulence in a number of phytopathogens 34, 36. Siderophore biosynthesis and uptake systems are the most common mechanisms used by phytopathogens to acquire nutrient Fe from their hosts 35. Beyond facilitating Fe uptake by pathogenic bacteria, siderophores have been shown to exert effects on Fe distribution and overall physiology within the plant host146, 147. These effects include induction of the salicylic acid signalling pathway in leaves and an Fe deficiency response in roots 146, 147. Interestingly, symbiotic bacteria within the rhizosphere also produce siderophores 148. These bacteria promote plant growth and their siderophores are thought to benefit the plants by defending against pathogenic fungal species as well as by enhancing Fe acquisition in roots 35, 148, 149. In addition to siderophores, Xanthomonas oryzae pv. oryzae, which causes bacterial blight in rice, expresses FeoB and this Fe acquisition system is necessary for pathogenesis 34. Clearly, Fe availability is a crucial component of both pathogenic and symbiotic relationships between plants and bacteria.
Mn and Zn in pathogen-host interactions
Nutritional immunity is not limited to strategies for withholding Fe 39. Mn and Zn also play vital roles in bacteria 40. Mn serves a catalytic role in many proteins and is important in oxidative stress resistance 41-44. In some bacteria, Mn2+ can replace the more reactive Fe2+ in Fe-containing proteins, reducing oxidative damage to these proteins 44. Furthermore, Mn-dependent superoxide dismutases are encoded by many pathogens, indicating that these organisms require Mn to defend against superoxide 45. Zn is the second most abundant transition metal in most living systems and can serve both catalytic and structural roles within proteins 46. In fact, it is estimated that Zn-binding proteins represent approximately 4-8 % of all proteins encoded in the genomes of prokaryotes 3. Given these crucial roles for Mn and Zn in bacterial physiology, it is not surprising that sequestration of these nutrient metals is an important innate defense strategy.
Chelating Mn2+ and Zn2+ at mucosal and epithelial surfaces
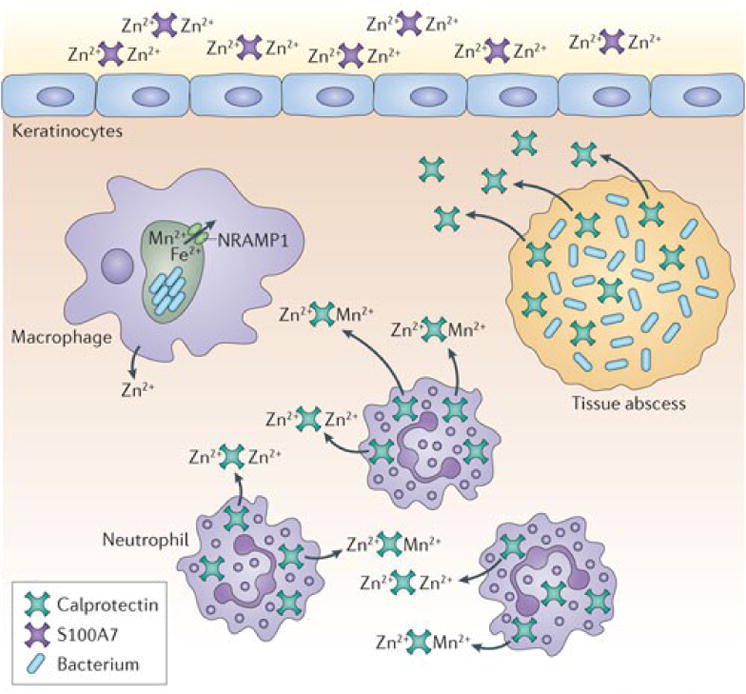
The S100 family of proteins is a large family of calcium binding proteins found in vertebrates, several of which have been implicated in defense against infection (Figure 2a). S100A7, also known as psoriasin, is secreted by keratinocytes and inhibits microbial growth through the chelation of nutrient Zn2+ 47. S100A12, also known as calgranulin C, binds both Zn2+ and Cu2+ in vitro and Cu-S100A12 is involved in the generation of superoxide species 48, 49. It is not yet known whether the antimicrobial properties of this protein result from the generation of superoxide or through nutrient metal sequestration. S100A8 and S100A9 function as a heterodimer known as calprotectin (also referred to as MRP 8/14 and calgranulin A/B). Calprotectin makes up approximately 40% of the protein composition of the neutrophil cytoplasm and is highly antimicrobial against a variety of bacterial and fungal pathogens 50-52. The antibacterial activity of calprotectin results from chelation of nutrient Mn2+ and Zn2+ (Box 2) 50. This chelation is mediated through two high-affinity binding sites both of which can bind Zn2+ with nanomolar affinity while only one binds Mn2+ with such affinity 45. Since the initial report defining a role for calprotectin in protection against Staphylococcus aureus infection, this protein has also been implicated in defence against infection by Salmonella Typhimurium and the fungal pathogens Aspergillus spp. and Candida spp. 51-54. However, as discussed below, some pathogens have evolved elegant mechanisms to counteract or exploit its antimicrobial properties.
Figure 2. Mn and Zn homeostasis at the pathogen-host interface.
a. Zn2+ and Mn2+ sequestration by S100 family proteins at epithelial surfaces and within tissue abscesses. S100A7 is released at epithelial surfaces, where it inhibits bacterial invasion through chelation of Zn2+. In deep tissues, infection leads to recruitment of neutrophils, which deliver calprotectin (S100A8/A9) to the infection site. Calprotectin inhibits bacterial growth through chelation of Mn2+ and Zn2+ and is thought to be transported away from the abscess by an as-yet-unknown mechanism. Engulfment of bacteria by macrophages leads to decreased Zn2+ uptake and increased Zn2+ efflux from the cytoplasm and efflux of Mn2+ and Fe from the phagosome by NRAMP1. b. Representative Mn2+ and Zn2+ uptake systems expressed by pathogenic bacteria. c. Proposed mechanisms of Zn2+ intoxication employed by the host and Zn2+ detoxification systems expressed by pathogens. Upon infection, Zn accumulates in the phagolysosome where it is toxic to bacteria. Gram-negative and Gram-positive bacteria primarily alleviate Zn toxicity through efflux of excess Zn from the cytoplasm. d. Proposed mechanism for Zn toxicity in bacteria. When the extracellular Zn2+: Mn2+ ratio is high, Zn2+ binds the SBP of Mn2+-specific transporters, preventing Mn2+ binding and uptake.
Box 2. Inhibition of bacterial processes through Mn2+ chelation by calprotectin.
Transition metal acquisition is crucial for pathogenesis yet the diverse roles for these metals in bacterial physiology are incompletely defined. The impact of limiting metals on bacterial processes is an area for active investigation. One recent example is the demonstration that Mn2+ chelation by calprotectin inhibits bacterial superoxide defences 45. Staphylococcus aureus encodes two Mn-dependent superoxide dismutases (SODs). Treatment with calprotectin sensitizes S. aureus to superoxide generating compounds by limiting Mn2+ availability and thus reducing SOD activity. Furthermore, S. aureus is more sensitive to neutrophil killing following exposure to calprotectin. These findings lead to a model in which neutrophils deliver a double hit to S. aureus by delivering calprotectin to the site of infection (i) where calprotectin chelates available Mn2+ and Zn2+ (ii). Mn2+ chelation by calprotectin reduces SOD activity (iii), thereby sensitizing S. aureus to ROS generated by the neutrophil (iv) (see Box Figure). Given the important role for Mn in resistance to oxidative stress in other bacteria, as well as the possibility that Zn2+ chelation by calprotectin could inhibit Cu/Zn-SODs, this model may be generally applicable to numerous pathogens 86.
In addition to their metal chelating properties, psoriacin, calprotectin and calgranulin C have proinflammatory properties and serve as markers for many inflammation-mediated pathologies 55. Given the multiple roles for these proteins in nutrient metal chelation and inflammation, it remains to be determined whether the inflammatory properties of these proteins are impacted by metal binding.
Bacterial Mn and Zn acquisition systems
The importance of Mn and Zn acquisition to bacterial pathogenesis has been demonstrated in many organisms including S. Typhimurium, C. jejuni, Yersinia spp., Brucella abortus, H. influenzae, Listeria monocytogenes and Streptococcus pneumoniae 56-66. The mechanisms of Mn2+ and Zn2+ transport across the outer membrane of Gram-negative bacteria are not completely defined. Although previously believed to diffuse through non-selective porins, designated transporters have recently been described suggesting that in some bacteria the outer membrane provides a selective barrier to these essential nutrients (Figure 2b). One such example is MnoP of Bradyrhizobium japonicum, which is a Mn2+-selective channel that facilitates transport of free Mn2+ across the outer membrane 67.
Although transport of Mn2+ through MnoP is thought to be passive, transport of Zn2+ across the outer membrane may be an energy-dependent process powered by the TonB-ExbB-ExbD system. A Zn-regulated TonB-dependent receptor designated ZnuD has been described in Neisseria meningitidis and orthologues of this receptor are encoded in the genomes of several other pathogens 68. ZnuD shares sequence similarity with the haem transporter HumA from Morexalla catarrhalis and facilitates haem acquisition when expressed in Escherichia coli 69. These findings, together with the dual regulation of ZnuD by Zur (Zn uptake regulator) and Fur (Fe uptake regulator), suggest that ZnuD may participate in both Zn2+ and haem acquisition 69. Alternatively, cross-regulation of ZnuD may stem from an increased need for exogenous haem under Zn-limiting conditions, as some endogenous haem biosynthetic enzymes require Zn 70.
