Abstract
Recent progress has been made in the identification of protein-coding genes and miRNAs that are expressed in and alter the behavior of colonic epithelia. However, the role of long non-coding RNAs (lncRNAs) in colonic homeostasis is just beginning to be explored. By gene expression profiling of post-mitotic, differentiated tops and proliferative, progenitor-compartment bottoms of microdissected adult mouse colonic crypts, we identified several lncRNAs more highly expressed in crypt bottoms. One identified lncRNA, designated non-coding Nras functional RNA (ncNRFR), resides within the Nras locus but appears to be independent of the Nras coding transcript. Stable overexpression of ncNRFR in non-transformed, conditionally immortalized mouse colonocytes results in malignant transformation, as determined by growth in soft agar and formation of highly invasive tumors in nude mice. Moreover, ncNRFR appears to inhibit the function of the tumor suppressor let-7. These results suggest precise regulation of ncNRFR is necessary for proper cell growth in the colonic crypt, and its misregulation results in neoplastic transformation.
Keywords: Progenitor cells, Non-coding RNA, Colon, Intestine, Colorectal cancer, let-7
1. Introduction
The intestinal epithelium is one of the most rapidly proliferating, self-renewing epithelia in the body [1]. Stem cells reside at the bottom of cellular invaginations called crypts of Lieberkühn. Stem cells continuously divide, giving rise to daughter cells that differentiate as they move up the crypt column and are ultimately shed into the lumen of the intestine [1].
Characterization of the intestinal progenitor cell compartment (PCC) has largely focused on identification of protein-coding genes that define the PCC and several of these have been shown by lineage labeling to mark stem cells [2–6]. The importance of non-coding RNAs in the PCC has been largely restricted to miRNA function [7,8], although long non-coding RNAs (lncRNAs) are now recognized as regulators of stem cell function in general [9]. The transcriptome consists of many lncRNAs that map mostly to intronic and extragenic regions of the genome [10] with estimates of total lncRNAs varying from 7% to greater than 50% of total transcripts [11–13]. The miRNAs are among the best characterized regulatory ncRNAs that bind through seed sequences, 2–8 base pairs in length at the 5′ end of the miRNA [14], which directly hybridize to target transcripts. Some miRNAs, like mir-17–92, may function as oncogenes, while others, like let-7, have tumor-suppressor functions [15,16]. LncRNAs have been found to participate in epigenetic regulation through chromatin remodeling and transcriptional regulation, with direct consequences for cancer progression [17,18]. In addition, non-coding pseudogenes can act as miRNA binding decoys that inhibit miRNA function, thus driving oncogenesis [19]. Herein, we have identified a colonic lncRNA that is localized to the PCC. This RNA is from the Nras locus of the mouse genome and we have designated it non-coding Nras Functional RNA (ncNRFR). Overexpression of ncNRFR in immortalized mouse colonocytes results in transformation in vitro and in vivo.
2. Materials and methods
2.1. Computer analysis
Programs used were the RNA-fold program (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) and the UCSC browser. BLASTX confirmed there are no homologous coding sequences in ncNRFR cDNA.
2.2. siRNA knockdown of ncNRFR
Oligonucleotides coding for ncNRFR siRNAs (see Supplementary Table 1) were cloned into pSilencer 4.1 hygro (ABI/Ambion, Austin, TX) and control non-targeting siRNAs in this vector were used as controls. Nucleofection (Lonza/Amaxa Gaithersburg, MD) was used to transfect with solution L with program X-005 (Lonza). Transfection was near 90–100% as measured by fluorescence with a pmaxGFP expression construct (Lonza).
2.3. RNA identification
Colonic crypt epithelium was isolated [20] and crypts were bisected with an opthalmic microscalpel. Approximately 7500 cells from 50 crypt fragments were pooled, yielding 0.5 µg of RNA that was linearly amplified (aRNA) yielding 30 µg [21]. These aRNAs were used to probe a microarray that identified a few candidate genes that had differential expression in the crypt epithelium. Three micrograms of aRNA was fluorescently labeled with Cy3 or Cy5 dyes and hybridized to a 5000-gene cDNA Research Genetics microarrray.
2.4. Northern blot analysis
Total RNA was purified with TRIzol reagent (Invitrogen) and RNAeasy (Qiagen, Valencia, CA). Nras cDNA probe (Thermo Fischer Open Biosystems clone ID: 6475312) was purified from an EcoRI 518 bp digested fragment, and ncNRFR probe fragment (bases 679–990, see Supplementary Table 1) was made by PCR amplification from a non-repeat region of the cDNA. These were used for Northern blots as described [22], using 10 µg of total RNA per lane.
2.5. Nude mouse cell injections
Three to six million cells were injected subcutaneously into the right flank of athymic nude mice. This study was carried out in strict accordance with animal care and use guidelines and approval of the Vanderbilt IACUC. Mice were monitored throughout the experiment for signs of distress and tumor growth greater than 2 cm.
