SUMMARY
X chromosome aneuploidies have long been associated with human cancers, but causality has not been established. In mammals, X chromosome inactivation (XCI) is triggered by Xist RNA to equalize gene expression between the sexes. Here we delete Xist in the blood compartment of mice and demonstrate that mutant females develop a highly aggressive myeloproliferative neoplasm and myelodysplastic syndrome (mixed MPN/MDS) with 100% penetrance. Significant disease components include primary myelofibrosis, leukemia, histiocytic sarcoma, and vasculitis. Xist-deficient hematopoietic stem cells (HSCs) show aberrant maturation and age-dependent loss. Reconstitution experiments indicate that MPN/MDS and myelofibrosis are of hematopoietic rather than stromal origin. We propose that Xist loss results in X reactivation and consequent genome-wide changes that lead to cancer, thereby causally linking the X chromosome to cancer in mice. Thus, Xist RNA not only is required to maintain XCI but also suppresses cancer in vivo.
INTRODUCTION
X chromosome inactivation (XCI) transcriptionally silences one X chromosome in the female mammal to balance gene expression between the sexes (Lyon, 1961; Payer and Lee, 2008; Lee, 2011; Wutz, 2011). The random form of XCI occurs only once during development, on embryonic days 4.5–5.5 (E4.5–5.5), when the epiblast consists of 10–20 cells. Beyond E5.5, the inactive X (Xi) enters into a “maintenance phase” in which the same X chromosome is propagated as Xi in subsequent cell divisions for the remainder of female life. Initiation of XCI depends on Xist, the 17 kb “X-inactive specific transcript” that targets and tethers Polycomb-repressive complexes to the X chromosome in cis (Brown et al., 1992; Clemson et al., 1996; Zhao et al., 2008; Jeon and Lee, 2011). Current views hold that, while Xist is essential for initiation of XCI both in an embryonic stem (ES) model (Penny et al., 1996) and in mice (Marahrens et al., 1997), Xist is dispensable once the Xi is established. Indeed, deleting Xist in vitro in post-XCI fibroblasts and somatic cell hybrids does not cause immediate X reactivation (Brown and Willard, 1994; Rack et al., 1994; Csankovszki et al., 1999). Furthermore, in ES models carrying autosomal Xist transgenes, switching off Xist after autosomal silencing does not lead to reactivation (Wutz and Jaenisch, 2000).
Nonetheless, Xist is continuously expressed throughout female life. Notably, recent studies have uncovered stochastic single-gene reactivation and a loss of Polycomb repression when Xist is conditionally deleted in mouse fibroblasts (Zhang et al., 2007). Moreover, inappropriate silencing of human XIST results in qualitatively aberrant stem cells (Shen et al., 2008; Silva et al., 2008; Anguera et al., 2012; Mekhoubad et al., 2012). Whereas Xist has been investigated extensively in cell culture, in vivo studies have been limited (Marahrens et al., 1997; Savarese et al., 2006; Kalantry et al., 2009; Namekawa et al., 2010). These findings and the limited exploration of in vivo models led us to suspect that Xist may have as yet undiscovered functions after XCI is established in the early embryo. Does X reactivation occur, and would potential X-overdosage have undesirable organismal effects?
Intriguingly, supernumerary X chromosomes have long been associated with human cancers (Moore and Barr, 1955; Liao et al., 2003; Pageau et al., 2007). For example, breast and ovarian cancer cells frequently lose the Barr body (the Xi) and duplicate the active X (Xa). The association between X and cancer also holds in men. For instance, XXY men have a 20- to 50-fold increased risk of breast cancer in a BRCA1 background (Fentiman et al., 2006), and testicular germ cell tumors often acquire supernumerary Xs (Kawakami et al., 2003). Nevertheless, an association between X and cancer has remained strictly correlative. Here, we explore the long-standing association in mice and demonstrate a direct causal link between X and cancer.
