Summary
The nature of the antigens recognized by γδ T cells and their potential recognition of major histocompatibility complex (MHC)-like molecules has remained unclear. The CD1 family of lipid-presenting molecules are suggested ligands for Vδ1 TCR-expressing γδ T cells, the major γδ lymphocyte population in epithelial tissues. We crystallized a Vδ1 TCR in complex with CD1d and the self-lipid sulfatide, revealing an unusual recognition of CD1d by germline Vδ1 residues spanning all complementarity determining region (CDR) loops, with sulfatide recognition separately encoded by non-germline CDR3δ residues. Binding and functional analysis showed that CD1d presenting self-lipids, including sulfatide, was widely recognized by gut Vδ1 + γδ T cells. These findings provide structural demonstration of MHC-like recognition of a self-lipid by γδ T cells and reveal the prevalence of lipid recognition by innate-like T cell populations.
Introduction
There are two major lineages of T lymphocytes, αβ and γδ T cells, which are defined by their T cell receptor (TCR) gene segment usage. While the αβ subset is the prominent T cell population in the circulation and lymph nodes devoted to the orchestration of the adaptive immune response, γδ T cells are particularly abundant in peripheral tissues, most notably the skin and intestinal epithelium, where they monitor early signs of tissue infection or stress (Vantourout and Hayday, 2013). The coexistence of two main T cell lineages has been conserved throughout vertebrate evolution, highlighting how each plays an important, non-redundant role in host defense and survival.
In humans there are two main populations of γδ T cells, Vδ1 and Vδ2 T cells, which predominate in the epithelium and circulation, respectively (McVay and Carding, 1999). It is hypothesized that the TCR-dependence of tissue localization may be linked to recognition of a conserved tissue-specific self-ligand, highlighting the importance of γδ TCR ligand characterization for our understanding of γδ T cell biology. However, these ligands remain poorly characterized in both the murine and human systems, and those that have been described exhibit strikingly little overlap between species (Vantourout and Hayday, 2013). Most identified ligands are self-derived and stress-induced, in accord with γδ T cells having an important role in the early detection of tissue damage. Such ligands include non-classical MHC-like molecules, such as murine T22, which has previously been the only ligand for which structural studies have revealed the mode of TCR recognition (Adams et al., 2005; Bonneville et al., 1989; Spada et al., 2000; Willcox et al., 2012). Other proposed ligands bear no resemblance to MHC molecules (Constant et al., 1994; Scotet et al., 2005; Zeng et al., 2012). In humans, intestinal epithelial Vδ1+ cells have been shown to respond to the stress-induced MHC-like molecules, MHC class I-related chain A (MICA) and MICB, though the role of the γδ TCR in this specific response has been controversial owing to the very low affinity of the interaction and competitive MICA recognition by other surface receptors such as natural killer group 2, member D (NKG2D) (Bauer et al., 1999; Groh et al., 1998; Xu et al., 2011).
One of the few human γδ TCR ligands reported in several independent studies is the lipid-presenting MHC-like molecule, CD1d. CD1d is a well-characterized as a TCR ligand for natural killer T (NKT) cells of the αβ T cell lineage, specializing in the presentation of both self-derived and foreign lipids (Rossjohn et al., 2012). However, NKT cells are over ten-fold less abundant in humans than in mice, and though the full extent of CD1d recognition by γδ T cells has yet to be determined, γδ T cells appear to be a substantial complement to NKT cells in the surveillance of lipid antigens (Bendelac et al., 2007). CD1d-mediated lipid antigen recognition has been described for γδ T cells in both the circulation and within the intestinal epithelium (Agea et al., 2005; Bai et al., 2012; Mangan et al., 2013; Russano et al., 2007). These cells predominantly use the Vδ1 TCR gene segment; however, recent studies have also identified CD1d-specific cells among the less prevalent Vδ3+ T cell population (Mangan et al., 2013). Lipid-specific Vδ1+ T cells exhibit heterogeneous phenotypes, which seem to vary according to tissue residence. Circulating phospholipid-specific γδ T cells have a Th2 polarized phenotype, whereas those of intestinal origin can mediate Th1-polarized or regulatory cytokine secretion (Agea et al., 2005; Russano et al., 2006; Russano et al., 2007). The lipid repertoire surveyed by Vδ1+ T cells comprises both exogenous pollen-derived phospholipids, and self-lipids including the glycolipid sulfatide, which is enriched in neuronal, kidney, and intestinal epithelial tissue (Bai et al., 2012; Breimer et al., 2012; Russano et al., 2006; Takahashi and Suzuki, 2012). The detection of self-ligands coincides with the roles of tissue-resident γδ T cells in tissue homeostasis and repair, yet conversely may explain their accumulation in tissues in the context of autoimmune pathology (Komano et al., 1995; Selmaj et al., 1991; Toulon et al., 2009; Wucherpfennig et al., 1992).
Despite these studies implicating γδ T cells as an important lipid reactive T cell population with diverse functional roles, the structural basis of CD1d-lipid recognition is unknown, as it is for all human γδ TCRs. In stark contrast, abundant structural studies have revealed the details of αβ TCR interaction with multiple ligand classes (Borg et al., 2007; Garcia et al., 1996; Lopez-Sagaseta et al., 2013). Here we provide the structural basis of human γδ TCR recognition and show how γδ TCRs detect ligands in the context of an antigen-presenting molecule, revealing a new paradigm for the antigen specificity of Vδ1+ T cells.
Results
Vδ1+ T cells recognize CD1d directly through the γδ TCR
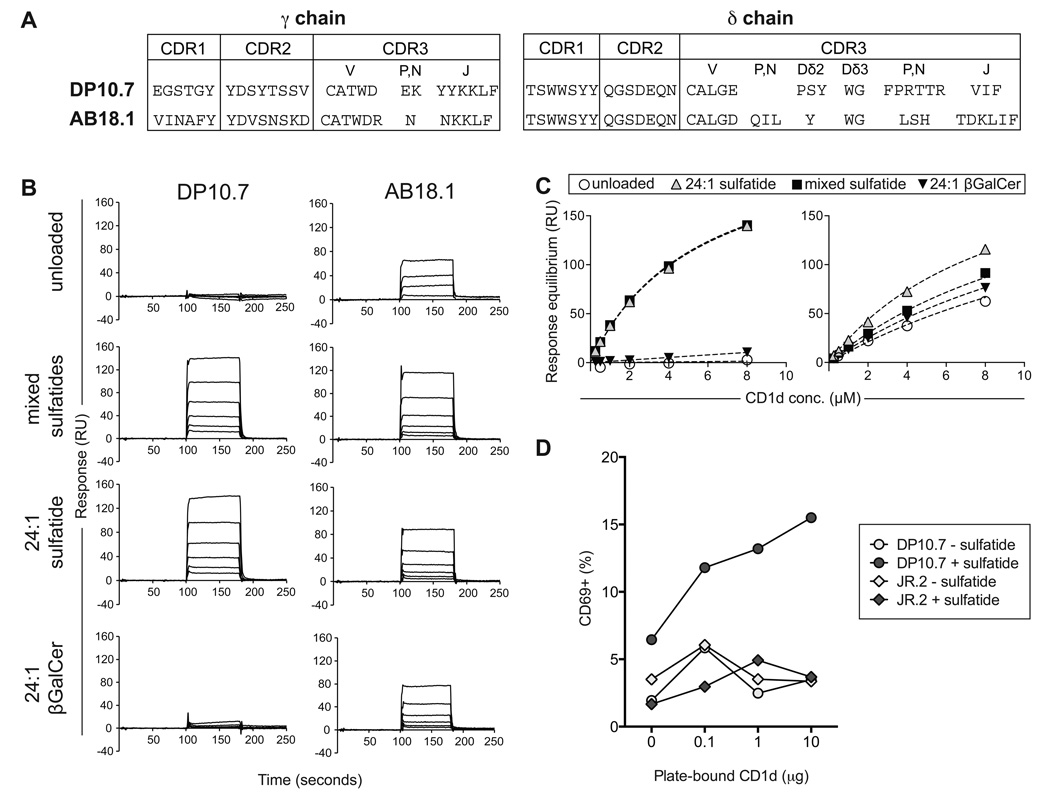
We have previously described two peripheral blood-derived CD1d-sulfatide reactive human Vδ1+ clones, the Vγ4Vδ1 DP10.7 and Vγ5Vδ1 AB18.1 TCRs (Figure 1A) (Bai et al., 2012). To determine whether this interaction is specifically mediated by the Vδ1+ γδ TCR, we used biochemical methods to measure the interaction between soluble, recombinant CD1d loaded with sulfatide and Vδ1+ γδ TCRs. CD1d loaded with endogenous, insect cell-derived lipids (unloaded) failed to bind to the DP10.7 TCR (equilibrium dissociation constant, Kd, not measurable), whereas it bound to the AB18.1 with moderate affinity (Kd: 20.6 µM) (Figure 1B,C). Loading with mixed bovine brain sulfatides (predominantly 24:0 and 24:1 acyl chains) dramatically enhanced CD1d binding to the DP10.7 TCR (Kd: 5.5 µM) and moderately affected binding to the AB18.1 TCR (Kd: 8.9 µM). Both clones bound to CD1d loaded with 24:1 sulfatide (acyl chain 24:1) at affinities comparable to that with mixed sulfatides (DP10.7 Kd: 5.6 µM; AB18.1 Kd: 13.9 µM). Comparing binding with 24:1 βGalCer-loaded CD1d, which lacks the 3’-galactose sulfate group characteristic of sulfatide lipids, showed that the sulfate moiety was essential for DP10.7 recognition (Kd >100µM). In line with sulfatide having a secondary role in recognition by the AB18.1 TCR, the absence of the 3’ sulfate group only moderately affected AB18.1 binding (Kd: 16.8 µM). Thus, the DP10.7 TCR makes energetically important contacts with the sulfatide antigen, whereas the AB18.1 TCR can recognize CD1d molecules largely indiscriminately of lipid antigens. Thus it is likely that variability in CDR3δ loop or γ chain sequences modulates Vδ1+ TCR engagement of CD1d-lipid surfaces (Figure 1A).