2.6. Isolated whole crypt RNA in situ hybridization (ISH)
RNA whole mount ISH was performed as described [23]. Fluorescent ISH was performed using the Tyramide Signal Amplification (TSA) kit with manufacturer’s instructions (Perkin Elmer, Waltham, MA).
3. Results and discussion
3.1. ncNRFR is expressed in the colonic PCC
To identify genes expressed in the mouse colonic PCC, we microdissected crypt tops (differentiated cells) and bottoms (PCC), and analyzed RNA expression from the two populations by microarray profiling. These results were validated for 10 randomly selected genes by ISH on isolated whole colonic crypts (Fig. 1, Supplementary Fig. 1 and gene list Supplementary Table 2). In general, ISH showed expression in discrete crypt regions rather than in a gradient along the crypt axis. Many of the genes expressed in the PCC corresponded to apparent lncRNAs.
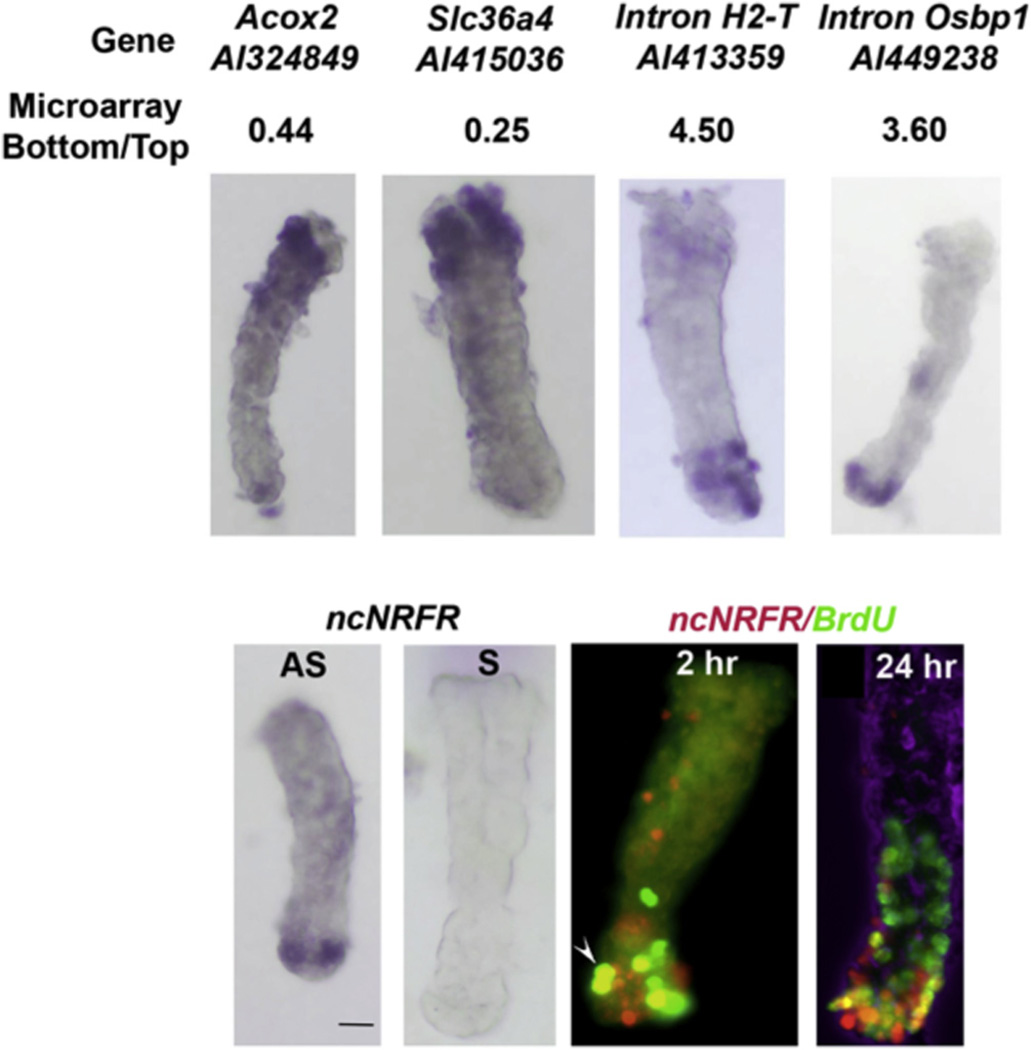
Fig. 1.
In situ hybridization (ISH) for genes expressed in the tops and bottoms of crypts. Top panels, examples of confirmatory RNA ISH with bottom to top microarray signal ratios and clone accession numbers shown. RNA ISH is shown for Acox2, Slc36a4, intronic lncRNA in H2-T, intronic lncRNA in Osbp1. Bottom panels from left to right, RNA ISH for ncNRFR and its negative sense control. Following a pulse of BrdU, colonic crypts were isolated and processed 2 and 24 h later and whole-mount staining of crypts for BrdU (green) and fluorescent ISH for ncNRFR (red) was performed. Shown are two representative crypts. The arrowhead points to a double stained cell. The far right panel has a DIC view overlaid in the blue channel to give an impression of the 3-D relationship of cells. The bottom to top microarray ratio for ncNRFR is 2.7. The scale bar is 50 uM.