RESULTS
Female-Specific Lethality
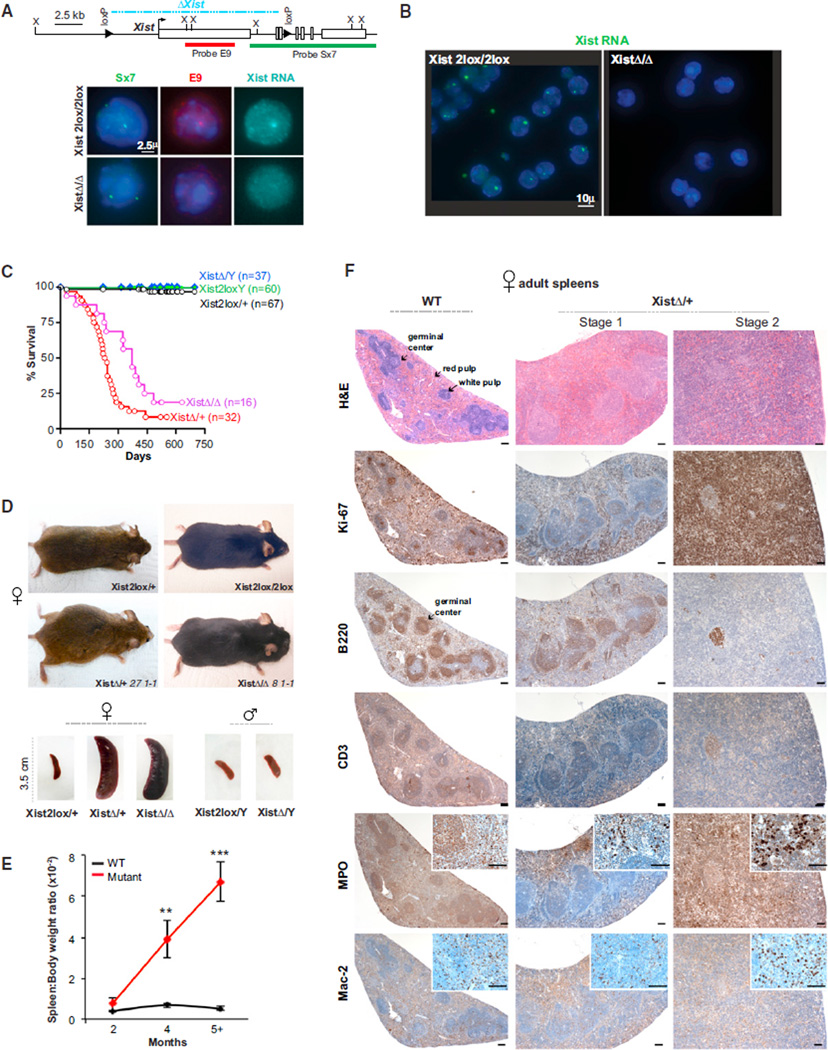
To address consequences of losing Xist in vivo, we used the Vav-Cre transgene to excise Xist conditionally in murine hematopoietic stem cells (HSC) as they emerge at E10.5, which is after the establishment of XCI (Müller et al., 1994; Csankovszki et al., 1999; de Boer et al., 2003). All XistΔ/+, XistΔ/XistΔ, and XistΔ/Y pups were born at expected frequencies and without obvious abnormalities. The Xist deletion was confirmed in splenocytes by RNA-DNA fluorescence in situ hybridization (FISH), in which the two alleles could be distinguished using probes Sx7 (present on both alleles) and E9 (absent in deleted allele) (Figure 1A). In XistΔ/XistΔ females, Xist was deleted from Xi in 100% of cells (Figure 1B). In XistΔ/+ females, Xist was deleted from Xi in ~50% of cells and from Xa in ~50%, consistent with the random nature of XCI. For two years, we monitored the mutant animals alongside of control littermates and performed experiments in accordance with the relevant regulatory standards of animal policies of MHG Institutional Animal Care and Use Committee (IACUC). Surprisingly, female mutants began to die at 1.5 months and, at the 2 year mark, only 10% of XistΔ/+ and XistΔ/XistΔ remained alive (Figure 1C). By contrast, XistΔ/Y males and control littermates (Xist+/+; Xist2lox/+; Xist2lox/Y; Vav-Cre) remained viable and healthy throughout this period. Thus, premature death was female specific, with XistΔ/+ and XistΔ/XistΔ females behaving indistinguishably. These data demonstrate a striking in vivo effect of deleting Xist after XCI is established.
Figure 1. Deleting Xist in the Blood Compartment Results in Female Specific Lethality.
(A) Map of the Xist 2lox and Xist Δ alleles and FISH probes used to distinguish the two alleles. Xist RNA is detected by using (Cy5-Sx9 probe, cyan). Representative RNA/DNA FISH are shown (n ≥ 50 each). Δ, deletion. X, XbaI site.
(B) Xist RNA FISH of splenocytes (n ≥ 100). Xist RNA: green, FITC-Sx9 probe.
(C) Female specific lethality: Kaplan-Meier kill curves plotted over 750 days were generated using Prism (GraphPad software). There were no differences between any control group: Vav-Cre, Xist2lox/+, or Xist2lox/Xist2lox females or corresponding male controls. These control genotypes were combined: females, “ Xist2lox/+”; males, “Xist2lox/Y”.
(D) Female-specific splenomegaly. Top panels: representative mutant females and age-matched controls are shown. Note abdominal swelling in 27-1-1 (XistΔ/+, 6.3 months old at death) and cervical mass in 8-1-1 (XistΔ/XistΔ, 7 months old at death). Animal ID numbers are shown with genotype throughout the figures. Bottom: representative spleens from each male and female genotype are shown.
(E) Age-dependent increase in the spleen-to-body weight ratio. Ratios taken at 2 (n = 2–6), 4 (n = 6), and 5 to 16 (n = 14–36) months of age. Means ± SEM are shown. Significance of the differences between mutant and WT spleen were calculated using the Student’s t test. **p ≤ 0.01; ***p ≤ 0.001.
(F) Temporal progression in spleen pathology. Immunostains are as indicated. Scale bars, 100 µm. See also Figures S1 and S2.