Figure 1. CD1d-sulfatide recognition by human Vδ1+ T cells is TCR-dependent.
(A) CDR loop amino acid sequences of two CD1d-sulfatide-specific Vδ1 clones (DP10.7 and AB18.1). (B) Surface Plasmon Resonance (SPR) binding responses of CD1d purified with endogenous lipids “unloaded” or loaded with indicated sulfatide variants, to C-terminally biotinylated DP10.7 and AB18.1 TCRs immobilized on a streptavidin Biacore© chip. Response units of CD1d binding (0.25 – 8 µM) after reference channel subtraction are shown. (C) Equilibrium affinity analysis of DP10.7 and AB18.1 TCR binding to indicated CD1d forms. (D) Percent CD69 upregulation on DP10.7 TCR transduced Jurkat J.RT3-T3.5 T cells stimulated by plate-bound CD1d purified with endogenous lipids (white circles) or loaded with sulfatide (grey circles). A control JR.2 TCR, which is CD1c specific, was similarly transduced and stimulated with CD1d presenting endogenous lipids (white diamonds) or CD1d-sulfatide (grey diamonds) (see also Figure S1).
The reactivity of Vδ1+ T cells towards CD1d molecules loaded with endogenous membrane lipids has been demonstrated previously (Russano et al., 2007), corroborating our biochemical characterization of endogenous CD1d-presented lipid recognition by the AB18.1 TCR. As the DP10.1 TCR exhibits exquisite sulfatide-specific reactivity, we confirmed the functional basis of this interaction by TCR transduction of Jurkat J.RT3-T3.5 T cells. A control Vδ1 TCR reported to recognize CD1c molecules, clone JR.2 (Spada et al., 2000), did not respond to plate-bound CD1d molecules with or without sulfatide loading. However, transduction of the DP10.7 TCR conferred a sulfatide-specific response correlating to the amount of CD1d immobilization, as measured by CD69 upregulation (Figure 1D). The higher basal expression of CD69 by DP10.7 TCR-transduced cells in the presence of sulfatide likely stems from the expression of CD1d on Jurkat cells themselves (Metelitsa et al., 2001). Together, these results demonstrate that Vδ1+ T cells recognize and respond to CD1d-sulfatide directly through the γδ TCR.
Functional characterization of CD1d-sulfatide-specific Vδ1+ T cells
To understand markers associated with CD1d-sulfatide-specific T cells, phenotypic studies were performed on Vδ1+ T cells isolated from the blood by CD1d-sulfatide tetramer MACS enrichment and FACS sorting (Bai et al., 2012). Like many γδ T cells, they expressed CD8αβ, CD8αα or were CD8-negative (Figure S1A). In agreement with the sequencing results demonstrating that they were the progeny of expanded T cell clones, the fresh cells exhibited a CD45RA+CD62L−CCR7− CD28− effector phenotype. Notably, given the preeminence of Vδ1+ T cells in the human gut, these sulfatide-specific T cells isolated from the blood did not express CD103, a marker for cells of intestinal origin. However, they expressed the myeloid markers CD16, CD11b and CD11c in the two individuals examined (Figure S1A).
The rarity of CD1d-sulfatide specific T cells in the blood precluded an analysis of their functional properties such as cytokine secretion or cytolytic functions directly ex vivo. Nevertheless, experiments carried out with clones DP10.7 and AB18.1 demonstrated predominant secretion of TNF-α and IFN-γ but little IL-4, IL-13 or IL-10, indicative of a Th1 phenotype, upon stimulation with ionomycin and PMA (Figure S1B).
The structural basis for Vδ1+ γδ TCR recognition of CD1d-sulfatide
To understand the molecular basis for Vδ1 TCR recognition of CD1d-sulfatide, we sought to crystallize the TCR-CD1d-sulfatide ternary complex. We developed a fusion human-mouse CD1d molecule (CD1dmα3) in which the native human ligand-binding α1 and α2 domains were fused with the murine α3 domain and heterodimerized with murine β2-microgobulin (β2m). These efforts were undertaken owing to the many published murine CD1d structures and the hypothesis that murine CD1d molecules may crystallize more readily (Rossjohn et al., 2012). This substitution did not affect DP10.7 TCR recognition and resulted in the successful crystallization of the unliganded CD1dmα3-sulfatide complex (Figure S2A, 2B, Table S1). This structure confirmed the identity of our fusion to that of the native human CD1d structure, particularly within the human α1 and α2 domains (Figure S2B). The electron density for sulfatide was unambiguous, revealing a stabilizing network of hydrogen bonds with CD1d as described for murine structures (Figure S2C, 2D) (Zajonc et al., 2005). In our structure, the sulfatide head-group was displaced by >4 Å when compared with the murine CD1d-sulfatide structure, owing to the presence of a tryptophan residue, rather than a glycine residue, in the human form (Figure S2E). This provides direct evidence that human and mouse CD1d molecules can differ in their presentation of the same lipid.
To crystalize the ternary complex of the DP10.7 TCR, human CD1d and sulfatide, we utilized the single-chain (sc) version of the TCR, in which the variable domains are expressed as a fusion protein. This strategy has been used previously for both αβ and γδ TCR crystallization (Maynard et al., 2005; Xu et al., 2011). We succeeded in crystallizing the ternary sc DP10.7 TCR-CD1dmα3-24:1 sulfatide complex and determined its structure to 3.0 Å resolution (Table S1).
Exclusive δ-chain mediated recognition of CD1d-sulfatide
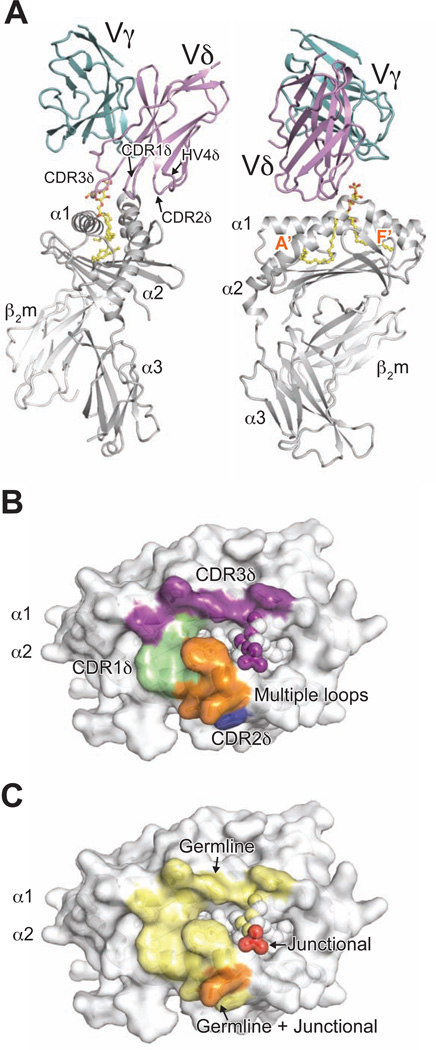
The DP10.7 TCR docks at a tilted angle, with all CDR loops from the δ chain contacting the CD1d-sulfatide complex and no contribution from the γ chain (Figure 2A). In fact, there are no γ chain residues within 5 Å of the CD1d-sulfatide surface. This dominance of the δ chain is consistent with our mutagenesis, discussed below, and the exclusive use of the Vδ1 domain in γδ T cells that respond to CD1d (Agea et al., 2005; Bai et al., 2012; Russano et al., 2007). The δ chain docks above the CD1d A’ pocket, which binds the acyl 24:1 chain of sulfatide (Figure 2A). The Vδ1-encoded residues of the CDR1, 2 and HV4δ loops make contacts nearly exclusively with the α2 helix of CD1d (Figure 2B). The CDR3δ loop largely contacts the α1 helix of CD1d and the sulfatide head group, but also shares some contacts with those of the other CDR and HV loops. The sulfatide antigen is contacted solely by junctional CDR3δ loop residues (Figure 2 C), suggesting that other CD1d-restricted Vδ1 T cells may share this common docking footprint and utilize recombined CDR3δ loops to detect different lipid antigens.
Figure 2. Complex structure of DP10.7 TCR and CD1d-sulfatide reveals exclusive δ chain dependence.
Overview of DP10.7 TCR-CD1d-sulfatide complex and contacts. (A) CD1d heterodimer, light grey; sulfatide, yellow; DP10.7 TCR γ-chain and δ-chain, light teal and violet, respectively. Side view (left) of complex shows tilted TCR docking angle, which restricts CD1dsulfatide contacts to the δ chain. Front view (right) shows orientation of TCR over the CD1d A’ pocket. (B) Surface of CD1d-sulfatide is shown in white, and residues that contact the TCR are colored according to CDR loop (residues ≤ 4.0 Å from TCR). CD1d residues contacted by CDR1δ, CDR2δ and CDR3δ shown in green, marine blue, and purple, respectively. CD1d residues contacted by multiple CDR/HV4 shown in orange. Atoms of sulfatide contacted by CDR3δ shown as purple spheres. (C) Surface of CD1d-sulfatide is shown in white, and residues (CD1d) or atoms (sulfatide) that contact the TCR are colored according to germline, non-germline, or both origin in light yellow, red, and light orange, respectively (see also Figure S2 and Table S1).