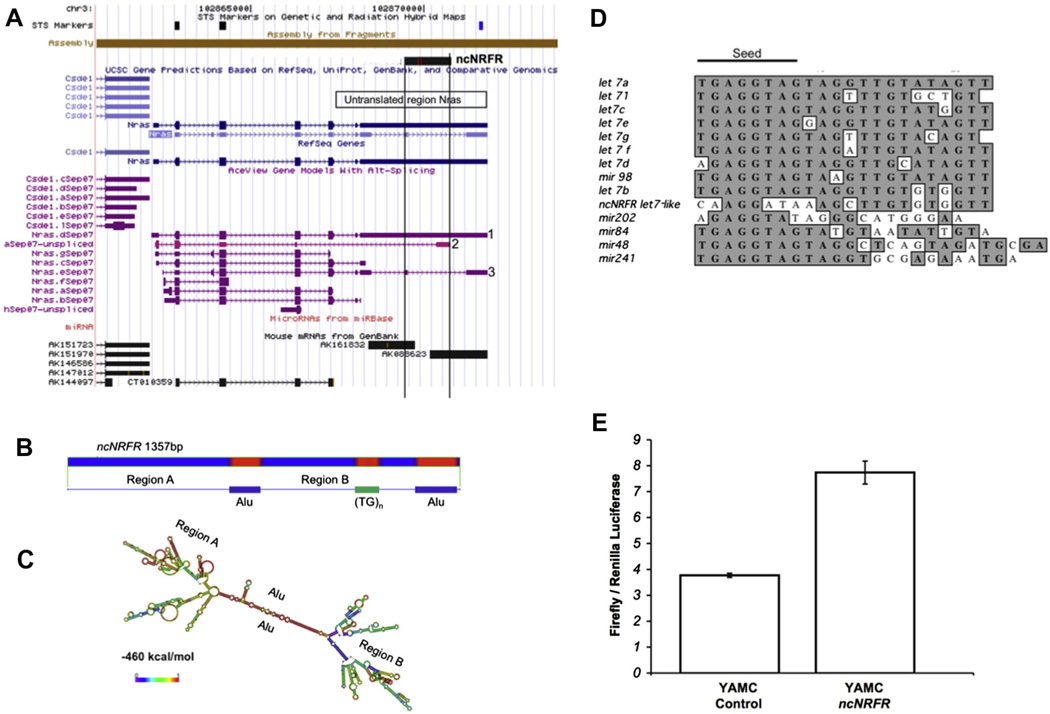
We identified a lncRNA 3′ to the coding portion of the Nras locus (Fig. 2A). This would represent a 1357 bp transcript, ncNRFR, which is transcribed in the same direction as Nras. It overlaps untranslated regions (UTRs) of some Nras splice variants and the intronic regions of others, but it does not overlap with other Nras transcripts (Fig. 2A). Two mouse Riken clones were found that overlap the 5′ and 3′ ends, respectively, of ncNRFR, suggesting transcriptional initiation occurs within this region. Based on annotated cDNA transcript information, there are numerous other ncRNAs in the ncNRFR locus that are independent from the Nras transcriptional locus, none of which map to known miRNAs. A BLASTX analysis showed no homologous coding sequences are present in ncNRFR; however, this analysis does not exclude the presence of short protein sequences that might code for unrecognized proteins in this transcript.
Fig. 2.
ncNRFR, coded for in the Nras locus, is predicted to fold into a complex secondary structure and has sequences similar to the let-7 family of miRNAs. (A) Shown is the blat search (http://genome.ucsc.edu/cgi-bin/hgBlat) for ncNRFR. Numerous transcripts map here, including two full-length polyadenylated Riken transcripts (AK161832 and AK088623) that overlap ncNRFR. (B) Repeat masker (http://www.repeatmasker.org) predicts two ALU-like repeats as well as a simple TG repeat sequence in ncNRFR. (C) Predicted RNA structure by the RNA-fold program (red overlay; http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) showing subregions; a free energy prediction is −460 kcal/mol. (D) A sequence within ncNRFR is compared to the sequence of let-7 family members and related miRNAs. Identical nucleotides are overlaid in gray. ncNRFR is most similar to let-7b. (E) A let-7 luciferase reporter consisting of let-7 binding sites present in the 3′-UTR of the luciferase gene was transfected into ncNRFR expressing and control YAMC cells. An increase in luciferase signal in ncNRFR cells indicates a decrease in let-7 activity. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
To confirm that ncNRFR was preferentially expressed in the PCC, we performed RNA ISH that showed ncNRFR is expressed most strongly in the crypt bottom (Fig. 1). This contrasts with Nras RNA ISH that shows an equal distribution along the crypt axis [24]. Fluorescent RNA ISH for ncNRFR was performed with immunostaining for BrdU in crypts from mice injected with BrdU. This shows ncNRFR is expressed in some cells labeled at 2 h. with BrdU, and ncNRFR expression mostly restricted to cells in the bottom of crypts as BrdU-labeled cells move up the crypt axis (Fig. 1). There are nonetheless a few cells in the upper part of the crypt that retain some ncNRFR expression (Supplementary Fig. 1). The Osbp1 and H2-T intronic RNAs showed similar expression patterns in the bottoms of crypts (Fig. 1).