It is not yet known whether Zn2+ is transported across the outer membrane as a free ion or as a Zn2+-chelate as is seen for Fe3+. The finding that ZnuD facilitates haem uptake suggests that Zn2+ may be translocated in a chelated form by this transporter. Moreover, transport of a Zn2+-chelate, rather than free Zn2+, might explain why Zn2+ transport is an energy-dependent process, unlike transport of free Mn2+ ions by the energy-independent MnoP channel. Zn2+-chelating compounds analogous to siderophores have not been identified in pathogenic bacteria. However, a putative tsinkosphore (“tsinkos” is greek for Zn) biosynthetic operon, the coelibactin gene cluster, has been identified in the antibiotic-producing bacterium Streptomyces coelicolor. In S. coelicolor this gene cluster is regulated by Zur in a Zn-dependent manner, supporting a role for this cluster and its putative product in the transport of Zn2+ 71. Moreover, pyochelin from Pseudomonas aeruginosa binds Zn2+ and Cu with high affinity in vitro, although transport of Zn2+-pyochelin has not been demonstrated in vivo 72. Nonetheless, the possibility remains that pathogenic bacteria secrete small molecule Zn2+ chelators as a strategy to acquire this nutrient during infection.
The import of Mn2+ and Zn2+ across the cytoplasmic membrane of both Gram-positive and Gram-negative bacteria is primarily facilitated by either ABC-family transporters or NRAMP family transporters 39, 73. These include high-affinity uptake systems such as ZnuABC, AdcBCA and MntABC as well as the NRAMP-family Mn2+ transporter MntH 41, 57, 58, 61, 62, 74, 75. Some FeoB orthologues may also transport Mn2+ 76. Translocation of Mn2+ and Zn2+ across the cytoplasmic membrane by ABC family transporters is analogous to that described above for siderophores and haem. Examples of Mn2+ and Zn2+ acquisition systems are depicted in Figure 2b and listed in Table 1.
Table 1.
Selected bacterial transition metal uptake systems.
| Substrate | Cell surface | Cell wall/periplasm | Cytoplasmic membrane | Pathogens |
|---|---|---|---|---|
| Haem, Haemoglobin, Hpt/haemoglobin | IsdA (haem) | IsdC | IsdDEF | S. aureus 6, 7 |
| IsdB (haemoglobin) | ||||
| IsdH (Hpt/haemoglobin) | ||||
| IsdXI/X221 (secreted hemophores) | B. anthracis 20 | |||
| Shp (haem) | HtsABC | Streptococcus pyogenes 8 | ||
| Shr (methaemoglobin) | ||||
| SvpA | HupDGC | Listeria monocytogenes 8 | ||
| HmuR | HmuT | HmuSUV | Yersinia pestis | |
| PhuR | PhuT | PhuSUV | Pseudomonas aeruginosa | |
| Transferrin (Fe3+) | TbpA (R/OMT), TbpB | Neisseria spp. 129, 130 | ||
| Siderophore | SirABC (Staphyloferrin B) 144 | S. aureus, S. pyogenes | ||
| HtsABC, Fhu (Staphyloferrin A) 145 | S. aureus | |||
| FepA (enterobactin) | FepB | FepCD (ABC-family) | Enterobacteriaceae | |
| FpvA (pyoverdine) | pyoverdine dissociates in periplasm | Pseudomonas aeruginosa | ||
| Fe2+ | FeoB (G protein) | Escherichia coli 32 | ||
| Helicobacter pylori 32 | ||||
| Xanthamonas oryzae pv. Oryzae 34 | ||||
| Campylobacter jejuni 33 | ||||
| Streptococcus suis 31 | ||||
| Mn2+ | MnoP | Bradyrhizobium japonicum 67 | ||
| MntABC | Neisseria gonorrhoeae 41 | |||
| Sit/YfeABCD | S. enterica sv. Typhimurium 56 Yersinia pestis 60, 62 | |||
| PsaABC | Streptococcus pneumoniae 86 | |||
| MntH | Brucella abortus 63 | |||
| Yersinia spp. 61, 62 | ||||
| Zn2+ | ZnuABC | Campylobacter jejuni 59 | ||
| Salmonella spp. 57, 58 | ||||
| ZnuD | ZnuABC* | Neisseria meningitidis 68 | ||
| ZevAB | H. influenzae 64 | |||
| AdcABC, AII | S. pneumoniae 66 | |||
| ZurAM | L. monocytogenes 65 | |||
based on homology
Several Mn2+ and Zn2+ transporters have demonstrated roles in pathogenesis, but a direct role in evading nutrient chelation by calprotectin or other host proteins has generally not been demonstrated. One exception is the recent evidence that S. Typhimurium expressing the ZnuABC zinc uptake system is resistant to calprotectin-mediated Zn2+ chelation 53. This system allows extracellular S. Typhimurium to resist the high levels of calprotectin that accumulate in the intestine following infection. Moreover, S. Typhimurium exploits calprotectin-mediated Zn-chelation in order to outcompete the host microbiota, which is less well adapted to the resulting nutrient-deficient environment 53.
Exploiting Mn and Zn toxicity to kill invading bacteria
In addition to mechanisms for withholding nutrient metals from invading bacteria, there is growing evidence to suggest that in mammals nutritional immunity harnesses the toxic properties of transition metals to kill bacteria 77. It was recently determined that following engulfment of either Mycobacterium tuberculosis or E. coli, macrophages release Zn from intracellular stores which then accumulates in the phagolysosome 78. Survival within the phagolysosome depends on the expression of a Zn2+ efflux system in M. tuberculosis, supporting the idea that bacteria encounter Zn toxicity in vivo and that the ability to resist this toxicity is important for pathogenesis 78. Given that Zn2+ uptake systems are required for the pathogenesis of other intracellular pathogens such as S. enterica, the relative contributions of Zn intoxication and Zn limitation by the host remain to be fully elucidated. The balance between these mechanisms is probably affected by the tissue and cell type, intracellular trafficking of the pathogen and the time point in the infectious cycle, as well as by the pathogen itself and its unique physiological requirements.
Bacterial Mn and Zn detoxification is primarily mediated by P-type ATPases (Figure 2c). These ATP-driven pumps have narrow substrate specificity, which is dictated by a membrane-embedded metal recognition site 79. Zn2+ can also be exported via RND-family transporters that span the inner membrane, periplasm and outer membrane of Gram-negative bacteria. In this case, energy from the proton motive force drives efflux from the cytoplasm or periplasm to the cell exterior. The requirement for Mn and Zn efflux systems in the pathogenesis of several bacteria suggests that bacteria encounter Mn and Zn toxicity in vivo 78, 80, 81. However, the mechanisms by which Mn and Zn cause toxicity are not fully known. Emerging data suggest that maintaining a defined ratio of transition metals is important for bacterial physiology 82, 83. For example, PsaA of S. pneumoniae, a pneumococcal surface protein, binds both Mn2+ and Zn2+ but only transports Mn2+84, 85. Increasing the extracellular ratio of Zn2+: Mn2+ leads to high-affinity binding of Zn2+ to PsaA, blocking Mn2+ uptake and thus potentiating the effects of Mn2+ depletion 83. Additionally, expression of czcD, which encodes a Zn efflux transporter, is up-regulated under Mn-limiting conditions, suggesting that Zn toxicity may be enhanced under Mn-deficient conditions 86.
Cu: new insights into an ancient antibacterial agent
Humans have recognized the antibacterial effects of Cu for millennia and have exploited this property for industrial and medical purposes 87. Despite the long history of Cu use as an antimicrobial, we have only recently begun to appreciate that Cu has a role in innate defence. The accumulation of Cu at sites of infection was first demonstrated in M. tuberculosis pulmonary infections where it was found that Cu resistance is necessary for M. tuberculosis virulence 88. In the mammalian host, bacteria encounter Cu within the phagolysosomes of macrophages. Interferon gamma induces expression of the Cu transporter Ctr1, which actively takes up Cu from the extracellular environment 89. Atox1 then shuttles Cu to ATP7A, a Cu transporter on the phagolysosomal membrane, facilitating Cu accumulation within this compartment 89-91 (Figure 3a).
Figure 3. New insights into the roles for Cu in innate immunity.
a. Mechanisms of Cu intoxication within macrophages. Following phagocytosis of bacteria, interferon gamma induces expression of the Cu+ importer Ctr1. Cu is bound by Atox1 and shuttled to the phagosomal Cu+ transporter, ATP7A. Accumulation of copper within the phagolysosome contributes to bacterial killing through multiple mechanisms including disruption of Fe-S cluster-containing bacterial proteins and possibly through the generation of reactive oxygen species. This leads to inhibition of bacterial metabolic processes and damage to DNA, proteins and lipids. b. Cu detoxification systems expressed by pathogenic bacteria. Bacteria encode multiple mechanisms to detoxify the cytoplasm or periplasm from excess Cu+ including expression of Cu+ efflux systems, periplasmic multicopper oxidases and cytoplasmic Cu chaperones. Many bacteria express several independently regulated Cu detoxification systems, which provide a graded response to Cu toxicity.