The buried surface area (BSA) of the DP10.7 TCR-CD1d-sulfatide interface is ~730Å2, with 600 Å2 from the TCR-CD1d interface and 130 Å2 from the TCR-sulfatide interface. This interface is the smallest reported among TCR-ligand structures, slightly less than that of iNKT TCR-CD1d-αGalCer complexes (Pellicci et al., 2009) and considerably smaller than that of the murine type II NKT TCR-CD1d-sulfatide complex, (~1010 Å2), though the affinity is about three times higher (Patel et al., 2012). Numerous ionic interactions in the DP10.7 TCR-CD1d-sulfatide interface likely compensate for the smaller interface area to facilitate a higher affinity interaction.
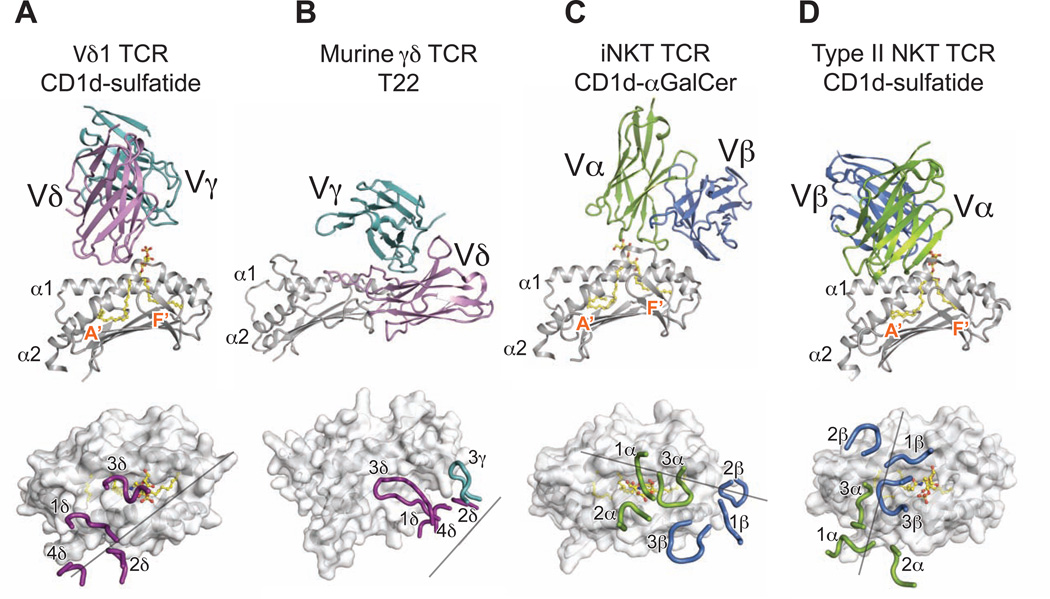
Comparisons with other TCR-ligand complex structures reveal that the DP10.7 TCR engages CD1d-sulfatide with a unique docking footprint. The murine G8-T22 complex is the only other structure of a γδ TCR with bound ligand, which is similarly dominated by δ chain interactions (Adams et al., 2005). However, the DP10.7 footprint is more evenly divided among all δ chain loops, rather than nearly exclusively relying on the CDR3δ loop as in G8-T22. (Figure 3A, 3B). The DP10.7 TCR footprint is also distinct from the recognition mode of CD1d-restricted Type I and II NKT TCRs. The most striking difference is the DP10.7 TCR’s sole reliance on just one chain for CD1d recognition, contrasting with that of the αβ TCRs. However, the DP10.7 TCR docks above the CD1d A’ pocket in an overall similar orientation to that of the Type II NKT TCR (Figure 3A,D) (Girardi et al., 2012; Patel et al., 2012). Similarly, both the DP10.7 and Type II NKT TCRs contact lipid antigens with their recombined CDR3 loops, in contrast to the invariant germline CDR1α loop-mediated lipid contacts by Type I NKT TCRs (Figure 3A,C,D) (Borg et al., 2007; Rossjohn et al., 2012). As CDR3δ loops have the most potential sequence diversity among all recombined immunoreceptors (Davis and Bjorkman, 1988), the central orientation of the CDR3δ over the CD1d antigen-binding pocket suggests the potential of Vδ1 TCRs to engage a diverse array of lipid antigens.
Figure 3. The Vδ1 DP10.7 TCR employs a unique docking mode.
View of TCR variable domains (top) in complex with α1,α 2 domains of ligands and CDR loop footprint/docking orientation (bottom) of TCR-ligand complexes. (A) Murine G8 γδ TCR-T22 complex (Protein Data Bank accession code 1YPZ) (B) Human iNKT TCR-CD1d-αGalCer complex (Protein Data Bank accession code 2PO6) (C) and Murine type II NKT TCR-CD1d-sulfatide complex (Protein Data Bank accession code 4EI5) (D). Surface of ligand α1,α2 domains shown in white; lipid ligands if present shown in yellow. TCR γ-chain shown in light teal, δ-chain in violet (A,B); TCR-α chain shown in green, β-chain in blue. (C,D) Grey lines represent vector connecting the conserved disulfide bond in each V domain (see also Table S2).
TCR contacts with CD1d are germline-encoded whereas CDR3δ junctional residues contact sulfatide
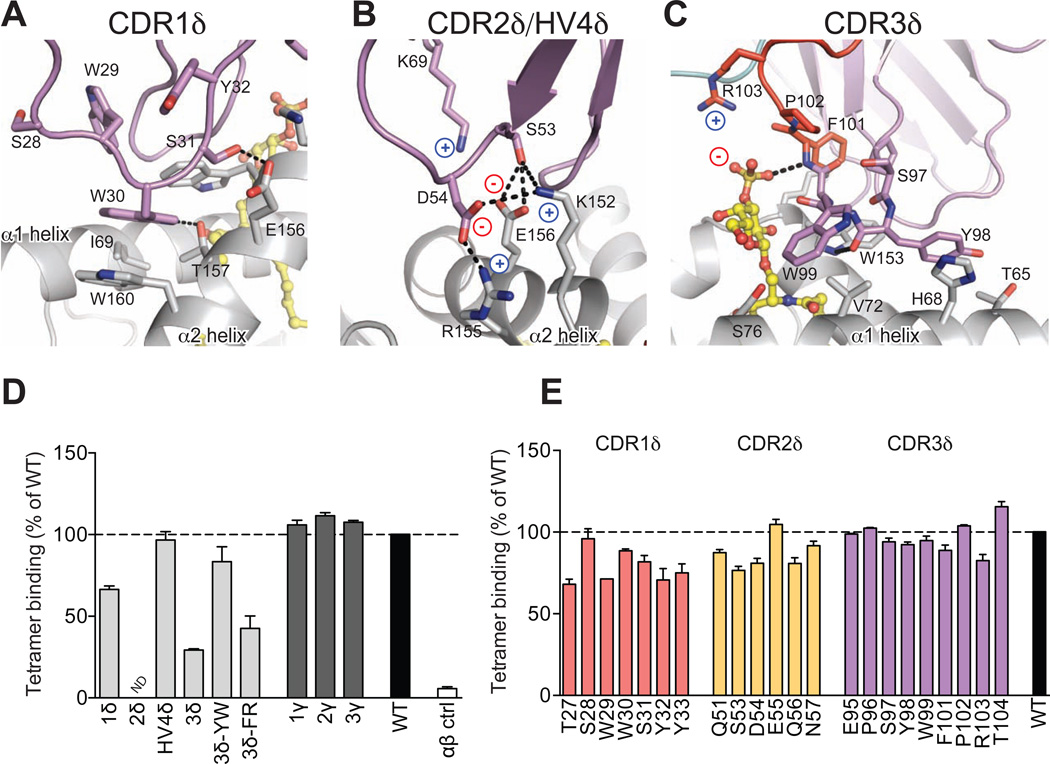
Electron density for all DP10.7 CDR loop residues and the sulfatide ligand were unambiguous, permitting detailed analysis of the TCR-CD1d-sulfatide interface (Figure S2, Table S2). We also performed TCR alanine mutagenesis and entire CDR loop-swapping experiments via a tetramer-binding assay to complement our structural analyses (Table S3). The CDR1δ accounts for 32% of the total BSA, most of which is contributed by Trp30 (Figure 4A). This residue makes extensive non-polar contacts, most prominently a π-stacking interaction with Trp160. Additional hydrogen bonds are formed by the CDR1δ Trp30 and Ser31 with CD1d Thr157 and Glu156, respectively (Figure 4A). The importance of this loop is validated by the observed reduction in binding when the unrelated Vδ2-encoded CDR1δ is grafted in its place. (Figure 4D, Table S3). Alanine mutagenesis revealed that several residues other than the key Trp30 and Ser31 were important for binding, which likely affect the overall loop conformation (Figure 4E). Though the CDR2δ loop is docked towards the outside of the CD1d α2 helix and contributes only 11% towards the total BSA, it forms many polar and ionic contacts with CD1d. Asp54 is at the center of a network of salt-bridges, making these contacts with Lys152 and Arg155 (Figure 4B). Ser53 is positioned to engage in several hydrogen bonds with Glu156 and Lys152. Alanine mutagenesis validates the contribution of these key CDR2δ residues (Figure 4E). Lys69 of the TCR HV4δ loop also contributes to this network of polar contacts, forming a salt-bridge with Glu156. The numerous specific Vδ1 interactions with CD1d highlight the strongly biased usage of this gene segment in described CD1d-reactive γδ TCRs (Agea et al., 2005; Bai et al., 2012; Russano et al., 2007), whereas CD1d-sulfatide reactive type II NKT TCRs can use several different Vα and Vβ genes for CD1d recognition (Park et al., 2001).