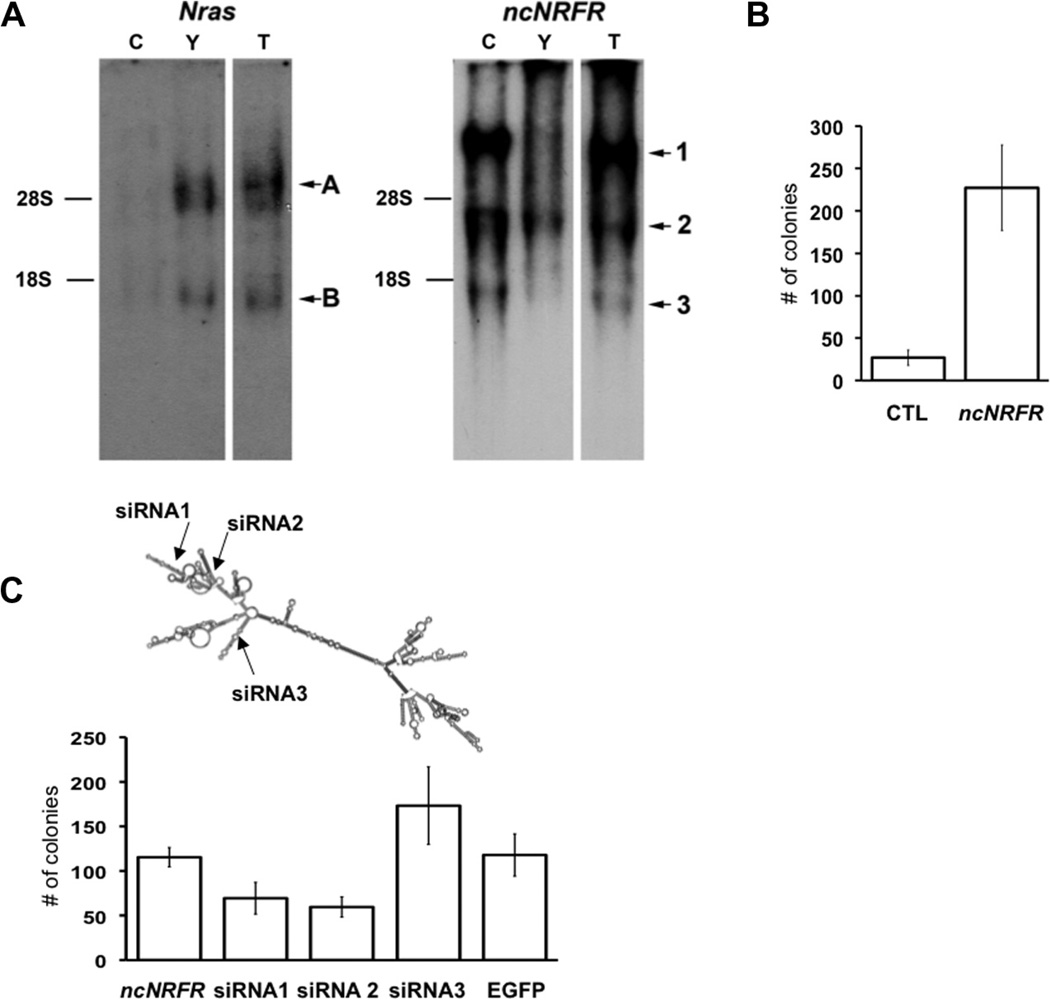
Northern blots using probes to Nras or ncNRFR confirmed that these loci appear to be independently transcribed in Young Adult Mouse Colon (YAMC) cells (Fig. 3A) with the two main transcripts (labeled A and B) for Nras not found in the ncNRFR blot (with major transcripts labeled 1, 2 and 3). YAMC cells are conditionally immortalized with a temperature sensitive T-antigen [25]; growth is permissive at 33 °C and restrictive at 37 °C. YAMC cells do not form colonies in soft agar or tumors in nude mice [26]. The ncNRFR transcription pattern is complicated, with very long transcripts (e.g. transcript 1) that do not appear to overlap the Nras coding region, since they are not present in the Nras Northern blot. An identical banding pattern was obtained with a probe to ncNRFR from another non-repeat region of the gene (42–216, not shown). The short Nras transcript (transcript B) is unlikely to correspond to the short ncNRFR transcript (transcript 3; approximately 1.3 kb); it is too small to encompass both Nras coding sequences and ncNRFR sequences based on current information of Nras splice variants. These two main clusters of Nras transcripts (A and B) correspond in size to previously identified Nras cDNAs (Supplementary Table 3). These results suggest the Nras and the ncNRFR loci contain independent transcripts, although it does not rule out longer stable processed Nras transcripts, yielding RNAs from the Nras 3′UTR, as has been observed in other cases throughout the human transcriptome (ENCODE project, [27]). Such processed non-coding 3′ UTR RNAs, like independently transcribed ncRNAs, can have important biological functions [28,29].