The mechanisms of Cu toxicity are not completely understood; however, accumulating evidence suggests it may be multifactorial involving both oxidative damage and disruption of Fe-S clusters. Like Fe, Cu can undergo Fenton chemistry in vitro, reacting with hydrogen peroxide (H2O2) to produce hydroxyl radicals, which in turn damage lipids, proteins and DNA (Figure 3a). Cu enhances the bactericidal capacity of macrophages in vitro, an effect which is further magnified by the addition of H2O2 and reversed by the addition of an antioxidant 89. It should be noted that the ability of Cu to mediate the Fenton reaction in vivo has been debated. Cu does not induce oxidative damage in E. coli, leaving open the possibility that oxidative damage observed in other organisms upon Cu exposure is the result of secondary mechanisms rather than a direct effect of Cu 92. Alternative mechanisms for Cu toxicity have been described. For example, recent characterization of a copA mutant of Neisseria gonorrhoeae revealed a role for Cu in potentiating nitrosative stress 93. CopA is a Cu+ exporter and CopA homologues are found in several other bacterial species. Loss of this protein in N. gonorrhoeae leads to increased sensitivity to Cu and nitrosative stress. Cervical epithelial cells produce nitric oxide (NO) in response to gonococcal infection 94; however, it is not yet known whether gonococci are exposed to extracellular Cu at this site of infection or whether Cu intoxication occurs following engulfment by immune cells. Finally, Cu+ also targets Fe-S clusters in dehydratases involved in processes such as branched-chain amino acid synthesis. A similar mechanism has also been demonstrated for metals such as Cd2+, Hg2+, Ag+ and Zn2+ 95. The resulting disruption of crucial metabolic processes can be reversed by addition of pathway end products in some bacteria 96, 97. Notably, this strategy does not reverse the toxicity associated with Hg2+ and Ag+, nor does it reverse Cu toxicity in an S. Typhimurium mutant lacking CueO, a Cu1+ oxidase 95, 96. As the addition of pathway end products does not reverse Cu toxicity in all bacteria, it is likely that there are multiple mechanisms of Cu toxicity 96. It is likely that the precise intracellular target for Cu depends on the bacterium and the physiological conditions in which Cu is encountered.
Cu acquisition and detoxification in bacteria
The mechanisms of Cu acquisition in bacteria are largely unknown. Methane-oxidizing bacteria (methanotrophs) utilize Cu in methane monooxygenases (MMOs) and Cu accumulation regulates the switch from soluble, Cu-independent sMMO to the membrane-bound, Cu-dependent pMMO form 98-100. Methanotrophs produce Cu-chelating compounds known as chalkophores (“chalko” is Greek for copper) or methanobactins (Mbs) 100-102. Mbs produced by Methylosinus trichosporium str. OB3b bind Cu+ with affinities in the range of 1019 – 1020 M-1 depending on pH 103, 104. The internalization of Mbs depends on the TonB/ExbB/ExbD system and is thus proposed to be analogous to siderophore uptake105. It remains to be determined whether pathogenic bacteria express compounds similar to methanobactin to acquire Cu. In addition to methanobactin-mediated transport, Cu can also be taken up by the cell through energy-independent channels, although the identity and selectivity of such channels is not currently known 105.
In contrast to the dearth of information regarding mechanisms of Cu uptake, the mechanisms of Cu detoxification have been characterized in multiple pathogenic bacteria and these systems are often necessary for pathogenesis 88, 96, 106-108. Bacteria possess several lines of defence against Cu toxicity, beginning with their relatively low physiological need for Cu and the physical localization of Cu-dependent proteins outside the cytoplasm. In addition, bacteria possess multiple mechanisms to detoxify the cytoplasm and periplasm in the presence of excess Cu. In general, this involves expression of cytoplasmic Cu chaperones, Cu exporters and periplasmic multicopper oxidases87. Mycobacterial Cu resistance involves expression of the cytoplasmic Cu chaperone, MymT, the Cu+ transporter, CtpV, and a mycomembrane transporter, MctB88, 108-110 (Figure 4b). Both CtpV and MctB are necessary for full virulence of M. tuberculosis 88, 108. In Gram-negative bacteria, Cu resistance is likewise mediated by the expression of Cu+ exporters. S. Typhimurium expresses two independently regulated P-type ATPases, CopA and GolT. Expression of copA is induced by the transcriptional regulator CueR/SctR in the presence of Cu+ together with genes encoding a putative periplasmic Cu+ chaperone, CueP, and the multicopper oxidase CuiD/CueO96, 111, 112. CuiD/CueO oxidizes Cu+ to Cu2+ in the periplasm and homologues of this protein are necessary for Cu resistance in several bacterial species 96, 111, 113. In addition to CopA, E. coli also expresses an RND-family Cu+ exporter known as CusABC. CopA and CusABC are independently regulated, and expression of these two systems provides a graded response to different levels of Cu toxicity 113.
Figure 4.
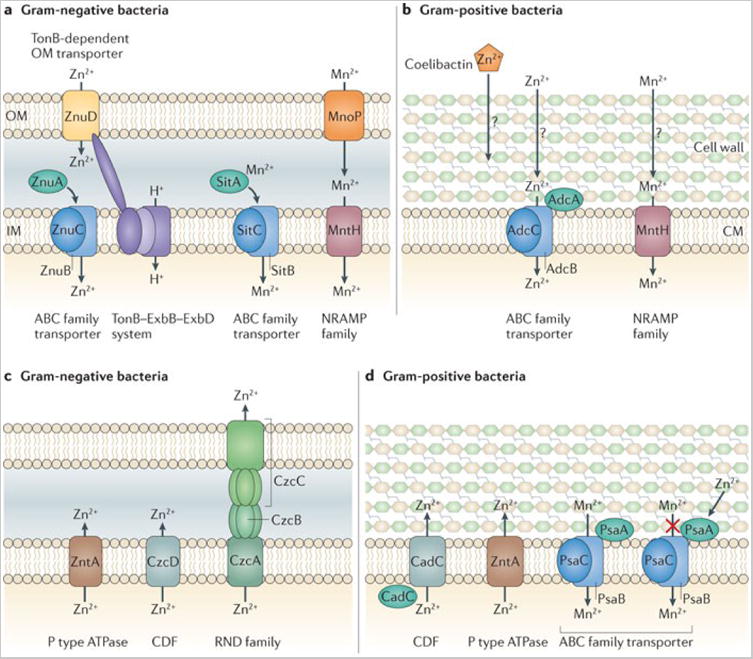
a | Mn2+ and Zn2+ uptake systems in pathogenic Gram-negative bacteria. b | Mn2+ and Zn2+ uptake systems in pathogenic Gram-positive bacteria. c | Proposed mechanisms of Zn2+ detoxification by Gram-negative pathogens. On infection, Zn2+ accumulates in the phagolysosome, where it is toxic to bacteria. Gram-negative bacteria alleviate this Zn2+ toxicity primarily through efflux of excess Zn2+ from the cytoplasm. d | Proposed mechanisms of Zn2+ and Mn2+ detoxification by Gram-positive pathogens. Zn2+ can be exported from the bacterial cytoplasm by dedicated transporters. Mn2+ is usually imported by the ABC family pneumococcal surface adhesin (Psa) system, but when the extracellular Zn2+/Mn2+ ratio is high, Zn2+ binds PsaA, the substrate-binding protein of this Mn2+-specific transporter, preventing Mn2+ binding and uptake. CDF, cation diffusion facilitator; CM, cytoplasmic membrane; IM, inner membrane; MntH, Mn transport H; OM, outer membrane; Znu, Zn uptake protein.
Cu detoxification strategies in Gram-positive bacteria are mostly analogous to those of Gram-negative bacteria. In some cases, a second P-type ATPase, CopB, as well as a putative cytoplasmic Cu chaperone, CopZ, are coexpressed with CopA 114. The multitude of Cu detoxification systems expressed by pathogenic bacteria highlights the importance of Cu intoxication as a host defence strategy and the applicability of Cu as an antimicrobial agent.
Evolutionary perspectives on nutritional immunity
The direct impact of nutritional immunity on human infectious diseases becomes clear when considering patients with inherited defects in transition metal homeostasis. To date, this is primarily restricted to defects in Fe homeostasis, although inherited disorders in Cu (Wilson's disease) and Zn (hyperzincemia and hypercalprotectinemia) homeostasis have also been described. Moreover, polymorphisms in the gene encoding NRAMP1, which may disrupt intracellular trafficking of both Mn and Fe, have been associated with increased susceptibility to intracellular pathogens such as Salmonella and Mycobacterium spp. 115-117 Hypercalprotectinemia, which is thought to be an inherited condition, is associated with autoimmunity but it remains to be determined whether these patients experience alterations in their ability to fight infections 118, 119. By contrast, it is well established that patients with Fe overload conditions are often susceptible to numerous infectious diseases. For example, frequent transfusions in patients with thalassemias and other chronic anaemias lead to excess Fe that predisposes these patients to infections 120, 121. Patients with inherited or acquired forms of the Fe storage disorder, haemochromatosis, are particularly susceptible to infections with enteric Gram-negative pathogens such as Vibrio vulnificus and Yersinia enterocolitica 122, 123. Interestingly, macrophages from patients with the inherited form of haemochromatosis resulting from the C282Y mutation in the gene HFE are very low in Fe. This observation led to the hypothesis that these patients are resistant to infection by intracellular pathogens such as Salmonella typhi, the causative agent of typhoid fever, and M. tuberculosis, the replication of which depends on intracellular Fe pools. If this is the case, resistance to some pathogens may provide evolutionary pressure to maintain this allele within the population 124.