Figure 4. Analysis of DP10.7 TCR-CD1d-sulfatide contacts.
(A–C) Polar contacts are shown in dashed black lines; charges are indicated to emphasize salt-bridge contacts. CD1d shown in light grey, sulfatide in yellow, TCR germline residues in violet, non-templated in red for CDR1δ (A); CDR2δ and HV4 (B); and CDR3δ (C) contacts. (D–E) TCR mutagenesis and binding measurements were determined by plate-binding assay. ELISA plate wells were coated with WT or mutant single-chain (sc) TCRs, and binding to CD1d-sulfatide-tetramers labeled with HRP was measured by colorimetric readout (A450). Non-specific binding to BSA was subtracted and binding was calculated as a % of WT. Shown are the mean and S.E.M. of at least two independent experiments. (D) Entire CDR loops were substituted with an unrelated CDR loop sequences and binding was compared with the WT DP10.7 TCR. Double-alanine mutations of indicted CDR3δ loop residues were also analyzed. (E) Single alanine point mutations of the DP10.7 TCR δ chain were made and binding vs WT TCR was measured as above (see also Figure S3 and Table S3).
In contrast, the extended CDR3δ loop forms fewer polar contacts with CD1d, though overall plays a key role in CD1d-sulfatide recognition as shown in our structure, and by a marked reduction in binding upon substitution of this loop with an unrelated CDR3δ loop sequence (Figure 4C, 4D, Table S3). The tip of the CDR3δ is comprised of Dδ segment-encoded residues, Tyr98 and Trp99, which make several VdW interactions with CD1d residues concentrated in the α1 helix (Figure 4C). Tyr98 contributes a main-chain hydrogen bond to Trp153 of the α2 helix via its backbone carbonyl, but otherwise no polar contacts are made by the five Dδ2 and Dδ3 residues of the CDR3δ loop. As such, alanine mutagenesis of Dδ-encoded residues minimally affected binding to CD1d-sulfatide (Figure 4D, 4E). All polar contacts with the sulfatide antigen are mediated by junctionally-encoded residues Phe101 and Arg103, which form a main-chain hydrogen-bond and salt-bridge, respectively, with the sulfate moiety (Figure 4C). Alanine mutagenesis of these residues shows an individually modest contribution, whereas dual Phe101 and Arg103 mutagenesis reduces CD1d-binding nearly as much as swapping of the entire CDR3δ loop (Figure 4D, 4E). This corroborates the key role of sulfatide interactions demonstrated by SPR measurements (Figure 1B, 1C). Overall, there were no residues that were absolutely required for CD1d-sulfatide recognition, in contrast to the conserved “hot-spot” residues characteristic of T22-reactive TCRs (Adams et al., 2008; Sandstrom et al., 2012). Our structure shows an interwoven network of contacts, and it is likely that compensatory interactions can substitute for single alanine mutations. Furthermore, our tetramer-based detection has an avidity affect, so smaller differences in binding may be partially masked. However, the reductions in binding observed, such as for alanine substitution of Asp54 and Arg103, were reproducible and correlate to their important in our complex structure.
In accordance with structural studies, swapping of entire γ chain CDR loops with unrelated sequences has no effect on CD1d-sulfatide binding (Figure 4D, Table S3). Though the γ chain lacks energetic contributions here, it remains possible that γ chain CDR loops could mediate contacts of bulkier CD1d-presented lipid antigens, such as the gangliosides common in nervous tissue (Haig et al., 2011; Tettamanti et al., 1973).
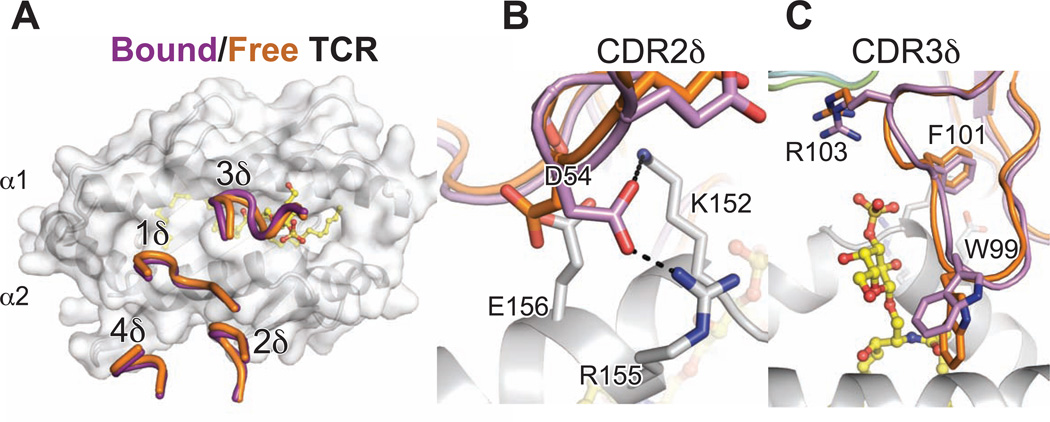
Minimal conformational change by the DP10.7 TCR upon CD1d-sulfatide ligation
Conformational change during binding can play a large role in αβ TCR recognition of peptide-MHC, particularly in the CDR3 loops that reorganize to optimally contact peptide antigens (Garcia and Adams, 2005); however, it is unknown whether γδ TCRs require the same flexibility in recognition of their ligand owing to the lack of structural data. To address this question, we solved the unliganded structure of the DP10.7 TCR, resolved to 3.3 Å resolution. Despite the moderate resolution limit, we were able to model the backbones for all δ chain CDR loops and most side-chains (Table S3, Figure S4A). Despite participating in crystal contacts, the CDR loop structures were essentially superimposable with those of the TCR in complex, indicating that they adopt similar rigid conformations (Figure 5A). Side chain conformations did vary, most notably for the CDR2δ residue D54, which flipped about 180° in order to form salt-bridges with CD1d α2 helix residues Arg155 and Lys152 in the complex structure (Figure 5B). Within the CDR3δ loop, the side-chain of Arg103 moves ~1Å closer to the sulfatide head group upon binding in order to fall within salt-bridge distance (Figure 5C). Otherwise, residues in the unliganded DP10.7 TCR are positioned to optimally form contacts as found in the complex structure, indicative of a more rigid-body type interaction.
Figure 5. Minimal conformation change required for CD1d-sulfatide recognition.
(A) Overall structure of the un-liganded DP10.7 TCR. The native DP10.7 δ and γ variable domains were fused to human α and β constant domains, respectively to facilitate protein expression and crystallization. Overall structure is shown with CDR loops indicated. Electron density for the CDR1γ loop was discontinuous, and represented as dashed line to show backbone connectivity. (B) Free TCR (chain B, orange) was aligned to bound TCR (purple) shown with CD1d (light grey)-sulfatide (yellow) complex. δ chain CDR loops of free and bound TCR are shown over CD1d surface. (C,D) Side-chains of CDR2δ,3δ loops that make rearrangements upon ligation are highlighted (see also Figure S4).
We also solved the structure of the AB18.1 TCR in the un-liganded form, which provides additional insight into how Vδ1 TCRs may be pre-oriented for CD1d recognition (Table S1). Superimposition of this structure with the liganded DP10.7 again reveals nearly identical conformation of the CDR1,2 and HV4δ loops (Figure S4B). However, the AB18.1 TCR CDR3δ loop contains an intervening residue before the conserved YWG motif (Figure 1A), and thus the loop is in a somewhat different conformation (Figure S4C). Accommodation of the extra residue in the CDR3δ loop would require conformational changes upon CD1d-sulfatide binding if this TCR were to dock in similar manner. However, the key salt bridge mediated by Arg103 in the DP10.7 structure would be lost if AB18.1 were docked in a similar fashion, as despite His104 potentially yielding a positive charge, its position is too far from the sulfate to form a salt bridge. This is in line with the AB18.1 TCR recognizing CD1d-sulfatide with only a slightly higher affinity than it does the “unloaded” CD1d molecule. The potentially closer proximity of several CDR3δ residues with the α1 and α2 helices of CD1d may explain the ability of the AB18.1 TCR to bind CD1d independently of sulfatide-loading, though a substantial shift in docking and γ chain contacts cannot be ruled out given it is unknown whether the DP10.7 docking mode is conserved among this and other Vδ1 TCRs. Overall, the conformations of the germline Vδ1 loops from two different un-bound TCRs match up nearly identically with the TCR in complex, despite different crystal packing, highlighting how these loops are pre-oriented for CD1d recognition.