Fig. 3.
Over-expression of ncNRFR leads to altered cell growth in soft agar. (A) Northern blot analysis of Nras and ncNRFR. Total RNA was isolated from mouse colonic crypt (C), YAMC (Y) and tumor-derived, ncNRFR-expressing YAMC cells (T). There are three major ncNRFR transcripts upregulated in (T) cells (as labeled in the figure). Transcript 1 is over 9 kb, transcript 2 is about 3 kb, and transcript 3 is about 1.3 kb. There are several Nras transcript bands apparent. These correspond in size with known Nras transcripts (see Table Supplementary 2). The positions for 28S (4.7 kb) and 18S (1.7 kb) ribosomal RNA bands are marked based on the ethidium bromide-stained loading control gel (not shown). (B) Bar graph depicting average colony numbers in soft agar formed by YAMC cells expressing different constructs. YAMC cells expressing ncNRFR (ncNRFR) show a significant increase in average colony number compared to YAMC with vector alone (CTL; p = 0.008; N = 3). (C) Bar graph depicting average colony numbers in soft agar from YAMC ncNRFR cells expressing different siRNAs. Two siRNAs (siRNA 1, p = 0.04 and 2, p = 0.002) inhibited soft agar growth, whereas the third siRNA (siRNA3, p = 0.21), and EGFP control (EGFP, p = 0.92) did not. All experiments were performed three times in triplicate.
Further analysis of ncNRFR showed it contains different repeat regions (Fig. 2B) that are often associated with known non-coding RNAs [30,31]. Two ALU-like sequences are present in the opposite orientation in ncNRFR and are predicted to form a highly stable stem-like structure yielding three distinct regions that all contain stem loop structures (Fig. 2C). Upon further analysis of the ncNRFR sequence, we found a short region that resembled the let-7 family of miRNA (Fig. 2D). The sequences similar to let-7 are outside of the seed region, but are most similar to the let-7b family member. The let-7 family of miRNAs acts as tumor suppressors and are downregulated in many cancers [32]. Given the important role of let-7 as a tumor suppressor any alteration of let-7 function would have important implications for regulation of cell growth. Therefore, we tested whether ncNRFR controls let-7 function. To do this, we used conditionally immortalized YAMC cells, which are useful to assay for oncogenic transformation in vitro and in vivo [26,33]. YAMC cells were transfected with puromycin-selectable lentivirus- containing ncNRFR or an empty control vector. Pools of transfected and puromycin-selected cells with ncNRFR or without ncNRFR were tested for altered let-7 activity using a reporter construct that contains let-7 binding sites in the 3′-UTR of a luciferase gene. YAMC ncNRFR cells relieve some repression of a let-7 reporter compared to control, suggesting ncNRFR expression down-regulates let-7 function (Fig 2E). Consistent with these results, a microarray comparison of genes expressed in ncNRFR versus CTL showed that let-7b targets were significantly upregulated due to ncNRFR expression representing a statistically significant trend using GSEA analysis (Supplementary Tables 4 and 5 and Supplementary data file 6).
3.2. ncNRFR transforms conditionally immortalized mouse colon cells in cell culture
YAMC cells with or without ncNRFR were then tested for altered growth in soft agar (Fig. 3B). Cells transfected with ncNRFR formed colonies in soft agar, whereas YAMC-vector controls had few colonies. We generated siRNAs to three regions of ncNRFR (Fig. 3C) to test the specificity of YAMC soft agar growth on ncNRFR expression. Stable expression of these siRNAs caused a reduction in soft agar growth in two of three constructs tested (Fig. 3C).