The existence of bacterial receptors that specifically recognize Fe or haem-binding proteins of their preferred or obligate hosts exemplifies the central role for nutritional immunity in the host-pathogen relationship. For example, Staphylococcus aureus IsdB preferentially binds human haemoglobin over haemoglobin from other species, demonstrating that S. aureus has evolved to recognize haemoglobin from its primary host with greater affinity 125, 126. This interaction plays an important role in pathogenesis as S. aureus preferentially utilizes human haemoglobin as an Fe source and transgenic mice expressing human haemoglobin are more susceptible to S. aureus infection in an IsdB-dependent manner 125. The structural basis for IsdB binding to human haemoglobin has not been determined. However, the co-crystal structure of IsdH NEAT domain 1 with human haemoglobin demonstrates interactions with several residues that differ between human and mouse haemoglobin and thus may mediate preferential recognition of the human protein 127. A preference for a particular host Fe source is not unique to S. aureus as other bacterial pathogens that preferentially colonize humans also grow better on human haemoglobin compared to haemoglobin from other species 125. In addition, the obligate human pathogens N. meningitidis and N. gonorrhoeae express the transferrin binding receptors TbpA and TbpB, which preferentially recognize human transferrin 128, 129. This species specificity appears to be dictated by interactions between either TbpA or TbpB with residues on transferrin that are unique to the human protein 130, 131. Both the examples of IsdB from S. aureus and TbpA from Neisseria spp. introduce the intriguing possibility that polymorphisms in human haemoglobin or transferrin may impact susceptibility or resistance to infection.
Conclusions and areas for further research
Advances in our understanding of nutritional immunity and the requirements of pathogens for transition metal homeostasis have led to numerous clinical and industrial applications. Cu has been used to prevent bacterial overgrowth on industrial surfaces and has been introduced into numerous medical devices to reduce the risk of bacterial infections 132-134. In addition, siderophores have been used therapeutically in patients with Fe overload disorders, and chalkophores show promise in treating Wilson's disease, an inherited copper storage disorder 135, 136.
It is clear that nutrient limitation by the host and nutrient acquisition by bacteria are crucial processes in the pathogenesis of infectious diseases. Likewise, transition metal intoxication has emerged as an important component of host defence while bacterial detoxification systems are necessary for pathogenesis. To date, much of the work in nutritional immunity has focused on transition metals. However, bacterial pathogens also rely on their hosts for additional nutrients such as carbon, nitrogen and sulphur. Emerging evidence suggests that successful adaptation to the host environment depends on the ability to take advantage of the available or predominant carbon sources 137-141. This is particularly true of intracellular pathogens which must utilize the nutrient pool in the host cell cytoplasm 142, 143. Unlike nutrient metal restriction by the host, it remains to be determined whether specific mechanisms to limit non-metal nutrients are components of nutritional immunity.
As the field of nutritional immunity progresses many important questions remain open. Bacterial transition metal acquisition systems have been extensively characterized for their roles in virulence. Despite this, the precise mechanisms through which nutrient metal starvation impact bacterial processes has not been clearly defined (Box 2). Bacterial genomes encode a multitude of predicted metal-dependent enzymes, however many of their functions and metal co-factor requirements have not been experimentally validated. Moreover, whether additional host proteins contribute to nutrient limitation, and the contribution of metalloproteins to processes such as immune cell recruitment, trafficking and activation, remain to be determined. The balance between the apparently contradictory strategies of nutrient metal limitation and metal intoxication by the host also remains to be fully elucidated. A more complete understanding of when, where and how these strategies are deployed will resolve this apparent paradox. Metals have a tremendous impact on the outcome of all host-microbe interactions. Therefore, defining the mechanisms and molecular machinery involved in the struggle for nutrient metal has the potential to uncover new therapeutic targets for the treatment of both plant and animal infections.
Figure 5.
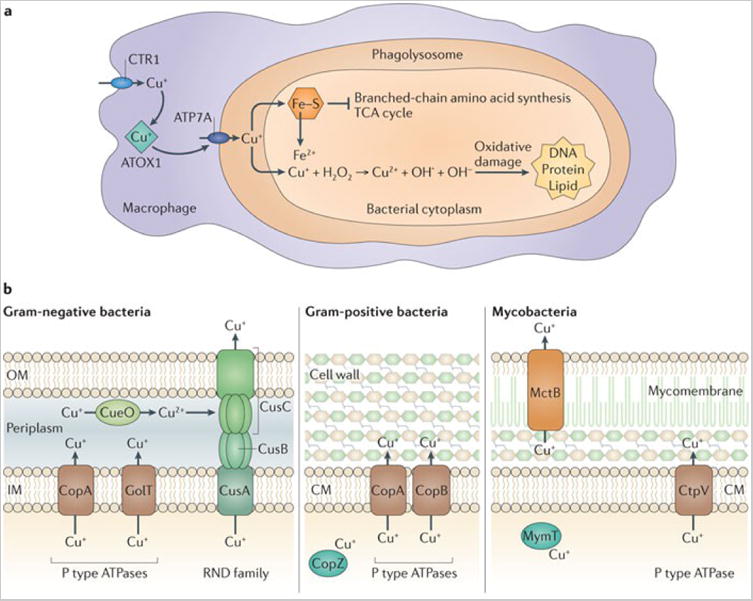
a | Mechanisms of Cu+ intoxication within macrophages. Following phagocytosis of bacteria, interferon-γ induces expression of the Cu+ importer Cu+ transport protein 1 (CTR1). Cu+ is bound by ATOX1 and shuttled to the phagosomal Cu+ transporter, ATP7A. Accumulation of Cu+ within the phagolysosome contributes to bacterial killing through multiple mechanisms, including the disruption of Fe–S cluster-containing bacterial proteins, and possibly the generation of reactive oxygen species. This leads to inhibition of bacterial metabolic processes and damage to DNA, proteins and lipids. b | Pathogenic bacteria encode multiple systems to detoxify the cytoplasm or periplasm when there is excess Cu+, including Cu+ efflux systems (such as the RND family transporter CusABC), periplasmic multicopper oxidases (such as blue Cu oxidase (CueO)) and cytoplasmic Cu+ chaperones (such as metallothionein (MymT)). Many bacteria express several independently regulated Cu+ detoxification systems, providing a graded response to Cu+ toxicity. CM, cytoplasmic membrane; IM, inner membrane; OM, outer membrane; TCA, tricarboxylic acid.
Microbial pathogens require nutrient metals in order to grow and cause disease. However, excess metals are toxic therefore metal levels must be tightly regulated during infection. Vertebrates have evolved to exploit these facts through strategies that either prevent access to nutrient metal or direct excess metal at invading pathogens. Collectively, these processes are known as nutritional immunity.
The struggle for nutrient metal between both host and pathogen is most well studied in the area of iron. In this case, iron is sequestered from invading pathogens intracellularly or in high-affinity iron binding proteins. To combat host-mediated iron sequestration, microbial pathogens elaborate a number of high-affinity iron acquisition systems.
Recently, vertebrate proteins of the innate immune system have been identified that prevent microbial infection through the chelation of nutrient manganese and zinc. These proteins are members of the S100 family of calcium binding proteins and are abundant at sites of inflammation. In addition to manganese and zinc sequestration, vertebrates can employ strategies to direct toxic levels of manganese and zinc at microbial pathogens. Bacterial measures to combat manganese and zinc sequestration as well as toxicity associated with excess manganese and zinc are beginning to be uncovered.
It is becoming increasingly evident that host-mediated direction of excess copper at microbial pathogens is a critical aspect of vertebrate defense against infection. This observation has provided an explanation for the broad conservation of copper detoxification systems across disease causing microbes.
The importance of nutritional immunity to defense against infection is highlighted by the observation that inherited defects in transition metal homeostasis dramatically impacts susceptibility to certain infectious diseases. This fact underscores the tremendous therapeutic potential of targeting bacterial metal acquisition systems.