Splenomegaly and Multilineage Hyperproliferation
The female mice typically perished after a period of illness characterized by wasting, shaking, rapid breathing, and lethargy. Nearly all mice showed distended abdomens and/or necks in the terminal stage (Figure 1D). Upon necropsy, XistΔ/+ and XistΔ/XistΔ females demonstrated massive splenomegaly (enlargement of spleen). Splenomegaly was progressive and specific to female mutants (Figures 1D and 1E, p < 0.01). Histologic analysis revealed that, prior to 2 months, the spleens were relatively normal (Figure S1, available online). But in stage 1 of the disease (2–5 months), staining for the proliferation marker, Ki67, showed general hyperplasia of all splenic compartments (Figures 1F and S2), including red pulp (where erythroids [Ter119+], myeloids [MPO+], monocytes/macrophages [Mac2+], and megakaryocytes reside [Schmitt et al., 2001]), and white pulp (where B cells [B220+] and T cells [CD3+] reside). In later stage 1, the germinal centers (B220+, a B cell marker) became enlarged (Figure S2). These changes suggested marked extramedullar hematopoiesis (EMH), the formation of new blood cells outside of the bone marrow. In stage 2 (beyond 4–5 months), myeloid cells (MPO+) and monocytes/macrophages (Mac2+) in the red pulp outproliferated other cell types and eradicated the white pulp and germinal centers (Figure 1F). XistΔ/+ and XistΔ/XistΔ females behaved similarly, whereas male mutants and female controls exhibited no pathology (Figure S2). Thus, deleting Xist resulted in hyperproliferation of all hematopoietic lineages, but myeloid cells have a competitive advantage.
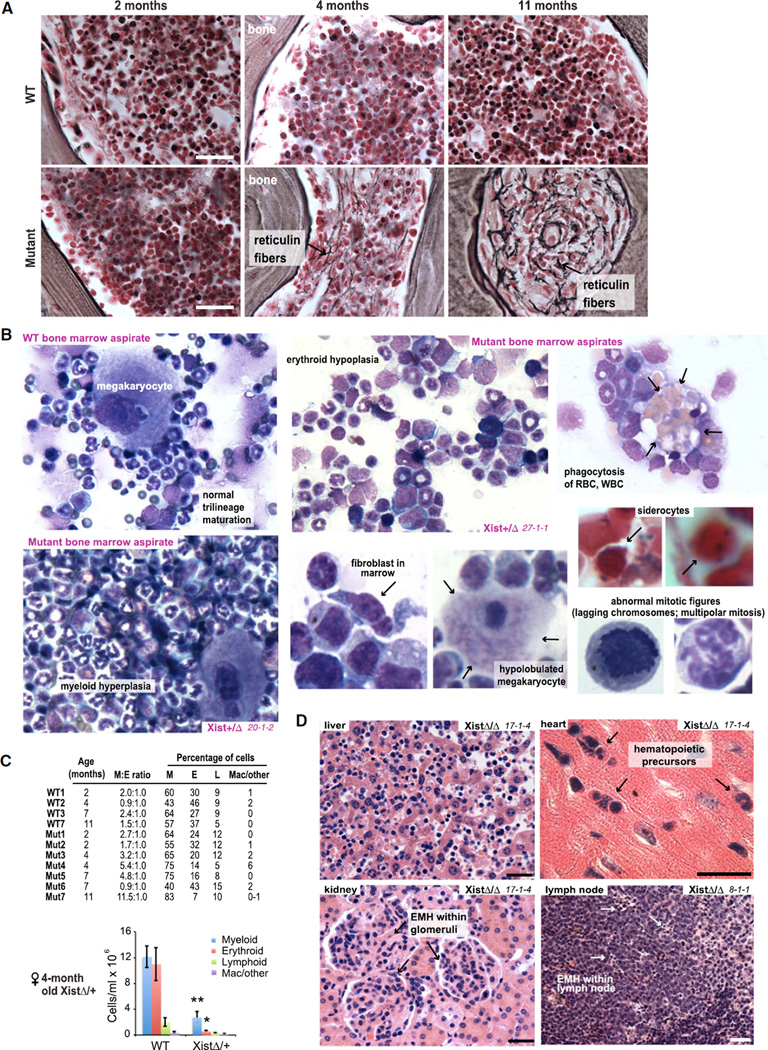
Bone Marrow Dysfunction: Myelofibrosis, Myeloproliferation, and Myelodysplasia
The adult mouse spleen may be an active site of hematopoiesis and account for ~30% of total hematopoiesis (Suttie, 2006; Percy and Barthold, 2007). Splenic EMH in the mutants, however, vastly exceeded physiological levels and was accompanied by defects in central hematopoiesis. Reticulin staining of the bone marrow revealed progressive myelofibrosis (Figure 2A), a pathological condition of unknown etiology in which the marrow is replaced by fibrotic tissue. In mutant females of ≤ 2 months of age, marrow cellularity and spleen size were normal. In stage 1, myeloid hyperplasia became a prominent feature (Figures 2B and 2C). In stage 2, marrow cellularity decreased dramatically as myelofibrosis became exuberant (Figure 2A). These changes paralleled development of splenomegaly (Figures 1E and 1F) and EMH not only in spleen but also in organs not normally associated with EMH, such as liver, kidney, lymph nodes, and cardiac muscle (Figure 2D). With concurrent marrow pathology, EMH might be partly compensatory.
Figure 2. Bone Marrow Insufficiency.
(A) Reticulin staining of bone sections reveals progressive hypocellularity and myelofibrosis (black stains representing reticulin fibers) in mutant females. XistΔ/+ cases are shown. Scale bars, 50 µm.
(B) Representative bone marrow cytology from WT and mutant females, with anomalies indicated. All were Wright-Giemsa stained except siderocytes stained by Prussian blue to reveal pathological presence of nonhemoglobin iron.
(C) Quantitation of bone marrow cells in seven mutant (Mut1–Mut7) and four WT (WT1–3 and 7) females in tabular (top) and histogram (bottom) form. M:E, myeloid to erythroid ratio. Mac, macrophages. Means ± SEM are shown. **p ≤ 0.01; *p ≤ 0.05.