3.3. ncNRFR transforms YAMC cells into highly invasive tumors in vivo
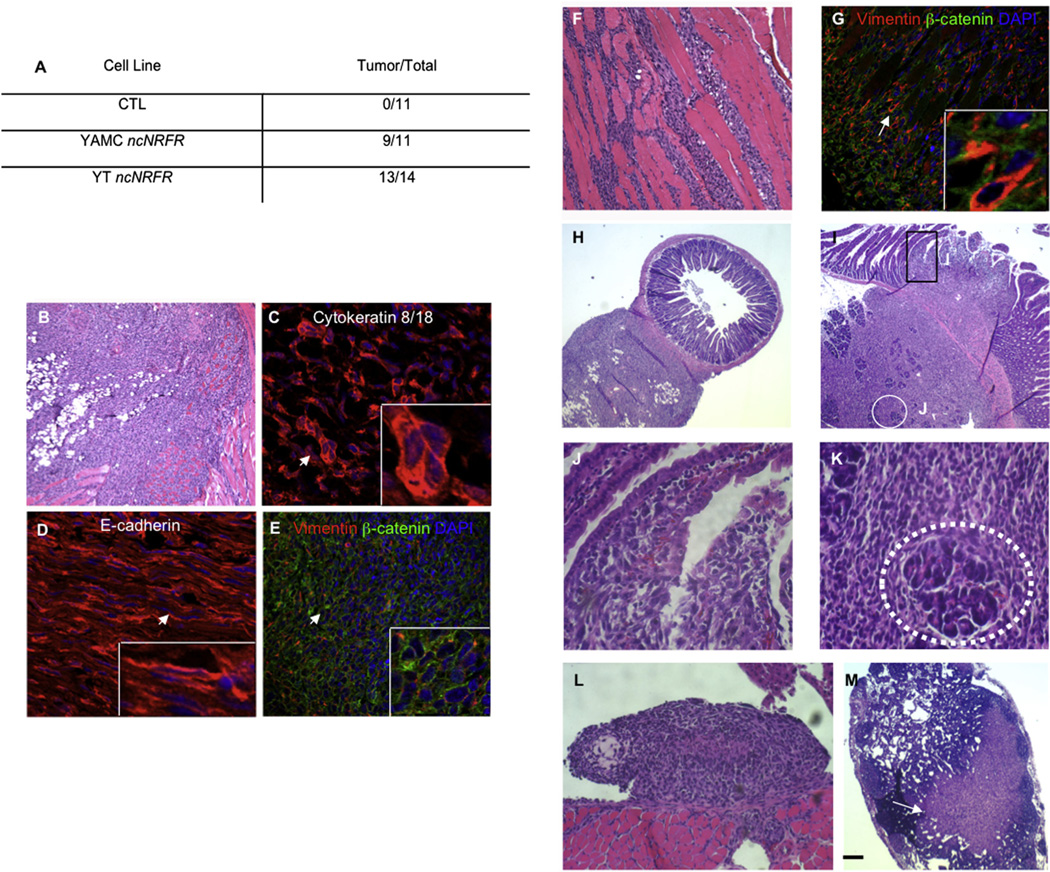
We then tested whether YMAC cells with ncNRFR formed tumors in athymic nude mice. Nine of 11 mice injected with YAMC ncNRFR cells developed tumors by 2 months of age, while none of those injected with the YAMC vector control cells formed tumors (Fig. 4A; CTL, 0 out of 11 mice).
Fig. 4.
ncNRFR-expressing YAMC cells are highly invasive when injected subcutaneously into nude mice. (A) The number of athymic nude mice injected subcutaneously with different cell lines and the numbers of tumors that formed (# tumors formed/total # injected). YT ncNRFR are re-derived from tumor cells isolated from YAMC ncNRFR tumors and selected with puromycin. (B) A re-derived YT ncNRFR mouse xenograft tumor (H&E-stained section). (C) Tumors sectioned and immunostained for keratins 8/18 (red) and (D) E-cadherin (red) with DNA DAPI stain (blue). (E) Immunostained section for vimentin (red), β-catenin (green) and DNA/DAPI (blue); there is cell surface β-catenin, despite cytoplasmic E-cadherin staining. (F) YT ncNRFR tumors invade through skeletal muscle (H&E-stained section). (G) Immunostained section for vimentin (red), β-catenin (green) and DNA/DAPI (blue) with vimentin immunoreactivity (arrow) increasing in cells that invade through the muscle (dark non-stained structures). Arrows in C, D, E and G mark the positions of magnified fields shown in the bottom right corner of each panel. H and E-stained sections, panels (H–M). (H) Tumor cells invade through the muscularis propria and into the lamina propria of the small intestine. (I) Tumor cells invade into the duodenum (black rectangle, high power field in J) and pancreas (white circle, high power field in K). (J) Invading cells entering into the luminal space and disrupting the villus epithelium. (K) A pancreatic acinus (dotted white line) surrounded by tumor cells. (L) YT ncNRFR cells invading into the diaphragm. (M) A lymph node containing tumor cells with the position of tumor cells marked by an arrow. Scale bar in (L) represents: 75 µm in B and F; 22 µm in C and D; 45 µM in E and G; 172 µM in H, I and M; 50 µM in L; 11 µM in J and K; in high power fields for C, D and G are 11 µM; in the E high power field it is 20 µM.
We generated multiple independent cell lines from an ncNRFRexpressing YAMC tumor (YT ncNRFR). These cultured cells survive selection with puromycin, indicating the original ncNRFR YAMC cells were still present in the tumor and could be re-isolated from it. Subcutaneous injection of these rederived cells in athymic nude mice resulted in shorter tumor latency, with tumors forming in one month (Fig. 4A, 13 out of 14 mice) compared to two months for the original YAMC ncNRFR cells.