Broad recognition of CD1d among Vδ1 clones
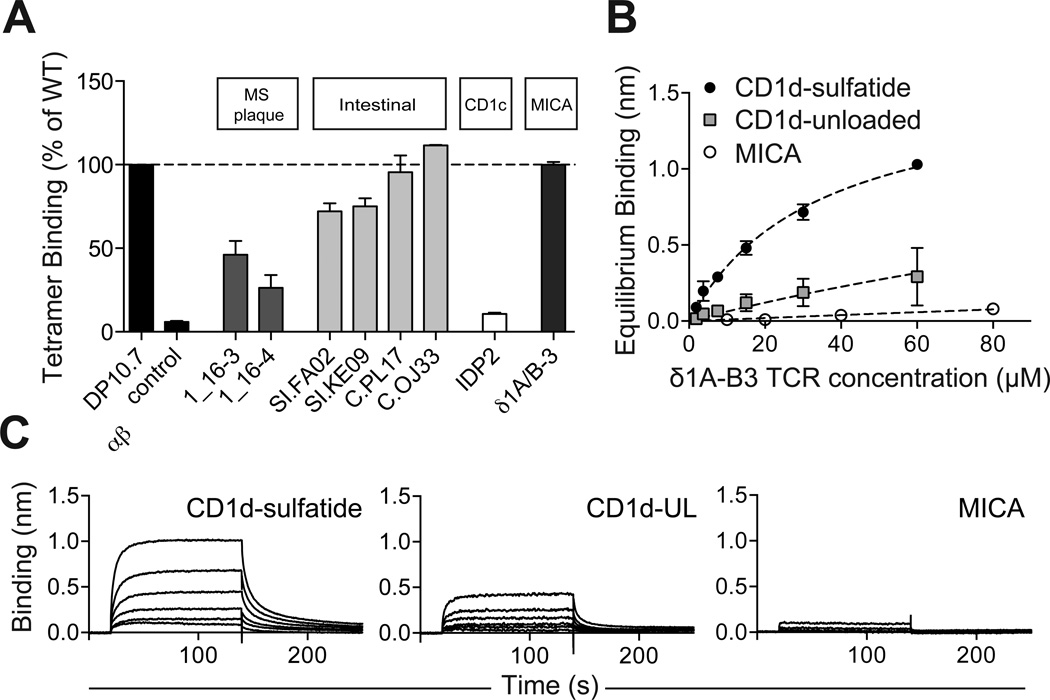
The prominent Vδ1 residue-mediated recognition of CD1d revealed by our complex structure led us to investigate the prevalence of CD1d-restriction among other human Vδ1+ T cells. We produced TCRs derived from published Vδ1 sequences of multiple sclerosis plaque-infiltrating and intestinal biopsy-derived γδ lymphocytes and measured their binding to CD1d-sulfatide tetramers (Figure S4A) (Chowers et al., 1994; Wucherpfennig et al., 1992). Vδ1+ T cells are a prominent population of both MS plaque-infiltrating γδ T cells, and within healthy intestinal epithelial tissues, though a definitive TCR ligand for these clones has yet to be identified. As only δ chain sequences were provided, we paired these with the DP10.7 TCR γ chain, which we showed to be irrelevant in CD1d recognition and thus allowing for an analysis of δ-chain exclusive interactions. This analysis does not, however, rule out contributions from the natively paired γ chains (advantageous or deleterious). Our analysis of two MS plaque clones demonstrated moderate to low binding to CD1d-sulfatide; while this is likely reflective of a low binding affinity, it establishes a basal recognition that may be enhanced by γ chain contributions, providing a possible link between Vδ1+ T cell infiltration of MS plaques, CD1d expression and the particular enrichment of sulfatide lipids in neuronal tissues (Takahashi and Suzuki, 2012).
In contrast to the moderate binding of TCRs from MS plaques, there was a striking prevalence of robust CD1d-sulfatide interactions among TCRs from intestinal-derived clones, which included some that bound as strongly as the DP10.7 TCR. Overall, those with shorter and more positively charged loops bound most strongly (Figure 6A, Figure S4A). We used biolayer interferometry (BLI) to measure a 25.8 µM CD1d-sulfatide affinity for the highest binding clone, OJ33, well within the 1–50 µM range of agonist TCR-ligand affinities (Figure S4B,C) (van der Merwe and Davis, 2003). This clone also bound unloaded CD1d with a moderate affinity (41.5 µM, Figure S4B,C). Despite our small sample size in this initial characterization, these results clearly show a potential for CD1d-restriction among intestinal Vδ1 clones, and specifically for recognition of sulfatide lipid antigens.
Figure 6. Widespread recognition of CD1d-sulfatide by Vδ1+ T cells.
(A) CD1d-sulfatide tetramer binding assay to plate-bound scTCRS. CD1d-sulfatide tetramers were labeled with HRP for colorimetric readout. Non-specific binding to BSA was subtracted and binding was calculated as a % of DP10.7 TCR. Shown are the mean and S.E.M. of at least two independent experiments. (B) BLI analysis of δ1A/B-3 sc TCR binding to immobilized CD1d-sulfatide, unloaded CD1d (CD1d-UL) or to MICA. TCR concentrations ranged from 1.88–60 µM (CD1d-sulfatide, CD1d-UL) or from 10–80 µM (MICA). Shown are representative data from one of two experiments. (C) Equilibrium affinity analysis of (B). Shown are the equilibrium binding mean and standard deviation from two experiments, as well as fits used to calculate Kd values (see also Figure S5).
We then examined the potential for CD1d recognition among clones (paired δ and γ chains) previously characterized as recognizing other non-classical MHC molecules. The clones IDP2 and δ1A/B-3 have been described as CD1c and MHC class I-related chain A (MICA)-specific, respectively (Brenner et al., 1986; Groh et al., 1998; Porcelli et al., 1989), though measurements of TCR interactions are either lacking or very weak (Xu et al., 2011). The δ1A/B-3 clone is the most well-studied among a population of intraepithelial lymphocytes (IELs) that are reactive to MICA (Groh et al., 1998). Using the above tetramer-based assay, we found that IDP2 bound negligibly to CD1d-sulfatide, but surprisingly, the δ1A/B-3 clones bound in a similar range as the DP10.7 TCR (Figure 6A). We validated this interaction by BLI affinity measurements and calculated a Kd of 33.9 µM (Figure 6B, C). The δ1A/B-3 TCR affinity for unloaded CD1d was almost 10-fold less (Kd = 241.3 µM), indicating that this TCR makes energetically important sulfatide-specific contacts (Figure 6B,C). Alignment with the complexed DP10.7 TCR demonstrates that δ1A/B-3 sulfatide specificity is likely conferred by the CDR3δ residue Arg99, which is essentially superimposable with that of the key DP10.7 sulfatide contact Arg103 and could readily form a salt-bridge with the sulfate moiety upon adoption of a different rotamer (Figure S5D). Validating published results showing that the δ1A/B-3 TCR binds MICA with very low affinity (Kd = 100–900 µM) (Xu et al., 2011), we observed very weak binding to MICA and were unable to accurately calculate an equilibrium affinity at the TCR concentrations used (Figure 6B,C). These results highlight the germline-encoded specificity of Vδ1 TCRs for CD1d molecules and demonstrate that this recognition is broadly distributed across blood and gut resident Vδ1 T cells.
Gut-derived Vδ1+ T cells are stimulated by CD1d and produce TNF-α
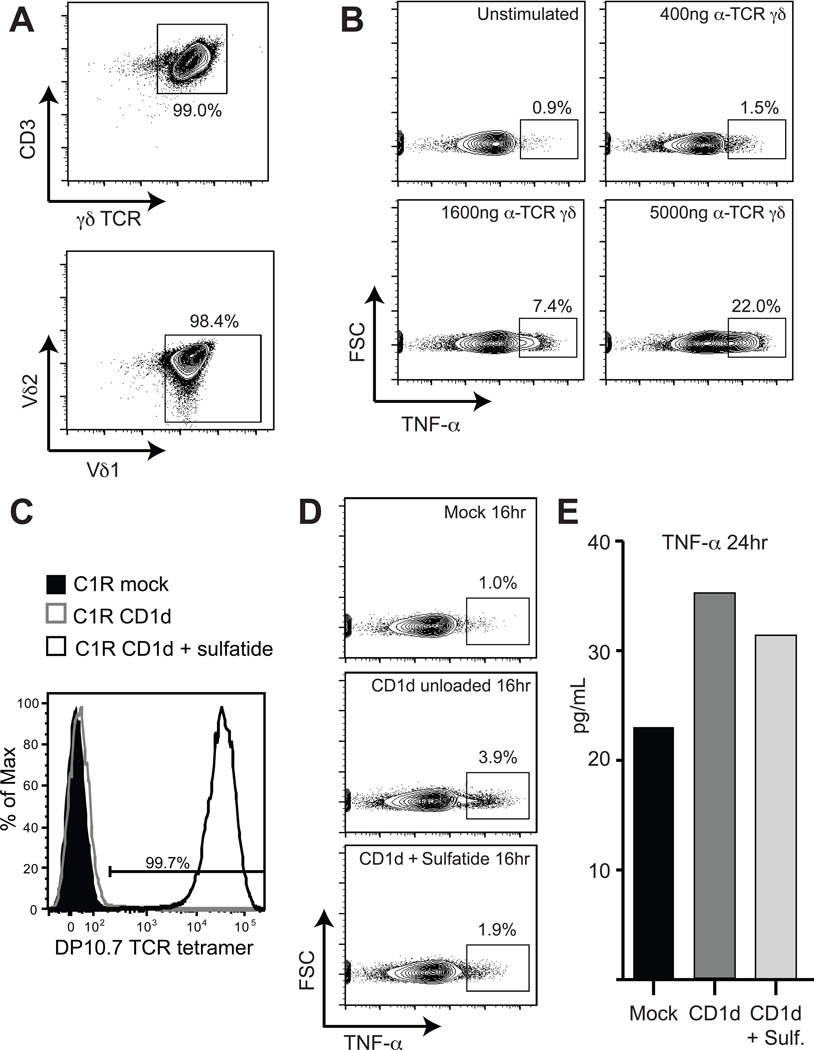
To further explore the recognition of CD1d in Vδ1 T cells in the gut, IEL were purified from jejunal biopsies (Jabri et al., 2000) and sorted based on expression of the Vδ1 TCR. These cells were expanded in an unbiased, CD1d-non-specific manner using irradiated feeder cells (Jabri et al., 2002) to generate enough cells for functional characterization. The expanded polyclonal cells uniformly expressed γδ TCRs containing a Vδ1 chain (Figure 7A). When stimulated with increasing concentrations of an anti-γδ TCR antibody, approximately one-fifth of the Vδ1 population produced TNF-α in response (Figure 7B), consistent with the TNF-α secretion noted in the characterization of blood Vδ1 by PMA-ionomycin treatment (Figure S1B). The effector activity of the remaining 80% of the Vδ1 polyclonal population is unknown, but suggests a diverse effector response to TCR triggering of this population.