Glossary of terms
- Transition metals
Elements within groups 3 through 12 of the periodic table whose atoms have an incomplete inner (penultimate) electron shell. These elements exhibit multiple valences due to their incomplete electron shell
- Protoporphyrin
a tetrapyrole ring containing two vinyl, four methyl and two propionic acid side chains. In the case of haem, the tetrapyrrole ring encircles a singular iron atom
- Haptoglobin
A serum protein that binds free haemoglobin and inhibits the oxidative activity of this protein
- Haemopexin
A heme-scavenging protein found in serum that binds heme with high affinity
- Natural resistance associated macrophage protein 1
Divalent cation transporter expressed on the phagosomal membrane
- Biliverdin
A green pigment that is a product of enzymatic heme catabolism
- Staphylobilin
The product of heme catabolism produced by the IsdG family of heme oxygenases
- Superoxide dismutase
An enzyme that catalyzes the formation of hydrogen peroxide from superoxide
- Pyochelin
A siderophore produced by Pseudomonas spp. that binds Fe3+ and some other metals ions with high affinity
- RND-family transporter
Efflux transporters that span the inner and outer membranes of Gram-negative bacteria. These transporters harness the proton gradient at the inner membrane to drive substrate efflux from the cytosol to the extracellular environment
- Fe-S cluster
Complexes of iron and bridging sulfides often found in metalloproteins. Fe-S clusters play structural or functional roles in proteins, most notably in electron transfer reactions and redox sensing
- Fenton reaction
The Fe2+-catalyzed production of hydroxyl radicals from peroxide. Fe2+ + H2O2→Fe3+ + OH• + OH−
- P-type ATPase
a class of autocatalytic ATP-hydrolyzing transporters found in bacteria, archaea and eukaryotes. Most members of this class transport cations
- Hemochromatosis
A condition of iron overload which can result from a primary defect in iron absorption or storage, or which can occur secondary to medical procedures such as blood transfusions
- SH3-domain protein
proteins containing the SRC homology 3 (SH3) domain consisting of five or six beta strands arranged as two tightly packed beta sheets. This domain typically mediates protein-protein interactions by binding to proline-rich regions on the binding partner
- Rhizosphere
The zone immediately surrounding the plant root where biological and chemical interactions occur between the plant, the soil itself and soil microorganisms
References
- 1.Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. Journal of Biological Inorganic Chemistry. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 2.Andreini C, Bertini I, Rosato A. Metalloproteomes: a bioinformatic approach. Accounts of chemical research. 2009;42:1471–1479. doi: 10.1021/ar900015x. [DOI] [PubMed] [Google Scholar]
- 3.Andreini C, Banci L, Bertini I, Rosato A. Zinc through the three domains of life. Journal of Proteome Research. 2006;5:3173–3178. doi: 10.1021/pr0603699. [DOI] [PubMed] [Google Scholar]
- 4.Weinberg ED. Nutritional immunity. Host's attempt to withold iron from microbial invaders. Journal ofthe American Medical Association. 1975;231:39–41. doi: 10.1001/jama.231.1.39. [DOI] [PubMed] [Google Scholar]
- 5.Weinberg ED. Iron availability and infection. Biochimica et Biophysica Acta. 2009;1790:600–605. doi: 10.1016/j.bbagen.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Cassat JE, Skaar EP, et al. Mion acquisition in Staphylococcus aureus: overcoming nutritional immunity. Seminars in Immunopathology. 2011:1–21. doi: 10.1007/s00281-011-0294-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haley KP, Skaar EP. A battle for iron: host sequestration and Staphylococcus aureus acquisition. Microbes and Infection. 2011:1–11. doi: 10.1016/j.micinf.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nobles CL, Maresso AW. The theft of host heme by Gram-positive pathogenic bacteria. Metallomics. 2011;3:788–796. doi: 10.1039/c1mt00047k. [DOI] [PubMed] [Google Scholar]
- 9.Ong ST, Shan Ho JZ, Ho B, Ding JL. Iron-withholding strategy in innate immunity. Immunobiology. 2006;211:295–314. doi: 10.1016/j.imbio.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Braun V, Hantke K. Recent insights into iron import by bacteria. Current Opinion in Chemical Biology. 2011;15:328–334. doi: 10.1016/j.cbpa.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Jabado N, et al. Natural resistance to intracellular infections: natural resistance-associated macrophage protein 1 (Nramp1) functions as a pH-dependent manganese transporter at the phagosomal membrane. The Journal of Experimental Medicine. 2000;192:1237–1248. doi: 10.1084/jem.192.9.1237. This paper uses a fluorescence-based assay to demonstrate that Nramp1 protects against infection by extrusion of divalent cations from the phagosome. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schaible UE, Collins HL, Priem F, Kaufmann SHE. Correction of the iron overload defect in beta-2-microglobulin knockout mice by lactoferrin abolishes their increased susceptibility to tuberculosis. The Journal of Experimental Medicine. 2002;196:1507–1513. doi: 10.1084/jem.20020897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Posey JE, Gherardini FC. Lack of a role for iron in the Lyme disease pathogen. Science. 2000;288:1651–1653. doi: 10.1126/science.288.5471.1651. An unprecedented demonstration of a human pathogen that has circumvented aspects of nutritional immunity by evolving to not require iron. [DOI] [PubMed] [Google Scholar]
- 14.Schalk IJ. Metal trafficking via siderophores in Gram-negative bacteria: specificities and characteristics of the pyoverdine pathway. Journal of Inorganic Biochemistry. 2008;102:1159–1169. doi: 10.1016/j.jinorgbio.2007.11.017. [DOI] [PubMed] [Google Scholar]
- 15.Flo TH, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–921. doi: 10.1038/nature03104. The primary demonstration that lipocalin-2-mediated binding of catecholate siderophores is critical to the innate immune response to bacterial infection. [DOI] [PubMed] [Google Scholar]
- 16.Abergel RJ, et al. Anthrax pathogen evades the mammalian immune system through stealth siderophore production. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18499–18503. doi: 10.1073/pnas.0607055103. A molecular explanation for the observation that the Bacillus anthracis siderophore petrobactin is required for infection whereas bacillibactin is not. The unusual 3,4-dihydroxybenzoyl chelating subunit of petrobactin prevents siderocalin binding establishing petrobactin as a “stealth siderophore”. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hantke K, Nicholson G, Rabsch W, Winkelmann G. Salmochelins, siderophores of Salmonella enterica and uropathogenic Escherichia coli strains, are recognized by the outer membrane receptor IroN. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:3677–3682. doi: 10.1073/pnas.0737682100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cornelis P. Iron uptake and metabolism in pseudomonads. Applied Microbiology and Biotechnology. 2010;86:1637–1645. doi: 10.1007/s00253-010-2550-2. [DOI] [PubMed] [Google Scholar]
- 19.Ratcliff-Griffin M, Wilks A, Stojiljkovic I. In: Iron transport in bacteria: molecular genetics, biochemistry, microbial pathogenesis and ecology. Crosa JH, Mey AR, Payne SM, editors. ASM Press; Washington, D.C.: 2004. pp. 86–94. [Google Scholar]
- 20.Honsa ES, Maresso AW. Mechanisms of iron import in anthrax. Biometals. 2011;24:533–545. doi: 10.1007/s10534-011-9413-x. [DOI] [PubMed] [Google Scholar]
- 21.Fabian M, Solomaha E, Olson JS, Maresso AW. Heme transfer to the bacterial cell envelope occurs via a secreted hemophore in the Gram-positive pathogen Bacillus anthracis. Journal of Biological Chemistry. 2009;284:32138–32146. doi: 10.1074/jbc.M109.040915. A functional analysis of the only known secreted hemophore produced by Gram-positive pathogens. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cescau S, et al. Heme acquisition by hemophores. Biometals. 2007;20:603–613. doi: 10.1007/s10534-006-9050-y. [DOI] [PubMed] [Google Scholar]
- 23.Wilks A. Heme oxygenase: evolution, structure, and mechanism. Antioxidants & Redox Signaling. 2002;4:603–614. doi: 10.1089/15230860260220102. [DOI] [PubMed] [Google Scholar]
- 24.Puri S, O'Brian MR. The hmuQ and hmuD genes from Bradyrhizobium japonicum encode heme-degrading enzymes. Journal of Bacteriology. 2006;188:6476–6482. doi: 10.1128/JB.00737-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chim N, Iniguez A, Nguyen TQ, Goulding CW. Unusual diheme conformation of the heme-degrading protein from Mycobacterium tuberculosis. Journal of Molecular Biology. 2010;395:595–608. doi: 10.1016/j.jmb.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haley KP, Janson EM, Heilbronner S, Foster TJ, Skaar EP. Staphylococcus lugdunensis IsdG liberates iron from host heme. Journal of Bacteriology. 2011;193:4749–4757. doi: 10.1128/JB.00436-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reniere ML, et al. The IsdG-family of haem oxygenases degrades haem to a novel chromophore. Molecular Microbiology. 2010;75:1529–1538. doi: 10.1111/j.1365-2958.2010.07076.x. Structural elucidation of staphylobilin, which is the enzymatic degradation product of the IsdG-family of heme degrading enzymes and the only product of enzymatic heme degradation that is distinct from biliverdin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang R, et al. Crystallization and preliminary crystallographic studies of Campylobacter jejuni ChuZ, a member of a novel haem oxygenase family. Acta crystallographica. 2011;67:1228–1230. doi: 10.1107/S1744309111026194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang R, et al. Crystal structure of Campylobacter jejuni ChuZ: a split-barrel family heme oxygenase with a novel heme-binding mode. Biochemical and Biophysical Research Communications. 2011;415:82–87. doi: 10.1016/j.bbrc.2011.10.016. [DOI] [PubMed] [Google Scholar]