(D) Nonphysiological EMH in multiple organs (17-1-4, XistΔ/XistΔ, 6 months old at death; 8-1-1, XistΔ/XistΔ, 7 months old at death). All sections were H&E stained. Scale bars, 50 µm.
Importantly, hematopoiesis at neither primary nor extramedullar sites was normal. In XistΔ/+ and XistΔ/XistΔ females (but not in any mutant males or control females), bone marrow aspirates showed multilineage proliferative and dysplastic changes (Figure 2B and data not shown). For example, findings included hypolobulated megakaryocytes, abnormal mitotic figures, binucleated erythroid precursors, siderocytes containing nonhemoglobin iron (revealed by Prussian blue staining), and increased phagocytosis of red and white blood cells (RBC, WBC) by macrophages, as well as increased numbers of fibroblasts consistent with myelofibrosis. Although hyperproliferative in early stages (increased Ki67 staining), end-stage animals demonstrated pancytopenia (loss of all lineages; Figure 2C) concurrent with exuberant myelofibrosis.
Circulating cells also demonstrated dysplastic changes (Figure 3A). In the myeloid lineage, we observed hypogranularity of neutrophils, atypical condensation of chromatin, and abnormal lobation in neutrophils and myelocytes (e.g., pseudo-Pelger-Huet anomaly). Marked leukocytosis or leukopenia, erythrophagocytosis, and increased numbers of circulating immature myelomonocytic cells (e.g., bands, metamyelocytes) were seen. In the platelet lineage, we noted megaplatelets with retained nuclei and clustered, atypical granulation in the cytoplasm occurring in the context of either thrombocytopenia or thrombocytosis (decreased or increased platelet counts, respectively) (Figures 3A and 3B). In spite of thrombocytopenia in some animals, megakaryocytic hyperplasia was evident in the marrow and at sites of EMH, suggesting aberrant platelet maturation.
Figure 3. Multilineage Dysplasia and Myeloproliferative Neoplasia.
(A) Representative peripheral blood smears from WT and mutant females, Wright-Giemsa stained unless noted.
(B) Hematologic analysis of end-stage mutants and age-matched control.
In the erythroid lineage, we observed poikilocytosis and anisocytosis (RBC of abnormal shape and variable size), circulating immature erythroids (binucleated rubricytes and metarubricytes; nucleated RBC, up to 40% circulating in stage 2) (Figure 3B), nuclear-cytoplasmic dysynchrony, dacrocytes (oddly shaped RBC characteristic of myelofibrosis), and increased numbers of Howell-Jolly bodies (condensed DNA remnants in mature RBC) (Figure 3A). Circulating erythroid precursors also had cytoplasmic vacuolation and nuclear irregularity. Nonhemoglobin iron revealed by Prussian blue staining (abnormal ring sideroblasts, siderocytes) and positive PAS staining (periodic acid-Schiff) suggested features of erythroleukemia. Anemia was characteristic, with the shift toward immature forms signaling an ineffectual regenerative response consistent with lethargy and rapid breathing in end-stage animals. We conclude that deleting Xist in the blood compartment results in multilineage defects, with features characteristic of myelofibrosis, myeloproliferation, and myelodysplasia.
Cancer
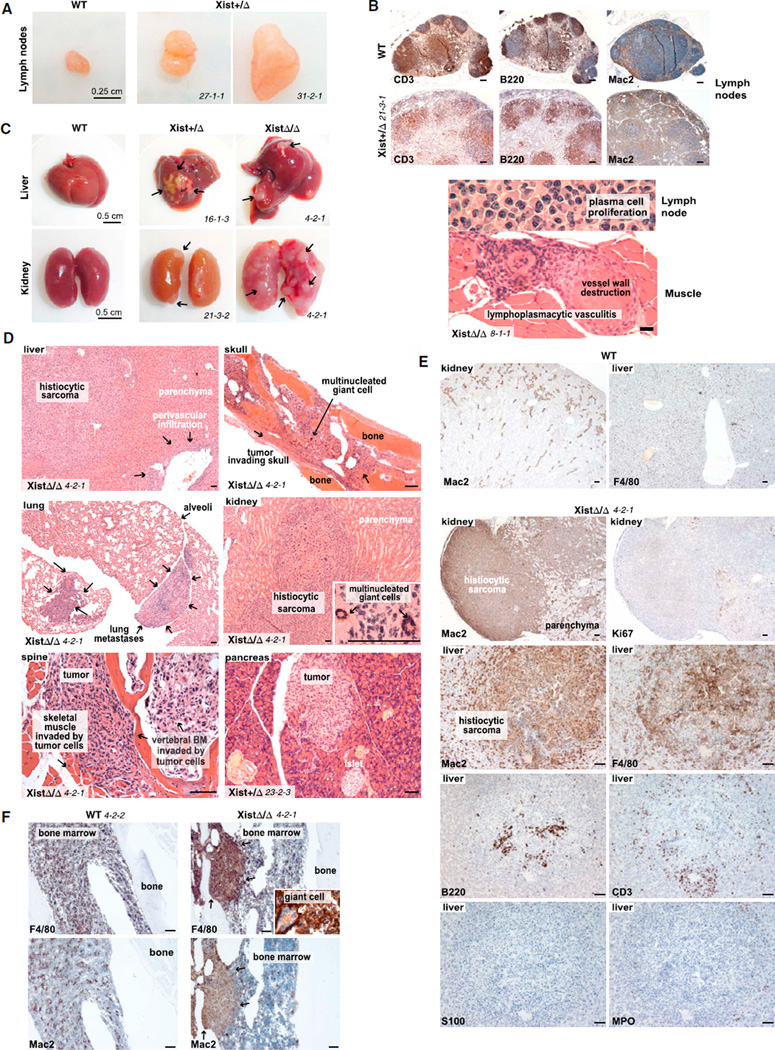
Myelofibrosis with myelodysplasia and myeloproliferation indicates a hematological cancer termed myeloproliferative neoplasm. Although the animal presentations varied, general trends were evident and were indistinguishable between XistΔ/+ and XistΔ/XistΔ mice. The possibility of cancer is supported by several coexisting conditions. Primary manifestations related to two types of leukemia, chronic myelomonocytic leukemia (CMML) and erythroleukemia. CMML features included persistent monocytosis, neutrophilia, pseudo-Pelger-Huet cells (0.5%–15.0% of circulating WBC), leukocytosis of up to 70,000 WBC/ml (Figures 3A and 3B), and increased numbers of