Figure 7. Reactivity of gut Vδ1+ T cells to CD1d presenting endogenous lipids including sulfatide.
(A) Flow cytometry contour plots showing CD3 and γδ TCR expression of a polyclonal Vδ1 IEL cell line (top) and Vδ1 versus Vδ2 TCR expression (bottom). (B) Flow cytometry contour plots of TNF-α intracellular-cytokine staining of a polyclonal Vδ1 IEL cell line stimulated with increasing concentrations of anti-γδ TCR antibody. The concentration of antibody used is indicated at the top of each plot and the percentages displayed indicate cells gated for TNF-α expression. (C) Flow cytometry histogram plot of DP10.7 TCR tetramer staining of C1R mock transfectant (shaded black), C1R-CD1d transfectant unloaded (grey line) or C1R-CD1d transfectant treated with 30 µg/ml sulfatide (black line). Percentage of tetramer positive cells is indicated above the drawn gate. (D) Flow cytometry contour plots of intracellular TNF-α staining after 16 hours of co-culture of a polyclonal Vδ1 IEL cell line with C1R mock transfectants (top), unloaded C1R-CD1d transfectants (middle) and C1R-CD1d transfectants loaded with sulfatide (bottom). (E) Bar graph showing TNF-α levels in the culture supernatant measured by ELISA after 24 hours of co-culture of a polyclonal Vδ1 IEL cell line with C1R-mock, C1R-CD1d unloaded and C1R-CD1d sulfatide transfectants.
We sought to determine whether CD1d could stimulate TNF-α production from this Vδ1 polyclonal population, using C1R transfected CD1d as targets. To first differentiate the Vδ1 response to CD1d presenting endogenous phospholipids (as documented in Russano et al. (Russano et al., 2007)) from that of CD1d presenting sulfatide on the C1R cells, we first established the loading efficiency of sulfatide in the C1R-CD1d transfectants used for these experiments. To do so, we took advantage of the exquisite specificity of the DP10.7 TCR to CD1d-sulfatide by generating tetramers of this TCR for use as a CD1d-sulfatide specific staining reagent. Staining of unloaded C1R-CD1d transfectants expressing endogenous antigens with the DP10.7 TCR tetramer showed no surface staining, consistent with our SPR experiments presented in Figure 1 showing no detectable binding between the DP10.7 TCR and CD1d presenting endogenous lipid (“unloaded”). In contrast, nearly 100% of C1R-CD1d transfectants loaded with sulfatide stained strongly positive for the DP10.7 TCR tetramer (Figure 7C). This suggests our protocol for loading C1R-CD1d transfectants with sulfatide effectively saturates the cell-surface CD1d with sulfatide, thus allowing us to discriminate the Vδ1 response against CD1d/sulfatide from that of CD1d presenting endogenous lipids.
We then measured the TNF-α response of the polyclonal Vδ1 cells to CD1d, either loaded with sulfatide or presenting endogenous lipids (“unloaded”). After 16 hours, approximately 4% of Vδ1 T cells were producing TNF-α in response to C1R cells transfected with CD1d presenting endogenous lipids, as measured by intracellular cytokine staining (Figure 7D), consistent with the reactivity noted by Spinozzi and colleagues (Russano et al., 2007). In response to C1R cells transfected with CD1d and loaded with sulfatide, approximately 2% of these cells stained positive for TNF-α production. Considering that ~20% of this polyclonal Vδ1 population produces TNF-α in response to TCR triggering and correcting for the background 1% of TNF-α detected in mock treated cells (Figure 7D, top), we can approximate that ~15% of TNF-α producing Vδ1 cells in this population respond to CD1d presenting endogenous lipids and ~5% respond to CD1d presenting sulfatide. After 24 hours the measured amount of TNF-α secreted in the supernatant of Vδ1 cells responding to the CD1d-endogenous versus CD1d-sulfatide samples trended similarly to the intracellular cytokine staining results (Figure 7E). Together, these results demonstrate that a population of Vδ1 T cells in the gut respond to CD1d presenting endogenous antigens including the lipid sulfatide.
Discussion
One of the prevalent ideas about γδ T cell recognition is that it is antibody-like in nature, contrasting with peptide-MHC-specific αβ TCRs that share an evolutionarily conserved, germline-encoded, footprint upon the MHC surface (Born et al., 2013; Garcia and Adams, 2005; Marrack et al., 2008). This observation about γδ TCR recognition has arisen from the diverse structural nature of previously described γδ TCR ligands, for which many have been ascribed to singular TCR clones (Vantourout and Hayday, 2013). However, given the limited number of V gene segments available for γδ TCR recombination, it is more likely that a few ligands dominate among the potential repertoire of recognizable antigens, such as the murine T22-reactive population (Adams et al., 2005; Shin et al., 2005) and in humans, MICA for Vδ1 TCRs and phosphorylated small molecules for Vδ2 TCRs. However, a clear γδ V segment-directed recognition of an antigen-presenting molecule has yet to be explicitly demonstrated. Our structure reveals the molecular basis for this association and shows how the recombined CDR3δ loop mediates fine antigen specificity in a manner akin to classical αβ TCRs.
γδ T cells are particularly abundant among intraepithelial T cells of the small intestine and colon. Numerous studies have demonstrated that this population exhibits a crucial role in the preservation of the epithelial barrier, via production of both tissue repair and inflammatory anti-microbial factors (Smith and Garrett, 2011). Given that the human γδ IEL population exhibits a substantial Vδ1 TCR cell bias, an important question remains as to whether these functions are controlled through TCR recognition of specific antigens. We examined a set of previously described human intestinal clones, finding that not only were these TCRs CD1d-specific, but also recognized the self-lipid sulfatide. In addition, we demonstrate that a sizable percent of jejunal derived Vδ1 T cells producing TNF-α (~15%) are responsive to CD1d presenting endogenous lipids and ~5% respond to CD1d presenting sulfatide. We cannot definitively conclude these cells are specific to sulfatide, as it is clear that some Vδ1 T cells can recognize CD1d independently of lipid (such as the AB18.1 T cell clone isolated from the blood), yet it is unlikely that all of these CD1d-sulfatide reactive T cells are lipid-independent. Furthermore, we have only assessed one effector output (TNF-α production); it is possible that other cytokines are being produced by some of the remaining 80% of Vδ1 T cells in this population in response to CD1d stimulation.
Among the TCRs we identified as CD1d-specific, most striking was the IEL clone δ1A/B-3 (Groh et al., 1998). This and other Vδ1+ IEL clones have been described as MICA-reactive for over a decade, with MICA a widely acknowledged Vδ1 TCR ligand (Vantourout and Hayday, 2013). Our results showing δ1A/B-3 TCR binds with appreciable affinity to CD1d-sulfatide, suggesting this may be an alternative stimulatory TCR ligand for MICA reactive T cells. Because the MICA-specific co-stimulatory receptor NKG2D is ubiquitously expressed among Vδ1+ γδ T cells and binds MICA with high affinity, it would effectively out-compete the low binding affinity of the δ1A/B-3 TCR for MICA recognition (Groh et al., 2001; Wu et al., 2002; Xu et al., 2011). MICA cell-surface expression and NKG2D engagement occurs only during cellular stress (Groh et al., 1999; Meresse et al., 2004); therefore, this signaling pathway may only be relevant during disease conditions. However the constitutive expression of CD1d in human intestinal epithelial cells (IECs) (Dougan et al., 2007; Perera et al., 2007), between which IEL stably reside, and the enrichment of endogenous lipids such as sulfatide in these tissues, (Breimer et al., 2012) may provide a range of signals to Vδ1+ IELs, homeostatic in healthy tissue and inflammatory when paired with MICA-NKG2D in a disease setting.
CD1 molecules were among the first described ligands for γδ T cells, yet the field of CD1-lipid recognition largely diverted to the study of αβ T cells with the discovery of NKT cells (Bendelac et al., 1995; Porcelli et al., 1989). IELs within the intestine are one of the largest T cell populations in humans, and of these γδ T cells comprise almost 40% (Cheroutre, 2005; Deusch et al., 1991). Our results and those from others have shown that human Vδ1+ T cells, and more recently, Vδ3+ T cells, can recognize CD1d-lipid antigens, implicating γδ T cells as the largest lipid-reactive population in humans (Agea et al., 2005; Bai et al., 2012; Mangan et al., 2013; Russano et al., 2007; Spada et al., 2000). While we have demonstrated sulfatide recognition by several Vδ1 clones, the potential for reactivity towards lipids with diverse chemical natures is clearly suggested by our structure, with the recombined CDR3δ loop positioned over the central lipid portal. Our results suggest the prevalence of lipid recognition among all Vδ1+ T cell populations should be re-examined.
Experimental procedures
CD1d, MICA, and TCR cloning, expression and purification
These protocols are described in detail in the Supplemental Methods.
Surface Plasmon Resonance
All SPR measurements were conducted using a Biacore 2000 instrument operated at 25°C. 800 response units (RU) of each TCR were immobilized on a streptavidin sensor chip, with one flow cell left blank for reference subtraction. All flow cells were then blocked with 1 µM biotin. Serial dilutions of CD1d (unloaded or loaded with indicated lipids) from 8 to 0.25 µM were flowed as analyte. Equilibrium dissociation constants were determined by nonlinear regression with GraphPad (Prism) software using a shared Bmax, given identical immobilizations of the TCRs.