- 30.Cornelissen C. Transferrin-iron uptake by Gram-negative bacteria. Frontiers in Bioscience. 2003 doi: 10.2741/1076. [DOI] [PubMed] [Google Scholar]
- 31.Aranda J, et al. Contribution of the FeoB transporter to Streptococcus suis virulence. International Microbiology. 2009;12:137–143. [PubMed] [Google Scholar]
- 32.Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. Feo – Transport of ferrous iron into bacteria. Biometals. 2006;19:143–157. doi: 10.1007/s10534-006-0003-2. [DOI] [PubMed] [Google Scholar]
- 33.Panciera R, Marlow D, Stintzi A. Major role for FeoB in Campylobacter jejuni ferrous iron acquisition, gut colonization, and intracellular survival. Infection and Immunity. 2006;74:5433–44. doi: 10.1128/IAI.00052-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pandey A, Sonti RV. Role of the FeoB protein and siderophore in promoting virulence of Xanthomonas oryzae pv. oryzae on rice. Journal of Bacteriology. 2010;192:3187–3203. doi: 10.1128/JB.01558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemanceau P, Expert D, Gaymard F, Bakker PAHM, Briat JF. Chapter 12 - Role of iron in plant-microbe interactions. Plant Innate Immunity. 2009;51:491–549. [Google Scholar]
- 36.Expert D. Withholding and exchanging iron: interactions between Erwinia spp. and their plant hosts. Annual Review of Phytopathology. 1999;37:307–334. doi: 10.1146/annurev.phyto.37.1.307. [DOI] [PubMed] [Google Scholar]
- 37.Geiser DL, Winzerling JJ. Insect transferrins: Multifunctional proteins. Biochimica et Biophysica Acta. 2012;1820:437–451. doi: 10.1016/j.bbagen.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 38.Watson RJ, Millichap P, Joyce SA, Reynolds S, Clarke DJ. The role of iron uptake in pathogenicity and symbiosis in Photorhabdus luminescens TT01. BMC microbiology. 2010;10:177. doi: 10.1186/1471-2180-10-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kehl-Fie TE, Skaar EP. Nutritional immunity beyond iron: a role for manganese and zinc. Current Opinion in Chemical Biology. 2010;14:218–224. doi: 10.1016/j.cbpa.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nies DH, Grass G. In: EcoSal—Escherichia coli and Salmonella: Cellular and Molecular Biology. Böck A, et al., editors. ASM Press; Washington, D.C.; [Google Scholar]
- 41.Tseng HJ, Srikhanta Y, McEwan AG, Jennings MP. Accumulation of manganese in Neisseria gonorrhoeae correlates with resistance to oxidative killing by superoxide anion and is independent of superoxide dismutase activity. Molecular Microbiology. 2001;40:1175–1186. doi: 10.1046/j.1365-2958.2001.02460.x. [DOI] [PubMed] [Google Scholar]
- 42.Anjem A, Varghese S, Imlay JA. Manganese import is a key element of the OxyR response to hydrogen peroxide in Escherichia coli. Molecular Microbiology. 2009;72:844–858. doi: 10.1111/j.1365-2958.2009.06699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin JE, Imlay JA. The alternative aerobic ribonucleotide reductase of Escherichia coli, NrdEF, is a manganese-dependent enzyme that enables cell replication during periods of iron starvation. Molecular Microbiology. 2011;80:319–334. doi: 10.1111/j.1365-2958.2011.07593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sobota JM, Imlay JA. Iron enzyme ribulose-5-phosphate 3-epimerase in Escherichia coli is rapidly damaged by hydrogen peroxide but can be protected by manganese. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:5402–5407. doi: 10.1073/pnas.1100410108. This manuscript demonstrates that critical enzymes that utilize iron as a cofactor are primary targets of hydrogen peroxide stress, and bacteria can protect against this stress by shifting from iron- to manganese-centered metabolism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kehl-Fie TE, et al. Nutrient metal sequestration by calprotectin inhibits bacterial superoxide defense, enhancing neutrophil killing of Staphylococcus aureus. Cell Host and Microbe. 2011;10:158–164. doi: 10.1016/j.chom.2011.07.004. This paper reveals that innate immune-mediated manganese chelation inactivates bacterial defenses against oxidative stress at the same time that the neutrophil attacks the invading pathogen with the oxidative burst. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hantke K. Bacterial zinc uptake and regulators. Current Opinion in Microbiology. 2005;8:196–202. doi: 10.1016/j.mib.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 47.Gläser R, et al. Antimicrobial psoriasin (S100A7) protects human skin from Escherichia coli infection. Nature Immunology. 2005;6:57–64. doi: 10.1038/ni1142. [DOI] [PubMed] [Google Scholar]
- 48.Moroz OV, et al. Structure of the human S100A12-copper complex: implications for host-parasite defence. Acta crystallographica Section D, Biological crystallography. 2003;59:859–867. doi: 10.1107/s0907444903004700. [DOI] [PubMed] [Google Scholar]
- 49.Moroz OV, et al. Both Ca2+ and Zn2+ are essential for S100A12 protein oligomerization and function. BMC Biochemistry. 2009;10:11. doi: 10.1186/1471-2091-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Corbin BD, et al. Metal chelation and inhibition of bacterial growth in tissue abscesses. Science. 2008;319:962–965. doi: 10.1126/science.1152449. This paper is the first demonstration that calprotectin protects against infection through nutrient metal chelation, establishing calprotectin as the only known manganese chelating protein of the innate immune system. [DOI] [PubMed] [Google Scholar]
- 51.McCormick A, et al. NETs formed by human neutrophils inhibit growth of the pathogenic mold Aspergillus fumigatus. Microbes and infection. 2010;12:928–936. doi: 10.1016/j.micinf.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 52.Urban CF, et al. Neutrophil extracellular traps contain calprotectin, a cytosolic protein complex involved in host defense against Candida albicans. PLoS Pathogens. 2009;5:e1000639. doi: 10.1371/journal.ppat.1000639. A seminal finding that calprotectin is an abundant component of neutrophil extracellular traps (NETs) and protects against fungal infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu JZ, et al. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host and Microbe. 2012;11:227–39. doi: 10.1016/j.chom.2012.01.017. The first demonstration of a bacterial pathogen that exploits calprotectin to provide a growth advantage over competing commensal bacteria. Specifically, Salmonella uses high affinity zinc acquisition systems to overcome calprotectin-mediated zinc chelation and thrive in the inflamed gut. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bianchi M, Niemiec MJ, Siler U, Urban CF, Reichenbach J. Restoration of anti-Aspergillus defense by neutrophil extracellular traps in human chronic granulomatous disease after gene therapy is calprotectin-dependent. The Journal of Allergy and Clinical Immunology. 2011;127:1243–52. e7. doi: 10.1016/j.jaci.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 55.Hsu K, et al. Anti-infective protective properties of S100 calgranulins. Antiinflammatory & Anti-allergy Agents in Medicinal Chemistry. 2009;8:290–305. doi: 10.2174/187152309789838975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kehres DG, Janakiraman A, Slauch JM, Maguire ME. SitABCD is the alkaline Mn (2+) transporter of Salmonella enterica serovar Typhimurium. Journal of Bacteriology. 2002;184:3159–3166. doi: 10.1128/JB.184.12.3159-3166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ammendola S, et al. High-affinity Zn2+ uptake system ZnuABC is required for bacterial zinc homeostasis in intracellular environments and contributes to the virulence of Salmonella enterica. Infection and Immunity. 2007;75:5867–5876. doi: 10.1128/IAI.00559-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Campoy S, et al. Role of the high-affinity zinc uptake znuABC system in Salmonella enterica serovar typhimurium virulence. Infection and Immunity. 2002;70:4721–4725. doi: 10.1128/IAI.70.8.4721-4725.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davis LM, Kakuda T, DiRita VJ. A Campylobacter jejuni znuA orthologue is essential for growth in low-zinc environments and chick colonization. Journal of Bacteriology. 2009;191:1631–1640. doi: 10.1128/JB.01394-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bearden SW, Perry RD. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Molecular Microbiology. 1999;32:403–414. doi: 10.1046/j.1365-2958.1999.01360.x. [DOI] [PubMed] [Google Scholar]
- 61.Champion OL, et al. Yersinia pseudotuberculosis mntH functions in intracellular manganese accumulation, which is essential for virulence and survival in cells expressing functional Nramp1. Microbiology. 2011;157:1115–1122. doi: 10.1099/mic.0.045807-0. [DOI] [PubMed] [Google Scholar]
- 62.Perry RD, et al. Manganese transporters Yfe and MntH are Fur regulated and important for the virulence of Yersinia pestis. Microbiology. 2012;158:804–15. doi: 10.1099/mic.0.053710-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anderson ES, et al. The manganese transporter MntH is a critical virulence determinant for Brucella abortus 2308 in experimentally infected mice. Infection and Immunity. 2009;77:3466–3474. doi: 10.1128/IAI.00444-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rosadini CV, Gawronski JD, Raimunda D, Argüello JM, Akerley BJ. A novel zinc binding system, ZevAB, is critical for survival of nontypeable Haemophilus influenzae in a murine lung infection model. Infection and Immunity. 2011;79:3366–3376. doi: 10.1128/IAI.05135-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Corbett D, et al. Two zinc uptake systems contribute to the full virulence of Listeria monocytogenes during growth in vitro and in vivo. Infection and Immunity. 2012;80:14–21. doi: 10.1128/IAI.05904-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bayle L, et al. Zinc uptake by Streptococcus pneumoniae depends on both AdcA and AdcAII and is essential for normal bacterial morphology and virulence. Molecular Microbiology. 2011;82:904–916. doi: 10.1111/j.1365-2958.2011.07862.x. [DOI] [PubMed] [Google Scholar]