circulating promonocytes (<20%) and immature cells with myelomonocytic features. In leukemic animals (e.g., 3-3-3 [XistΔ/Δ, 13.7 months], 27-1-1 [XistΔ/+, 6.3 months], and 23-2-1 [XistΔ/+, 9.6 months]), leukocytosis occurred together with splenomegaly and general lymphadenopathy (Figure 4A). Large germinal centers showed increased numbers of B cells, disorganized T cells, histiocytes, and mitotic rates (Figure 4B). A coexisting erythroleukemia-like syndrome was supported by erythrodysplasia, anemia, and expanded circulating erythroid precursors with positive cytoplasmic PAS and Prussian-Blue staining (e.g., 24-1-4, XistΔ/+, 11.9 months; 4-2-3, XistΔ/XistΔ, 12 months) (Figures 3A and 3B).
Figure 4. Histiocytic Sarcoma and Lymphoplasmacytic Vasculitis.
(A) Enlarged lymph nodes from end-stage mutants shown with a WT control. Cervical lymph nodes of 27-1-1 (XistΔ/+, 6.3 months old) and brachial lymph node of 31-2-1 (XistΔ/+, 5.8 months old) are shown.
(B) Top: sections of enlarged lymph node from 21-3-1 (XistΔ/+, 8.8 months old at death) with follicular B and T cell and intra- and interfollicular histiocytic expansion are shown. Bottom: sections of enlarged lymph node from 8-1-1 (XistΔ/XistΔ, 7 months old at death) with a plasmacytoma-like infiltrate are shown. Lymphoplasmacytic vasculitis is also shown. See also Figure S2O.
(C) Masses containing metastatic histiocytic sarcoma (arrows) in end-stage liver and kidney.
(D) H&E stains of metastatic histiocytic sarcoma in multiple organs. Scale bars represent 100 µm.
(E) Immunohistochemistry confirms histiocytic sarcoma. Scale bars represent 100 µm.
(F) Histiocytic sarcoma in bone marrow. Scale bars represent 100 µm. Note positive F4/80 staining for intralesional giant cells (inset).
Mutant animals also demonstrated a histiocytic sarcoma (HS, also called malignant histiocytosis), a rapidly fatal cancer of unknown but presumed hematologic origin. The HS was female specific (Figure 4C) and characterized by atypical spindloid cells, multinucleated giant cells, marked hemophagocytosis, and a high mitotic rate (Figure 4D; see inset). Approximately 20% of animals showed widespread HS in bone marrow, pancreas, lung, liver, kidney, and/or lymph nodes (Figure 4D). Local invasion led to spinal compression and limb paralysis (animal 4-2-1, XistΔ/XistΔ, 9.4 months); renal failure resulted from kidney infiltration. Immunohistochemistry demonstrated a histiocytic but nonLangerhans origin, confirming the diagnosis of metastatic HS (Figures 4E and 4F: Mac2+, F4/80+, S100−, MPO−, B220−, and CD3−).
A third manifestation was an unusual vasculitis, comprised not of neutrophils but of lymphocytes and plasma cells (Figures 4B and S2O). For example, in animal 8-1-1 (XistΔ/XistΔ, 7 months), a massive cervical lymph node (>1 cm diameter, Figure 1D) exhibited necrosis, dystrophic calcification, a disorganized capsule, and a striking lymphoplasmacytic infiltrate around blood vessels (Figure 4B, bottom panel). Multiple organs were usually affected (Figures 4D and S2O), with accompanying vessel destruction and organ damage (e.g., glomerulonephritis). This rare vasculitis can be associated with lymphoma, though no lymphoid malignancy was obvious.
Leukemia, HS, and vasculitis coexisted to varying degrees and accounted for different clinical presentations and outcomes. CMML was the dominant component in ~20% of mutants, metastatic masses consistent with HS in ~22%, and vasculitis in ~40%. These features suggested a diagnosis of ‘myeloproliferative neoplasm’ (MPN) with features of ‘myelodysplastic syndrome’ (MDS), classified by the World Health Organization (WHO) as “mixed MPN/MDS” (Tefferi, 2011; Vainchenker et al., 2011). Indeed, primary myelofibrosis, CMML, and erythroleukemia are significant components of human MPN/MDS. We conclude that deleting Xist results in a fulminant, highly lethal neoplasia.