Jurkat transductions and plate-stimulation assays
The full-length δ and γ chains of the DP10.7 TCR, and a control Vδ1 TCR JR.2, were cloned into pMSCV vectors (a gift of M. Kuhns) and retrovirally transduced into TCRβ- Jurkat J.RT3-T3.5 cells. To measure CD1d-sulfatide specific stimulation, 96-wel flat bottom plates were coated with purified WT human CD1d at the indicted amounts (0–10 µg/well) at 37 C for 2 hours. Wells were washed and bovine brain sulfatides (Maytreya, 100 µg/mL), or DMSO control, were added for 3 hours at 37 C. Wells were washed and 5×104 DP10.7 or JR.2 TCR J.RT3-T3.5 transductants were added for a 12-hour incubation. Activation was measured by CD69 expression (FN50, Biolegend).
Crystallization, structure determination, refinement and analysis
These protocols are described in detail in the Supplemental Methods.
CD1d tetramer production, plate binding and BLI measurements of TCRs
These protocols are described in detail in the Supplemental Methods.
Vδ1 IEL cell line characterization and stimulation
Intraepithelial lymphocytes were purified from a jejunal biopsy as described (Jabri et al., 2000) (details in Supplemental Methods).
Supplementary Material
Highlights.
γδ T cells recognize CD1d-sulfatide directly through the TCR
Structure of a γδ TCR-CD1d-sulfatide complex shows exclusive δ chain role
Germline residues contact CD1d; junctional CDR3δ residues contact sulfatide
CD1d-sulfatide recognition is prevalent among human gut Vδ1 T cells
Acknowledgements
We thank the staff of the Advanced Proton Source at GM/CA-CAT (23-ID) and LS-CAT (21-ID) for their use and assistance with X-ray beamlines; Ruslan Sanishvili and Craig Ogata in particular for help and advice during data collection. We thank Mike Kuhns for the gift of the P2-pMSCV vector. This work was supported by National Institutes of Health Grants R56_AI097386 and R01_AI073922 to E.J.A., R01_DK67180 to B.J., RO1 AI038339 and PO1 AI053725 to A.B., and P30_DK42086 to the DDRCC (University of Chicago Digestive Diseases Research Core Center) and.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers The coordinates and structure factors for the CD1d-sulfatide, svDP10.7 TCR-CD1d-sulfatide complex, DP10.7 TCR, and AB18.1 TCR have been deposited in the Protein Data Bank under the accession codes 4MQ7, 4MNG, 4MNH, 4NDM respectively.
References
- Adams EJ, Chien YH, Garcia KC. Structure of a gammadelta T cell receptor in complex with the nonclassical MHC T22. Science. 2005;308:227–231. doi: 10.1126/science.1106885. [DOI] [PubMed] [Google Scholar]
- Adams EJ, Strop P, Shin S, Chien YH, Garcia KC. An autonomous CDR3delta is sufficient for recognition of the nonclassical MHC class I molecules T10 and T22 by gammadelta T cells. Nat Immunol. 2008;9:777–784. doi: 10.1038/ni.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agea E, Russano A, Bistoni O, Mannucci R, Nicoletti I, Corazzi L, Postle AD, De Libero G, Porcelli SA, Spinozzi F. Human CD1-restricted T cell recognition of lipids from pollens. J Exp Med. 2005;202:295–308. doi: 10.1084/jem.20050773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai L, Picard D, Anderson B, Chaudhary V, Luoma A, Jabri B, Adams EJ, Savage PB, Bendelac A. The majority of CD1d-sulfatide-specific T cells in human blood use a semiinvariant Vdelta1 TCR. Eur J Immunol. 2012;42:2505–2510. doi: 10.1002/eji.201242531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, Spies T. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–729. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- Bendelac A, Lantz O, Quimby ME, Yewdell JW, Bennink JR, Brutkiewicz RR. CD1 recognition by mouse NK1+ T lymphocytes. Science. 1995;268:863–865. doi: 10.1126/science.7538697. [DOI] [PubMed] [Google Scholar]
- Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. [DOI] [PubMed] [Google Scholar]
- Bonneville M, Ito K, Krecko EG, Itohara S, Kappes D, Ishida I, Kanagawa O, Janeway CA, Murphy DB, Tonegawa S. Recognition of a self major histocompatibility complex TL region product by gamma delta T-cell receptors. Proc Natl Acad Sci U S A. 1989;86:5928–5932. doi: 10.1073/pnas.86.15.5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, Rossjohn J. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- Born WK, Kemal Aydintug M, O'Brien RL. Diversity of gammadelta T-cell antigens. Cell Mol Immunol. 2013;10:13–20. doi: 10.1038/cmi.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breimer ME, Hansson GC, Karlsson KA, Larson G, Leffler H. Glycosphingolipid composition of epithelial cells isolated along the villus axis of small intestine of a single human individual. Glycobiology. 2012;22:1721–1730. doi: 10.1093/glycob/cws115. [DOI] [PubMed] [Google Scholar]
- Brenner MB, McLean J, Dialynas DP, Strominger JL, Smith JA, Owen FL, Seidman JG, Ip S, Rosen F, Krangel MS. Identification of a putative second T-cell receptor. Nature. 1986;322:145–149. [PubMed] [Google Scholar]
- Cheroutre H. IELs: enforcing law and order in the court of the intestinal epithelium. Immunol Rev. 2005;206:114–131. doi: 10.1111/j.0105-2896.2005.00284.x. [DOI] [PubMed] [Google Scholar]
- Chowers Y, Holtmeier W, Harwood J, Morzycka-Wroblewska E, Kagnoff MF. The V delta 1 T cell receptor repertoire in human small intestine and colon. J Exp Med. 1994;180:183–190. doi: 10.1084/jem.180.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constant P, Davodeau F, Peyrat MA, Poquet Y, Puzo G, Bonneville M, Fournie JJ. Stimulation of human gamma delta T cells by nonpeptidic mycobacterial ligands. Science. 1994;264:267–270. doi: 10.1126/science.8146660. [DOI] [PubMed] [Google Scholar]
- Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- Deusch K, Luling F, Reich K, Classen M, Wagner H, Pfeffer K. A major fraction of human intraepithelial lymphocytes simultaneously expresses the gamma/delta T cell receptor, the CD8 accessory molecule and preferentially uses the V delta 1 gene segment. Eur J Immunol. 1991;21:1053–1059. doi: 10.1002/eji.1830210429. [DOI] [PubMed] [Google Scholar]
- Dougan SK, Kaser A, Blumberg RS. CD1 expression on antigen-presenting cells. Curr Top Microbiol Immunol. 2007;314:113–141. doi: 10.1007/978-3-540-69511-0_5. [DOI] [PubMed] [Google Scholar]
- Garcia KC, Adams EJ. How the T cell receptor sees antigen--a structural view. Cell. 2005;122:333–336. doi: 10.1016/j.cell.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, Teyton L, Wilson IA. An alphabeta T cell receptor structure at 2.5 A and its orientation in the TCR-MHC complex. Science. 1996;274:209–219. doi: 10.1126/science.274.5285.209. [DOI] [PubMed] [Google Scholar]
- Girardi E, Maricic I, Wang J, Mac TT, Iyer P, Kumar V, Zajonc DM. Type II natural killer T cells use features of both innate-like and conventional T cells to recognize sulfatide self antigens. Nat Immunol. 2012;13:851–856. doi: 10.1038/ni.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh V, Rhinehart R, Randolph-Habecker J, Topp MS, Riddell SR, Spies T. Costimulation of CD8alphabeta T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat Immunol. 2001;2:255–260. doi: 10.1038/85321. [DOI] [PubMed] [Google Scholar]
- Groh V, Rhinehart R, Secrist H, Bauer S, Grabstein KH, Spies T. Broad tumor-associated expression and recognition by tumor-derived gamma delta T cells of MICA and MICB. Proc Natl Acad Sci U S A. 1999;96:6879–6884. doi: 10.1073/pnas.96.12.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh V, Steinle A, Bauer S, Spies T. Recognition of stress-induced MHC molecules by intestinal epithelial gammadelta T cells. Science. 1998;279:1737–1740. doi: 10.1126/science.279.5357.1737. [DOI] [PubMed] [Google Scholar]
- Haig NA, Guan Z, Li D, McMichael A, Raetz CR, Xu XN. Identification of self-lipids presented by CD1c and CD1d proteins. J Biol Chem. 2011;286:37692–37701. doi: 10.1074/jbc.M111.267948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabri B, de Serre NP, Cellier C, Evans K, Gache C, Carvalho C, Mougenot JF, Allez M, Jian R, Desreumaux P, et al. Selective expansion of intraepithelial lymphocytes expressing the HLA-E-specific natural killer receptor CD94 in celiac disease. Gastroenterology. 2000;118:867–879. doi: 10.1016/S0016-5085(00)70173-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabri B, Selby JM, Negulescu H, Lee L, Roberts AI, Beavis A, Lopez-Botet M, Ebert EC, Winchester RJ. TCR specificity dictates CD94/NKG2A expression by human CTL. Immunity. 2002;17:487–499. doi: 10.1016/s1074-7613(02)00427-2. [DOI] [PubMed] [Google Scholar]