- 67.Hohle TH, Franck WL, Stacey G, O'Brian MR. Bacterial outer membrane channel for divalent metal ion acquisition. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:15390–15395. doi: 10.1073/pnas.1110137108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stork M, et al. An outer membrane receptor of Neisseria meningitidis involved in zinc acquisition with vaccine potential. PLoS Pathogens. 2010;6:e1000969. doi: 10.1371/journal.ppat.1000969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumar P, Sannigrahi S, Tzeng YL. The Neisseria meningitidis ZnuD zinc receptor contributes to interactions with epithelial cells and supports heme utilization when expressed in Escherichia coli. Infection and Immunity. 2012;80:657–667. doi: 10.1128/IAI.05208-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li JM, Russell CS, Cosloy SD. The structure of the Escherichia coli hemB gene. Gene. 1989;75:177–184. doi: 10.1016/0378-1119(89)90394-6. [DOI] [PubMed] [Google Scholar]
- 71.Kallifidas D, et al. The zinc-responsive regulator Zur controls expression of the coelibactin gene cluster in Streptomyces coelicolor. Journal of Bacteriology. 2010;192:608–611. doi: 10.1128/JB.01022-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brandel J, et al. Pyochelin, a siderophore of Pseudomonas aeruginosa: Physicochemical characterization of the iron(iii), copper(ii) and zinc(ii) complexes. Dalton Transactions. 2012;41:2820–2834. doi: 10.1039/c1dt11804h. [DOI] [PubMed] [Google Scholar]
- 73.Klein JS, Lewinson O. Bacterial ATP-driven transporters of transition metals: physiological roles, mechanisms of action, and roles in bacterial virulence. Metallomics. 2011;3:1098. doi: 10.1039/c1mt00073j. [DOI] [PubMed] [Google Scholar]
- 74.Andresen E, et al. S100A7/psoriasin expression in the human lung: unchanged in patients with COPD, but upregulated upon positive S. aureus detection. BMC pulmonary medicine. 2011;11:10. doi: 10.1186/1471-2466-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nielubowicz GR, Smith SN, Mobley HLT. Zinc uptake contributes to motility and provides a competitive advantage to Proteus mirabilis during experimental urinary tract infection. Infection and Immunity. 2010;78:2823–2833. doi: 10.1128/IAI.01220-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dashper SG, et al. A novel Porphyromonas gingivalis FeoB plays a role in manganese accumulation. The Journal of Biological Chemistry. 2005;280:28095–28102. doi: 10.1074/jbc.M503896200. [DOI] [PubMed] [Google Scholar]
- 77.Botella H, Stadthagen G, Lugo-Villarino G, de Chastellier C, Neyrolles O. Metallobiology of host-pathogen interactions: an intoxicating new insight. Trends in microbiology. 2012;20:106–112. doi: 10.1016/j.tim.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 78.Botella H, et al. Mycobacterial P1-type ATPases mediate resistance to zinc poisoning in human macrophages. Cell Host and Microbe. 2011;10:248–259. doi: 10.1016/j.chom.2011.08.006. This paper reports the surprising finding that zinc is directed at Mycobacterium tuberculosis within the phagosome and M. tuberculosis neutralizes the toxic effects of zinc accumulation through efflux. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hou ZJ, Narindrasorasak S, Bhushan B, Sarkar B, Mitra B. Functional analysis of chimeric proteins of the Wilson Cu(I)-ATPase (ATP7B) and ZntA, a Pb(II)/Zn(II)/Cd(II)-ATPase from Escherichia coli. The Journal of Biological Chemistry. 2001;276:40858–40863. doi: 10.1074/jbc.M107455200. [DOI] [PubMed] [Google Scholar]
- 80.Veyrier FJ, Boneca IG, Cellier MF, Taha MK. A novel metal transporter mediating manganese export (MntX) regulates the Mn to Fe intracellular ratio and Neisseria meningitidis virulence. PLoS Pathogens. 2011;7:e1002261. doi: 10.1371/journal.ppat.1002261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rosch JW, Gao G, Ridout G, Wang YD, Tuomanen EI. Role of the manganese efflux system mntE for signalling and pathogenesis in Streptococcus pneumoniae. Molecular Microbiology. 2009;72:12–25. doi: 10.1111/j.1365-2958.2009.06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jacobsen FE, Kazmierczak KM, Lisher JP, Winkler ME, Giedroc DP. Interplay between manganese and zinc homeostasis in the human pathogen Streptococcus pneumoniae. Metallomics. 2011;3:38–41. doi: 10.1039/c0mt00050g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McDevitt CA, et al. A molecular mechanism for bacterial susceptibility to zinc. PLoS Pathogens. 2011;7:e1002357. doi: 10.1371/journal.ppat.1002357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dintilhac A, Alloing G, Granadel C, Claverys JP. Competence and virulence of Streptococcus pneumoniae: Adc and PsaA mutants exhibit a requirement for Zn and Mn resulting from inactivation of putative ABC metal permeases. Molecular Microbiology. 1997;25:727–739. doi: 10.1046/j.1365-2958.1997.5111879.x. [DOI] [PubMed] [Google Scholar]
- 85.Lawrence MC, et al. The crystal structure of pneumococcal surface antigen PsaA reveals a metal-binding site and a novel structure for a putative ABC-type binding protein. Structure. 1998;6:1553–1561. doi: 10.1016/s0969-2126(98)00153-1. [DOI] [PubMed] [Google Scholar]
- 86.Ogunniyi AD, et al. Central role of manganese in regulation of stress responses, physiology, and metabolism in Streptococcus pneumoniae. Journal of Bacteriology. 2010;192:4489–4497. doi: 10.1128/JB.00064-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Samanovic MI, Ding C, Thiele DJ, Darwin KH. Copper in microbial pathogenesis: meddling with the metal. Cell Host and Microbe. 2012;11:106–115. doi: 10.1016/j.chom.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wolschendorf F, et al. Copper resistance is essential for virulence of Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:1621–1626. doi: 10.1073/pnas.1009261108. This paper reports that copper transport proteins are critical for Mycobacterial Cu resistance and infection in animal models. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.White C, Lee J, Kambe T, Fritsche K, Petris MJ. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. Journal of Biological Chemistry. 2009;284:33949–33956. doi: 10.1074/jbc.M109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim BE, Nevitt T, Thiele DJ. Mechanisms for copper acquisition, distribution and regulation. Nature Chemical Biology. 2008;4:176–185. doi: 10.1038/nchembio.72. [DOI] [PubMed] [Google Scholar]
- 91.Kim HW, et al. Human macrophage ATP7A is localized in the trans-Golgi apparatus, controls intracellular copper levels, and mediates macrophage responses to dermal wounds. Inflammation. 2011 doi: 10.1007/s10753-011-9302-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Macomber L, Rensing C, Imlay JA. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. Journal of bacteriology. 2007;189:1616–1626. doi: 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Djoko KY, et al. Phenotypic characterization of a copA mutant of Neisseria gonorrhoeae identifies a link between copper and nitrosative stress. Infection and Immunity. 2012;80:1065–1071. doi: 10.1128/IAI.06163-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Edwards JL. Neisseria gonorrhoeae survival during primary human cervical epithelial cell infection requires nitric oxide and is augmented by progesterone. Infection and Immunity. 2010;78:1202–1213. doi: 10.1128/IAI.01085-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu FF, Imlay JA. Silver(I), mercury(II), cadmium(II), and zinc(II) target exposed enzymic iron-sulfur clusters when they toxify Escherichia coli. Applied and environmental microbiology. 2012 doi: 10.1128/AEM.07368-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Achard MES, et al. The multi-copper-ion oxidase CueO of Salmonella enterica serovar Typhimurium is required for systemic virulence. Infection and Immunity. 2010;78:2312–2319. doi: 10.1128/IAI.01208-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Macomber L, Imlay JA. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8344–8349. doi: 10.1073/pnas.0812808106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Knapp CW, Fowle DA, Kulczycki E, Roberts JA, Graham DW. Methane monooxygenase gene expression mediated by methanobactin in the presence of mineral copper sources. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12040–12045. doi: 10.1073/pnas.0702879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hakemian AS, Rosenzweig AC. The biochemistry of methane oxidation. Annual Review of Biochemistry. 2007;76:223–241. doi: 10.1146/annurev.biochem.76.061505.175355. [DOI] [PubMed] [Google Scholar]
- 100.Kenney GE, Rosenzweig AC. Chemistry and biology of the copper chelator methanobactin. ACS Chemical Biology. 2011:120124121712008. doi: 10.1021/cb2003913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim HJ. Methanobactin, a copper-acquisition compound from methane-oxidizing bacteria. Science. 2004;305:1612–1615. doi: 10.1126/science.1098322. [DOI] [PubMed] [Google Scholar]
- 102.Balasubramanian R, Rosenzweig AC. Copper methanobactin: a molecule whose time has come. Current Opinion in Chemical Biology. 2008;12:245–249. doi: 10.1016/j.cbpa.2008.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hakemian AS, et al. The copper chelator methanobactin from Methylosinus trichosporium OB3b binds copper(I) Journal of the American Chemical Society. 2005;127:17142–17143. doi: 10.1021/ja0558140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.El Ghazouani A, et al. Copper-binding properties and structures of methanobactins from Methylosinus trichosporium OB3b. Inorganic Chemistry. 2011;50:1378–1391. doi: 10.1021/ic101965j. [DOI] [PubMed] [Google Scholar]
- 105.Balasubramanian R, Kenney GE, Rosenzweig AC. Dual pathways for copper uptake by methanotrophic bacteria. Journal of Biological Chemistry. 2011;286:37313–37319. doi: 10.1074/jbc.M111.284984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shafeeq S, et al. The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Molecular Microbiology. 2011;81:1255–1270. doi: 10.1111/j.1365-2958.2011.07758.x. [DOI] [PubMed] [Google Scholar]