Origin in Blood Cells Rather Than Stroma
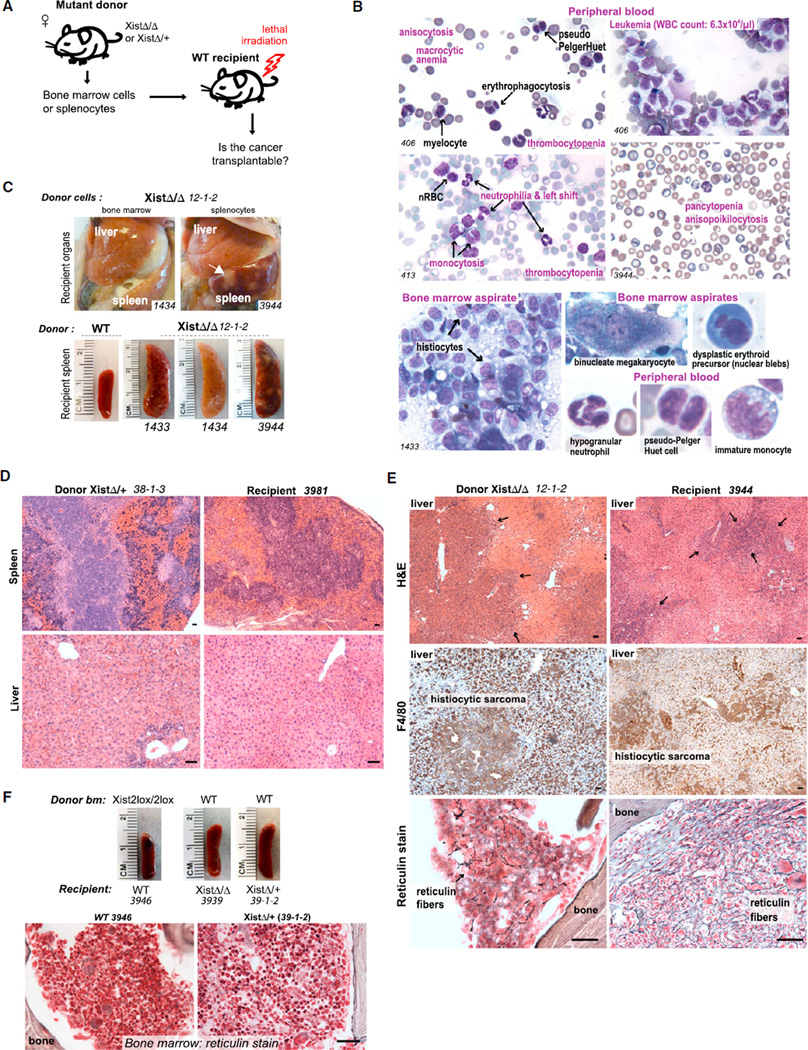
To distinguish between a primary defect of stromal versus hematopoietic cells, we performed three types of transplantation (Tp) experiments: (1) mutant into wild-type (mutant-to-WT) Tp, where lethally irradiated WT recipients were reconstituted with bone marrow (n = 12 mice) or splenic cells (n = 6 mice) from mutant females with overt disease, to determine whether the disease could be conferred by hematopoietic cells alone (hematopoietic cell autonomous) (Figure 5A); (2) reciprocal WT-to-mutant Tp (n = 3 mice), to determine whether the mutant host could induce disease in genetically WT hematopoietic cells (hematopoietic cell nonautonomous); and (3) WT-to-WT Tp to control for effects of transplantation (n = 5 mice). At 1 month post-Tp with either mutant bone marrow or splenic cells, recipients showed full bone marrow reconstitution by donor cells, as determined by Ly-5.2+ state of donor cells (FACS analysis, not shown). Significantly, between 1 and 4 months post-Tp, multiple mice in the mutant-to-WT Tp group became ill and recapitulated mixed MPN/MDS (n = 11 mice) (Figures 5D, 5E, S3A, and S4).
Figure 5. Transplantability of MPN/MDS Suggests a Hematopoietic Rather Than Stromal Origin.
(A) Schematic of transplantation experiments.
(B) Dysplasia in peripheral blood and bone marrow of mutant-to-WT transplants. See also Figures S3A–S3C.
(C) Histiocytic sarcoma recapitulated in WT recipients of mutant bone marrow or splenocytes. Recipient livers and spleen (1434) were pale from anemia. White masses in recipient spleen contain histiocytic sarcoma (arrow). Mice succumbed within 40 days after transplantation.
(D) H&E stain of EMH in matched donor and recipient livers and spleens, as indicated.
(E) Immunohistochemistry confirms histiocytic sarcoma in donor and recipient (middle panels). Reticulin stain of bone marrow shows exuberant myelofibrosis in donor (at necropsy) and recipient (40 days after transplantation).
(F) WT-to-mutant transplantations reversed disease. WT-to-WT transplantation controls were normal. Scale bars represent 100 µm. See also Figure S4.