- Komano H, Fujiura Y, Kawaguchi M, Matsumoto S, Hashimoto Y, Obana S, Mombaerts P, Tonegawa S, Yamamoto H, Itohara S, et al. Homeostatic regulation of intestinal epithelia by intraepithelial gamma delta T cells. Proc Natl Acad Sci U S A. 1995;92:6147–6151. doi: 10.1073/pnas.92.13.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Sagaseta J, Dulberger CL, Crooks JE, Parks CD, Luoma AM, McFedries A, Van Rhijn I, Saghatelian A, Adams EJ. The molecular basis for Mucosal-Associated Invariant T cell recognition of MR1 proteins. Proc Natl Acad Sci U S A. 2013;110:E1771–E1778. doi: 10.1073/pnas.1222678110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan BA, Dunne MR, O'Reilly VP, Dunne PJ, Exley MA, O'Shea D, Scotet E, Hogan AE, Doherty DG. Cutting edge: CD1d restriction and Th1/Th2/Th17 cytokine secretion by human Vdelta3 T cells. Journal of immunology. 2013;191:30–34. doi: 10.4049/jimmunol.1300121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard J, Petersson K, Wilson DH, Adams EJ, Blondelle SE, Boulanger MJ, Wilson DB, Garcia KC. Structure of an autoimmune T cell receptor complexed with class II peptide-MHC: insights into MHC bias and antigen specificity. Immunity. 2005;22:81–92. doi: 10.1016/j.immuni.2004.11.015. [DOI] [PubMed] [Google Scholar]
- McVay LD, Carding SR. Generation of human gammadelta T-cell repertoires. Crit Rev Immunol. 1999;19:431–460. [PubMed] [Google Scholar]
- Meresse B, Chen Z, Ciszewski C, Tretiakova M, Bhagat G, Krausz TN, Raulet DH, Lanier LL, Groh V, Spies T, et al. Coordinated induction by IL15 of a TCRindependent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity. 2004;21:357–366. doi: 10.1016/j.immuni.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Metelitsa LS, Naidenko OV, Kant A, Wu HW, Loza MJ, Perussia B, Kronenberg M, Seeger RC. Human NKT cells mediate antitumor cytotoxicity directly by recognizing target cell CD1d with bound ligand or indirectly by producing IL-2 to activate NK cells. Journal of immunology. 2001;167:3114–3122. doi: 10.4049/jimmunol.167.6.3114. [DOI] [PubMed] [Google Scholar]
- Park SH, Weiss A, Benlagha K, Kyin T, Teyton L, Bendelac A. The mouse CD1d-restricted repertoire is dominated by a few autoreactive T cell receptor families. J Exp Med. 2001;193:893–904. doi: 10.1084/jem.193.8.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel O, Pellicci DG, Gras S, Sandoval-Romero ML, Uldrich AP, Mallevaey T, Clarke AJ, Le Nours J, Theodossis A, Cardell SL, et al. Recognition of CD1d-sulfatide mediated by a type II natural killer T cell antigen receptor. Nat Immunol. 2012;13:857–863. doi: 10.1038/ni.2372. [DOI] [PubMed] [Google Scholar]
- Pellicci DG, Patel O, Kjer-Nielsen L, Pang SS, Sullivan LC, Kyparissoudis K, Brooks AG, Reid HH, Gras S, Lucet IS, et al. Differential recognition of CD1d-alphagalactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity. 2009;31:47–59. doi: 10.1016/j.immuni.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera L, Shao L, Patel A, Evans K, Meresse B, Blumberg R, Geraghty D, Groh V, Spies T, Jabri B, Mayer L. Expression of nonclassical class I molecules by intestinal epithelial cells. Inflamm Bowel Dis. 2007;13:298–307. doi: 10.1002/ibd.20026. [DOI] [PubMed] [Google Scholar]
- Porcelli S, Brenner MB, Greenstein JL, Balk SP, Terhorst C, Bleicher PA. Recognition of cluster of differentiation 1 antigens by human CD4-CD8-cytolytic T lymphocytes. Nature. 1989;341:447–450. doi: 10.1038/341447a0. [DOI] [PubMed] [Google Scholar]
- Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI. Recognition of CD1d-restricted antigens by natural killer T cells. Nat Rev Immunol. 2012;12:845–857. doi: 10.1038/nri3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russano AM, Agea E, Corazzi L, Postle AD, De Libero G, Porcelli S, de Benedictis FM, Spinozzi F. Recognition of pollen-derived phosphatidyl-ethanolamine by human CD1d-restricted gamma delta T cells. J Allergy Clin Immunol. 2006;117:1178–1184. doi: 10.1016/j.jaci.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Russano AM, Bassotti G, Agea E, Bistoni O, Mazzocchi A, Morelli A, Porcelli SA, Spinozzi F. CD1-restricted recognition of exogenous and self-lipid antigens by duodenal gammadelta+ T lymphocytes. J Immunol. 2007;178:3620–3626. doi: 10.4049/jimmunol.178.6.3620. [DOI] [PubMed] [Google Scholar]
- Sandstrom A, Scharf L, McRae G, Hawk AJ, Meredith SC, Adams EJ. gammadelta T cell receptors recognize the non-classical major histocompatibility complex (MHC) molecule T22 via conserved anchor residues in a MHC peptide-like fashion. The Journal of biological chemistry. 2012;287:6035–6043. doi: 10.1074/jbc.M111.333153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotet E, Martinez LO, Grant E, Barbaras R, Jeno P, Guiraud M, Monsarrat B, Saulquin X, Maillet S, Esteve JP, et al. Tumor recognition following Vgamma9Vdelta2 T cell receptor interactions with a surface F1-ATPase-related structure and apolipoprotein A-I. Immunity. 2005;22:71–80. doi: 10.1016/j.immuni.2004.11.012. [DOI] [PubMed] [Google Scholar]
- Selmaj K, Brosnan CF, Raine CS. Colocalization of lymphocytes bearing gamma delta T-cell receptor and heat shock protein hsp65+ oligodendrocytes in multiple sclerosis. Proc Natl Acad Sci U S A. 1991;88:6452–6456. doi: 10.1073/pnas.88.15.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S, El-Diwany R, Schaffert S, Adams EJ, Garcia KC, Pereira P, Chien YH. Antigen recognition determinants of gammadelta T cell receptors. Science. 2005;308:252–255. doi: 10.1126/science.1106480. [DOI] [PubMed] [Google Scholar]
- Smith PM, Garrett WS. The gut microbiota and mucosal T cells. Front Microbiol. 2011;2:111. doi: 10.3389/fmicb.2011.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spada FM, Grant EP, Peters PJ, Sugita M, Melian A, Leslie DS, Lee HK, van Donselaar E, Hanson DA, Krensky AM, et al. Self-recognition of CD1 by gamma/delta T cells: implications for innate immunity. J Exp Med. 2000;191:937–948. doi: 10.1084/jem.191.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi T, Suzuki T. Role of sulfatide in normal and pathological cells and tissues. J Lipid Res. 2012;53:1437–1450. doi: 10.1194/jlr.R026682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettamanti G, Bonali F, Marchesini S, Zambotti V. A new procedure for the extraction, purification and fractionation of brain gangliosides. Biochim Biophys Acta. 1973;296:160–170. doi: 10.1016/0005-2760(73)90055-6. [DOI] [PubMed] [Google Scholar]
- Toulon A, Breton L, Taylor KR, Tenenhaus M, Bhavsar D, Lanigan C, Rudolph R, Jameson J, Havran WL. A role for human skin-resident T cells in wound healing. J Exp Med. 2009;206:743–750. doi: 10.1084/jem.20081787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Merwe PA, Davis SJ. Molecular interactions mediating T cell antigen recognition. Annu Rev Immunol. 2003;21:659–684. doi: 10.1146/annurev.immunol.21.120601.141036. [DOI] [PubMed] [Google Scholar]
- Vantourout P, Hayday A. Six-of-the-best: unique contributions of gammadelta T cells to immunology. Nat Rev Immunol. 2013;13:88–100. doi: 10.1038/nri3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willcox CR, Pitard V, Netzer S, Couzi L, Salim M, Silberzahn T, Moreau JF, Hayday AC, Willcox BE, Dechanet-Merville J. Cytomegalovirus and tumor stress surveillance by binding of a human gammadelta T cell antigen receptor to endothelial protein C receptor. Nat Immunol. 2012;13:872–879. doi: 10.1038/ni.2394. [DOI] [PubMed] [Google Scholar]
- Wu J, Groh V, Spies T. T cell antigen receptor engagement and specificity in the recognition of stress-inducible MHC class I-related chains by human epithelial gamma delta T cells. Journal of immunology. 2002;169:1236–1240. doi: 10.4049/jimmunol.169.3.1236. [DOI] [PubMed] [Google Scholar]
- Wucherpfennig KW, Newcombe J, Li H, Keddy C, Cuzner ML, Hafler DA. Gamma delta T-cell receptor repertoire in acute multiple sclerosis lesions. Proc Natl Acad Sci U S A. 1992;89:4588–4592. doi: 10.1073/pnas.89.10.4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu B, Pizarro JC, Holmes MA, McBeth C, Groh V, Spies T, Strong RK. Crystal structure of a gammadelta T-cell receptor specific for the human MHC class I homolog MICA. Proc Natl Acad Sci U S A. 2011;108:2414–2419. doi: 10.1073/pnas.1015433108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajonc DM, Maricic I, Wu D, Halder R, Roy K, Wong CH, Kumar V, Wilson IA. Structural basis for CD1d presentation of a sulfatide derived from myelin and its implications for autoimmunity. J Exp Med. 2005;202:1517–1526. doi: 10.1084/jem.20051625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Wei YL, Huang J, Newell EW, Yu H, Kidd BA, Kuhns MS, Waters RW, Davis MM, Weaver CT, Chien YH. gammadelta T cells recognize a microbial encoded B cell antigen to initiate a rapid antigen-specific interleukin-17 response. Immunity. 2012;37:524–534. doi: 10.1016/j.immuni.2012.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.