Moreover, YT ncNRFR cells were highly invasive, often invading through muscle into the peritoneal cavity (Fig. 4B–M) with vimentin more highly expressed in cells migrating through the muscle layer (Fig. 4G). Immunohistochemical staining of tumors showed that cells are positive for Keratin 8 and 18 and for E-cadherin (Figs. 4C and D respectively), although E-cadherin staining was largely cytoplasmic. Cells are vimentin-positive and contain cell surface β-catenin (Fig. 4E), suggesting there may be another cell surface cadherin with which β-catenin interacts. Thus, YT ncNRFR tumors exhibit features of epithelial-to-mesenchymal transition (reviewed in [34]).
Remarkably, YT ncNRFR tumor cells invaded into intra-abdominal organs. The tumor invaded into the wall of the intestine and into the lumen of the gut (Fig. 4H–J), as well as pancreas (Fig. 4I and K), diaphragm (Fig. 4L) and stomach (not shown). In addition, tumor cells were seen in lymph nodes (Fig. 4M), which could be due either to lymphatic spread of the YT ncNRFR cells or direct invasion into the lymph node.
Northern blot analysis of RNA from YAMC-control (Y) and YT-ncNRFR (T) cells show a specific 1.3 kb transcript (Fig. 3A, transcript 3) that corresponds in size to ncNRFR and a greater than 9 kb and a 3 kb transcript induced by ncNRFR expression, which are significantly upregulated in ncNRFR-expressing cells (transcripts 1 and 2 respectively). Transcripts 1 and 2 are probably either overlapping existing ncNRFR transcripts stabilized by expression of ncNRFR or an alternatively processed from of ncNRFR. YT-ncNRFR upregulated transcripts resemble total crypt RNAs (C) from the ncNRFR locus. Expression of Nras coding transcripts remains approximately the same in all these cell lines, although relatively much less in mouse colonic crypts. These results suggest the transformed nature of YAMC cells is associated with re-establishing a crypt-like transcription pattern of ncNRFR in YAMC cells.
The control of normal PCC growth is dependent on multiple layers of iterative feedback that direct patterning of the stem cell environment, allowing it to balance proliferation and differentiation. In cancer, this balance is often perturbed, leading to a short-circuiting of this process, causing uncontrolled proliferation and loss of a restricted progenitor niche [1]. The role of lncRNAs in cancer progression, both as growth suppressors and as oncogenes, is a highly active area of research, with the lncRNAs being shown to regulate transcription, chromatin structure, multi-protein complex formation and miRNA function [17,18]. When overexpressed ncNRFR appears to perturb the growth of normal colonic epithelial cells, leading to a more transformed state, perhaps through inhibition of let-7 function.
Supplementary Material
Acknowledgments
This work was supported by NCI CA 46413, NCI CA 151566, GI Special Program of Research Excellence (P50 95103), Mouse Models of Human Cancers Consortium (U01 084239) to RJC. JLF has a career development award from the GI Special Program of Research Excellence and is recipient of a DDRC pilot grant (P30 DK58404); Core Services performed through Vanderbilt University Medical Center’s Digestive Disease Research Center supported by NIH Grant P30DK058404 Core Scholarship. This includes the Novel Cell Line Development and Tissue Morphology Subcores and Cell Imaging Core. We thank Ramona Deal, Dexter Duncan, Coe Foutch, Pamela Robinson, and Loice Jeyakumar for their assistance and Lynne Lapierre for advice and discussions.
Abbreviations
- lncRNAs
long non-coding RNAs
- ncNRFR
non-coding Nras functional RNA
- PCC
progenitor cell compartment
- ISH
in situ hybridization
- UTRs
untranslated regions
- YAMC
Young adult mouse colon
- PCR
polymerase chain reaction
- dpc
days postcoitus
- aRNA
amplified RNA
- BrdU
bromodeoxyuridine
- siRNA
small interfering RNAs
- GSEA
gene set enrichment analysis
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bbrc.2013.09.040.