First, monthly monitoring of peripheral blood demonstrated progressive anemia and thrombocytopenia (Figures 5A and S3A). Second, leukemia (17,000–64,000 WBC/µl) with high monocyte and granulocyte counts was observed in multiple donor-recipient combinations in the mutant-to-WT group (n = 7 mice), matching the CMML-like disease of Xist mutants (Figure S3A). Third, dysplastic changes identical to those in mutant donors occurred in recipient blood and bone marrow, including hypogranular neutrophils, binucleated RBC, pseudo-Pelger-Huet cells, erythroid and megakaryocytic dysplasia, histiocytosis, and multinucleated giant cells (n = 10 mice) (Figure 5B). Fourth, necropsies of all dead and/or moribund animals revealed splenomegaly in the mutant-to-WT Tp group (n = 8 mice) (Figure 5C). Fifth, metastatic HS was recapitulated in multiple recipients (n = 3 mice). For example, bone marrow and splenic cells from XistΔ/XistΔ donor 12-1-2 separately gave rise to HS in three different recipients, all reaching terminal stage within 40 days of Tp from widespread metastases (1433,1434, and 3944; Figures 5C, 5E, S4B, and S4C). Marrow effacement by histiocytes and multinucleated giant cells and immunohistochemistry of infiltrative masses were consistent with HS (Figures 5E and S4). Finally, exuberant myelofibrosis in the recipient occurred in multiple donor-recipient combinations (n = 8 mice) (Figures 5E and S4A), indicating that myelofibrosis could be induced solely and rapidly (<2 months) by hematopoietic cells. None of the WT-to-WT Tp controls showed these abnormalities (n = 5 mice) (Figures 5F and S3A).
Intriguingly, in the reverse Tp experiments (WT-to-mutant), mutant phenotypes were rescued by WT donor hematopoietic cells (n = 2). There was a normalization of spleen-to-body mass ratios (Figure 5F) and peripheral blood profiles (Figure S3C). Moreover, myelofibrosis, tumor infiltration, and other organ pathologies were ameliorated (Figures 5F and S4D). These findings indicate that myelofibrosis and MPN/MDS could be corrected by transplanting WT blood cells after radiation conditioning of recipients. Thus, the Xist-deficient host environment is not sufficient to confer disease. We conclude that blood cells– rather than stroma–play the primary role in the pathogenesis of MPN/MDS.
A Primary HSC Defect
The multilineage hematopoietic phenotypes pointed to an HSC anomaly. FACS analysis of primitive (Lin−SCA-1+c-KIT+; “LSK+” cells) and committed progenitor HSCs (Lin− SCA-1− c-KIT+; “LSK−” cells) (Okada et al., 1992) revealed both qualitative and quantitative defects (Figures 6A–6C). In mutant bone marrow cells, an increased LSK+/LSK− ratio (Figures 6A and 6B) suggested differentiation defects. Although myeloid, lymphoid, and erythroid lineages were present (Figures S5C and S5D), dysplastic changes (Figures 2 and 3) indicated that maturation was incomplete. To test the differentiation capacity of Xist-deficient HSCs, we performed competitive repopulation assays (Harrison, 1980), in which we mixed mutant (n = 5) or WT bone marrow cells (n = 4) with WT congenic bone marrow cells (Ly-5.1+) at 1:1 ratio and transplanted them into lethally irradiated recipients to test relative reconstitution potential. Intriguingly, in all experiments, WT cells showed a statistically significant competitive advantage over mutant cells in peripheral blood (Figure 6D) and bone marrow (Figure 6E). Thus, mutant cells were compromised in their ability to repopulate irradiated hosts. Consistent with this, the numbers of each cell type were generally decreased in mutant marrows (Figure 6F). These results demonstrated a defect of mutant HSC to engraft or mature. Interestingly, whereas none in the control group died, three of five animals receiving mutant competitors perished within 4 months. Thus, the maturation defect paralleled the lethal tumorigenic potential of mutant cells, even in the presence of WT cells.
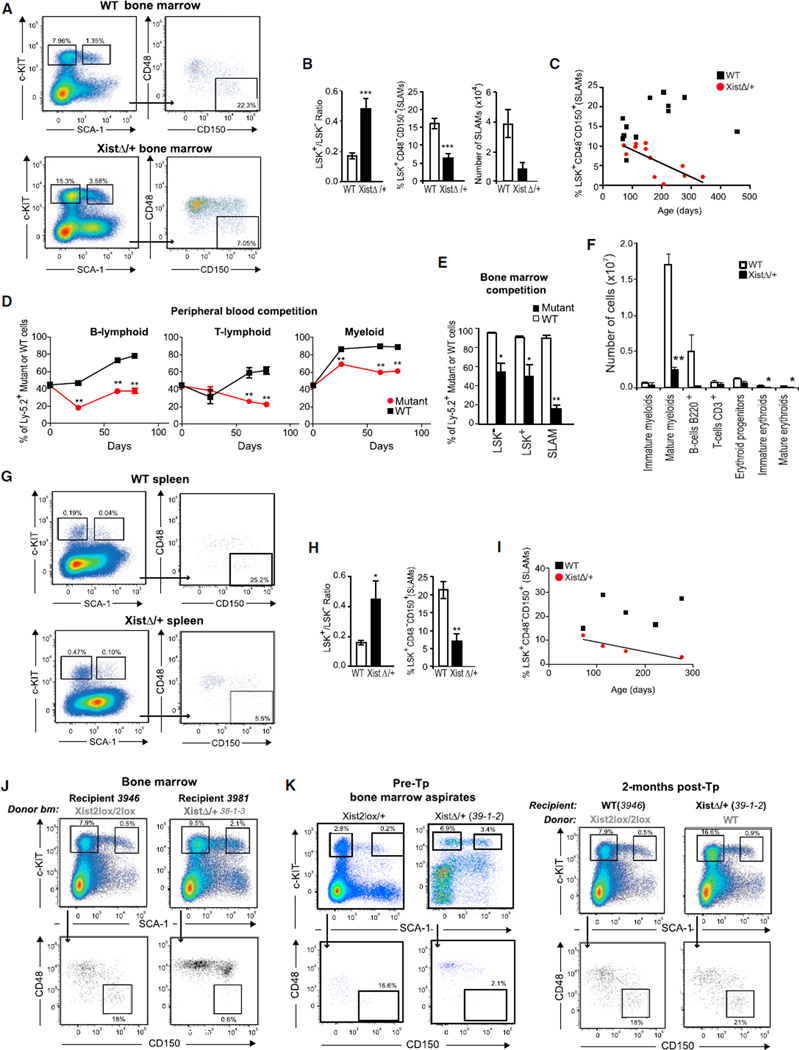
Figure 6. A Primary Defect in the HSC.