- 107.Sharan R, Chhibber S, Reed RH. A murine model to study the antibacterial effect of copper on infectivity of Salmonella enterica serovar Typhimurium. International Journal of Environmental Research and Public Health. 2011;8:21–36. doi: 10.3390/ijerph8010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ward SK, Abomoelak B, Hoye EA, Steinberg H, Talaat AM. CtpV: a putative copper exporter required for full virulence of Mycobacterium tuberculosis. Molecular Microbiology. 2010;77:1096–1110. doi: 10.1111/j.1365-2958.2010.07273.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu T, et al. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nature Chemical Biology. 2006;3:60–68. doi: 10.1038/nchembio844. A seminal paper describing the identification of a large, previously uncharacterized family of transcriptional regulators highlighted by the copper-specific repressor CsoR. [DOI] [PubMed] [Google Scholar]
- 110.Festa RA, et al. A novel copper-responsive regulon in Mycobacterium tuberculosis. Molecular Microbiology. 2010;79:133–148. doi: 10.1111/j.1365-2958.2010.07431.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kim JS, et al. The sctR of Salmonella enterica serova Typhimurium encoding a homologue of MerR protein is involved in the copper-responsive regulation of cuiD. FEMS Microbiology Letters. 2002;210:99–103. doi: 10.1111/j.1574-6968.2002.tb11166.x. [DOI] [PubMed] [Google Scholar]
- 112.Osman D, et al. Copper homeostasis in Salmonella is atypical and copper-CueP is a major periplasmic metal complex. Journal of Biological Chemistry. 2010;285:25259–25268. doi: 10.1074/jbc.M110.145953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Outten FW, Huffman DL, Hale JA, O'Halloran TV. The independent cue and cus systems confer copper tolerance during aerobic and anaerobic growth in Escherichia coli. The Journal of Biological Chemistry. 2001;276:30670–30677. doi: 10.1074/jbc.M104122200. [DOI] [PubMed] [Google Scholar]
- 114.Lu ZH, Dameron CT, Solioz M. The Enterococcus hirae paradigm of copper homeostasis: copper chaperone turnover, interactions, and transactions. Biometals. 2003;16:137–143. doi: 10.1023/a:1020709307589. [DOI] [PubMed] [Google Scholar]
- 115.Hsu YH, et al. Association of NRAMP 1 gene polymorphism with susceptibility to tuberculosis in Taiwanese aboriginals. Journal of the Formosan Medical Association = Taiwanyizhi. 2006;105:363–369. doi: 10.1016/S0929-6646(09)60131-5. [DOI] [PubMed] [Google Scholar]
- 116.Huang JH, et al. Analyses of the NRAMP1 and IFN-gammaR1 genes in women with Mycobacterium avium-intracellulare pulmonary disease. American journal of respiratory and critical care medicine. 1998;157:377–381. doi: 10.1164/ajrccm.157.2.9706012. [DOI] [PubMed] [Google Scholar]
- 117.Tanaka G, et al. Pulmonary Mycobacterium avium complex infection: association with NRAMP1 polymorphisms. The European respiratory journal: official journal of the European Society for Clinical Respiratory Physiology. 2007;30:90–96. doi: 10.1183/09031936.00042506. [DOI] [PubMed] [Google Scholar]
- 118.Isidor B, et al. Hyperzincemia and hypercalprotectinemia: unsuccessful treatment with tacrolimus. Acta paediatrica (Oslo, Norway: 1992) 2009;98:410–412. doi: 10.1111/j.1651-2227.2008.01092.x. [DOI] [PubMed] [Google Scholar]
- 119.Saito Y, et al. Hyperzincemia with systemic inflammation: a heritable disorder of calprotectin metabolism with rheumatic manifestations? The Journal of pediatrics. 2002;140:267–269. doi: 10.1067/mpd.2002.121699. [DOI] [PubMed] [Google Scholar]
- 120.Lee ACW, Li CH. Age as a factor in severe bacterial infection in transfusion-dependent patients with thalassemia major. Clinical Infectious Diseases. 2004;38:1194–5. doi: 10.1086/382683. author reply 1195. [DOI] [PubMed] [Google Scholar]
- 121.Wang SC, et al. Severe bacterial infection in transfusion-dependent patients with thalassemia major. Clinical Infectious Diseases. 2003;37:984–988. doi: 10.1086/378062. [DOI] [PubMed] [Google Scholar]
- 122.Gerhard GS, et al. Vibrio vulnificus septicemia in a patient with the hemochromatosis HFE C282Y Mutation. Archives of Pathology and Laboratory Medicine. 2001;125:1107–9. doi: 10.5858/2001-125-1107-VVSIAP. [DOI] [PubMed] [Google Scholar]
- 123.Höpfner M, N R, R A, H D, S S, F UR. Yersinia enterocolitica infection with multiple liver abscesses uncovering a primary hemochromatosis. Scandinavian Journal of Gastroenterology. 2001;36:220–4. doi: 10.1080/003655201750066004. [DOI] [PubMed] [Google Scholar]
- 124.Weinberg ED. Survival advantage of the hemochromatosis C282Y mutation. Perspectives in Biology and Medicine. 2008;51:98–102. doi: 10.1353/pbm.2008.0001. [DOI] [PubMed] [Google Scholar]
- 125.Pishchany G, et al. Specificity for human hemoglobin enhances Staphylococcus aureus infection. Cell Host and Microbe. 2010;8:544–550. doi: 10.1016/j.chom.2010.11.002. This manuscript provides evidence that polymorphisms within hemoglobin impact susceptibility to Staphylococcus aureus infections. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Torres VJ, Pishchany G, Humayun M, Schneewind O, Skaar EP. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. Journal of Bacteriology. 2006;188:8421–8429. doi: 10.1128/JB.01335-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Krishna Kumar K, et al. Structural basis for hemoglobin capture by Staphylococcus aureus cell-surface protein, IsdH. Journal of Biological Chemistry. 2011;286:38439–38447. doi: 10.1074/jbc.M111.287300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zarantonelli ML, et al. Transgenic mice expressing human transferrin as a model for meningococcal infection. Infection and Immunity. 2007;75:5609–5614. doi: 10.1128/IAI.00781-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Schryvers AB, Morris LJ. Identification and characterization of the transferrin receptor from Neisseria meningitidis. Molecular Microbiology. 1988;2:281–288. doi: 10.1111/j.1365-2958.1988.tb00029.x. [DOI] [PubMed] [Google Scholar]
- 130.Noinaj N, et al. Structural basis for iron piracy by pathogenic Neisseria. Nature. 2012 doi: 10.1038/nature10823. The crystal structure of the Neisseria TbpA in complex with transferrin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Calmettes C, Alcantara J, Yu RH, Schryvers AB, Moraes TF. The structural basis of transferrin sequestration by transferrin-binding protein B. Nature structural & molecular biology. 2012 doi: 10.1038/nsmb.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Borkow G, Gabbay J. Copper as a biocidal tool. Current Medicinal Chemistry. 2005;12:2163–2175. doi: 10.2174/0929867054637617. [DOI] [PubMed] [Google Scholar]
- 133.Mikolay A, et al. Survival of bacteria on metallic copper surfaces in a hospital trial. Applied Microbiology and Biotechnology. 2010;87:1875–1879. doi: 10.1007/s00253-010-2640-1. [DOI] [PubMed] [Google Scholar]
- 134.Casey AL, et al. Role of copper in reducing hospital environment contamination. The Journal of Hospital Infection. 2010;74:72–77. doi: 10.1016/j.jhin.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 135.Zhou T, Ma Y, Kong X, Hider RC. Design of iron chelators with therapeutic application. Dalton transactions. 2012 doi: 10.1039/c2dt12159j. [DOI] [PubMed] [Google Scholar]
- 136.Summer KH, et al. The biogenic methanobactin is an effective chelator for copper in a rat model for Wilson disease. Journal of Trace Elements in Medicine and Biology. 2011;25:36–41. doi: 10.1016/j.jtemb.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 137.Thiennimitr P, et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Winter SE, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hoffman LR, et al. Nutrient availability as a mechanism for selection of antibiotic tolerant Pseudomonas aeruginosa within the CF airway. PLoS Pathogens. 2010;6:e1000712. doi: 10.1371/journal.ppat.1000712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ponton F, Wilson K, Cotter SC, Raubenheimer D, Simpson SJ. Nutritional immunology: A multi-dimensional approach. PLoS Pathogens. 2011;7:e1002223. doi: 10.1371/journal.ppat.1002223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rohmer L, Hocquet D, Miller SI. Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends in microbiology. 2011;19:341–348. doi: 10.1016/j.tim.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Eisenreich W, Dandekar T, Heesemann J, Goebel W. Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nature Reviews Microbiology. 2010;8:401–412. doi: 10.1038/nrmicro2351. [DOI] [PubMed] [Google Scholar]
- 143.Price CTD, Al-Quadan T, Santic M, Rosenshine I, Abu Kwaik Y. Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science. 2011;334:1553–1557. doi: 10.1126/science.1212868. [DOI] [PubMed] [Google Scholar]
- 144.Cheung J, Beasley FC, Liu S, Lajoie GA, Heinrichs DE. Molecular characterization of staphyloferrin B biosynthesis in Staphylococcus aureus. Molecular Microbiology. 2009;74:594–608. doi: 10.1111/j.1365-2958.2009.06880.x. [DOI] [PubMed] [Google Scholar]
- 145.Beasley FC, et al. Characterization of staphyloferrin A biosynthetic and transport mutants in Staphylococcus aureus. Molecular Microbiology. 2009;72:947–963. doi: 10.1111/j.1365-2958.2009.06698.x. [DOI] [PubMed] [Google Scholar]
- 146.Segond D, et al. NRAMP genes function in Arabidopsis thaliana resistance to Erwinia chrysanthemi infection. The Plant Journal. 2009;58:195–207. doi: 10.1111/j.1365-313X.2008.03775.x. [DOI] [PubMed] [Google Scholar]
- 147.Dellagi A, et al. Microbial siderophores exert a subtle role in Arabidopsis during infection by manipulating the immune response and the iron status. PLANT PHYSIOLOGY. 2009;150:1687–1696. doi: 10.1104/pp.109.138636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Rokhbakhsh-Zamin F, et al. Characterization of plant-growth-promoting traits of Acinetobacter species isolated from rhizosphere of Pennisetum glaucum. Journal of Microbiology and Biotechnology. 2011;21:556–566. [PubMed] [Google Scholar]
- 149.Lemanceau P, Bauer P, Kraemer S, Briat JF. Iron dynamics in the rhizosphere as a case study for analyzing interactions between soils, plants and microbes. Plant and Soil. 2009;321:513–535. [Google Scholar]