References
- 1.Brittan M, Wright NA. Stem cell in gastrointestinal structure and neoplastic development. Gut. 2004;53:899–910. doi: 10.1136/gut.2003.025478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, Higginbotham JN, Juchheim A, Prasad N, Levy SE, Guo Y, Shyr Y, Aronow BJ, Haigis KM, Franklin JL, Coffey RJ. The Pan-ErbB negative regulator lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149:146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker N, Bartfeld S, Clevers H. Tissue-resident adult stem cell populations of rapidly self-renewing organs. Cell Stem Cell. 2010;7:656–670. doi: 10.1016/j.stem.2010.11.016. [DOI] [PubMed] [Google Scholar]
- 4.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat. Genet. 2008;40:915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334:1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snippert HJ, van Es JH, van den Born M, Begthel H, Stange DE, Barker N, Clevers H. Prominin-1/CD133 marks stem cells and early progenitors in mouse small intestine. Gastroenterology. 2009;136:2187–2194. doi: 10.1053/j.gastro.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, Barad O, Bentwich Z, Szafranska AE, Labourier E, Raymond CK, Roberts BS, Juhl H, Kinzler KW, Vogelstein B, Velculescu VE. The colorectal microRNAome. Proc. Natl. Acad. Sci. USA. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monzo M, Navarro A, Bandres E, Artells R, Moreno I, Gel B, Ibeas R, Moreno J, Martinez F, Diaz T, Martinez A, Balague O, Garcia-Foncillas J. Overlapping expression of microRNAs in human embryonic colon and colorectal cancer. Cell. Res. 2008;18:823–833. doi: 10.1038/cr.2008.81. [DOI] [PubMed] [Google Scholar]
- 9.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, Yang X, Amit I, Meissner A, Regev A, Rinn JL, Root DE, Lander ES. LincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattick JS, Makunin IV. Non-coding RNA. Hum. Mol. Genet. 2006;15(1):R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 11.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 12.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. BioEssays. 2007;29:288–299. doi: 10.1002/bies.20544. [DOI] [PubMed] [Google Scholar]
- 14.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 15.Zhang B, Pan X, Cobb GP, Anderson TA. MicroRNAs as oncogenes and tumor suppressors. Dev. Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 16.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 17.Nie L, Wu HJ, Hsu JM, Chang SS, Labaff AM, Li CW, Wang Y, Hsu JL, Hung MC. Long non-coding RNAs: versatile master regulators of gene expression and crucial players in cancer. Amer. J. Trans. Res. 2012;4:127–150. [PMC free article] [PubMed] [Google Scholar]
- 18.Gutschner T, Diederichs S. The Hallmarks of Cancer: a long non-coding RNA point of view. RNA Biol. 2012;9 doi: 10.4161/rna.20481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whitehead RH, Brown A, Bhathal PS. A method for the isolation and culture of human colonic crypts in collagen gels. In Vitro Cell Dev. Biol. 1987;23:436–442. doi: 10.1007/BF02623860. [DOI] [PubMed] [Google Scholar]
- 21.Wang E, Miller LD, Ohnmacht GA, Liu ET, Marincola FM. High-fidelity mRNA amplification for gene profiling. Nat. Biotechnol. 2000;18:457–459. doi: 10.1038/74546. [DOI] [PubMed] [Google Scholar]
- 22.Thomas PS. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc. Natl. Acad. Sci. USA. 1980;77:5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westerfield M. The Zebrafish Book. Eugene, Oregon: University of Oregon Press; 1995. [Google Scholar]
- 24.Keller JW, Haigis KM, Franklin JL, Whitehead RH, Jacks T, Coffey RJ. Oncogenic K-RAS subverts the antiapoptotic role of N-RAS and alters modulation of the N-RAS:gelsolin complex. Oncogene. 2007;26:3051–3059. doi: 10.1038/sj.onc.1210103. [DOI] [PubMed] [Google Scholar]
- 25.Whitehead RH, VanEeden PE, Noble MD, Ataliotis P, Jat PS. Establishment of conditionally immortalized epithelial cell lines from both colon and small intestine of adult H-2Kb-tsA58 transgenic mice. Proc. Natl. Acad. Sci. USA. 1993;90:587–591. doi: 10.1073/pnas.90.2.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D’Abaco GM, Whitehead RH, Burgess AW. Synergy between Apc min and an activated ras mutation is sufficient to induce colon carcinomas. Mol. Cell. Biol. 1996;16:884–891. doi: 10.1128/mcb.16.3.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Post-transcriptional processing generates a diversity of 5’-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jupe ER, Liu XT, Kiehlbauch JL, McClung JK, Dell’Orco RT. Prohibitin in breast cancer cell lines: loss of antiproliferative activity is linked to 3′ untranslated region mutations. Cell Growth Differ. 1996;7:871–878. [PubMed] [Google Scholar]
- 29.Rastinejad F, Blau HM. Genetic complementation reveals a novel regulatory role for 3′ untranslated regions in growth and differentiation. Cell. 1993;72:903–917. doi: 10.1016/0092-8674(93)90579-f. [DOI] [PubMed] [Google Scholar]
- 30.Hasler J, Samuelsson T, Strub K. Useful ‘junk’: Alu RNAs in the human transcriptome. Cell Mol. Life Sci. 2007;64:1793–1800. doi: 10.1007/s00018-007-7084-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amaral PP, Dinger ME, Mercer TR, Mattick JS. The eukaryotic genome as an RNA machine. Science. 2008;319:1787–1789. doi: 10.1126/science.1155472. [DOI] [PubMed] [Google Scholar]
- 32.Bussing I, Slack FJ, Grosshans H. Let-7 microRNAs in development, stem cells and cancer. Trends Mol. Med. 2008;14:400–409. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 33.McMurray HR, Sampson ER, Compitello G, Kinsey C, Newman L, Smith B, Chen SR, Klebanov L, Salzman P, Yakovlev A, Land H. Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype. Nature. 2008;453:1112–1116. doi: 10.1038/nature06973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savagner P. The epithelial-mesenchymal transition (EMT) phenomenon. Ann. Oncol. J. Eur. Soc.Med. Oncol. 2010;21(Suppl. 7):89–92. doi: 10.1093/annonc/mdq292. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.