(A) Representative FACS analysis of bone marrow cells from a 5-month-old female mutant and a WT littermate mouse showing increased LSK+/LSK− ratio and decreased number of SLAM-enriched long-term HSCs (LSK+CD48−CD150+) in mutant mice.
(B) Histograms of bone marrow LSK+/LSK− ratios and SLAM-enriched long-term HSCs suggest, respectively, a failure of maturation and loss of progenitors in mutant (n = 12) female mice in comparison to WT female mice (n = 13). Means ± SEM shown; Student’s t test, ***p ≤ 0.001.
(C) Progressive loss of SLAM-enriched long-term HSCs in bone marrow of mutant mice (n = 12) over time as compared to WT mice (n = 13). R2= 0.5317.
(D and E) Competitive repopulation assays reveal maturation defects in mutant cells. WT (Xist2lox/+) or mutant (XistΔ/+) cells were mixed at 1:1 ratio with cells isolated from congenic WT Ly-5.1+ mice and transplanted into WT hybrid Ly-5.1 +/Ly-5.2+ recipients (n = 4–5 per group). Peripheral blood was sampled for FACS analysis repeatedly for 2.5 months (D), and bone marrow was sampled at 2.5 months (E). Means ± are SEM shown; *p < 0.05, **p ≤ 0.01.
(F) FACS analysis of bone marrow cells in 4-month-old mutant and WT mice (n = 4 mice per group). Means ± SEM shown; *p < 0.05, **p ≤ 0.01.
(G) Representative FACS analysis of splenocytes from a 5-month-old female mutant and a WT littermate mouse showing an increased LSK+/LSK− ratio and a decreased number of SLAM-enriched long-term HSCs in mutant mice.
(H) Histograms of splenic LSK+/LSK− ratios and SLAM-enriched long-term HSCs suggest, respectively, a failure of maturation and loss of progenitors over time in mutant females (n = 4) as compared to WT females (n = 6). Means ± SEM shown. Student’s t tests show *p ≤ 0.05 and **p ≤ 0.01.
(I) Progressive loss of SLAM-enriched long-term HSCs in spleen of mutant mice (n = 4) over time as compared to WT mice (n = 6). R2 = 0.846.
(J) FACS analyses show that HSC maturation defects (elevated LSK+/LSK− ratios) and loss of SLAM-enriched long-term HSCs are recapitulated in the WT recipients of mutant bone marrow cells at 2 months post-Tp.
(K) FACS analyses of reverse transplantation (WT-to-mutant), with WT-to-WT control. At 2 months post-Tp with WT bone marrow cells, mutant 39-1-2 (XistΔ/+, 8.2 months) shows a correction of LSK+/LSK− ratio and number of SLAM-enriched long-term HSCs (right panels), in comparison to pre-Tp profiles (left panels). See also Figures S3, S5, and S6.
On further evaluation, time-course analysis showed an age-dependent loss of SLAM-enriched long-term HSCs, as evidenced by a decrease in the LSK+CD48−CD150+ (SLAMs [Kiel et al., 2005]) and LSK+CD34−Flt3− (Adolfsson et al., 2005) populations (Figures 6A, 6B, and S5B), while the population remained stable or even increased 2.5-fold over time in WT mice (Figure 6C). This loss was consistent with progressive myelofibrosis and pancytopenia in late-stage disease (Figures 2 and 3). Although the deficiencies were most apparent in late stages, the increased LSK+/LSK− ratio and maturation defects were already evident in predisease and early-disease mice (Figure S5). Furthermore, the defects were recapitulated in HSCs derived from the spleen, a site of EMH (Figures 6G–6I, S6A, and S6C).
Finally, reciprocal transplantation experiments showed that WT recipients of mutant bone marrow cells recapitulated the maturation defects (elevated LSK+/LSK− ratios) and loss of long-term HSCs at 2 months post-Tp (Figures 6J and S3D). By contrast, mutant recipients of WT bone marrow cells showed a correction of the maturation defect and number of long-term HSCs at 2 months post-Tp, in comparison to their pre-Tp profiles (Figures 6K and S3E). We conclude that there are intrinsic quantitative and qualitative defects in the mutant HSCs.
Genetic Pathway Reconstruction
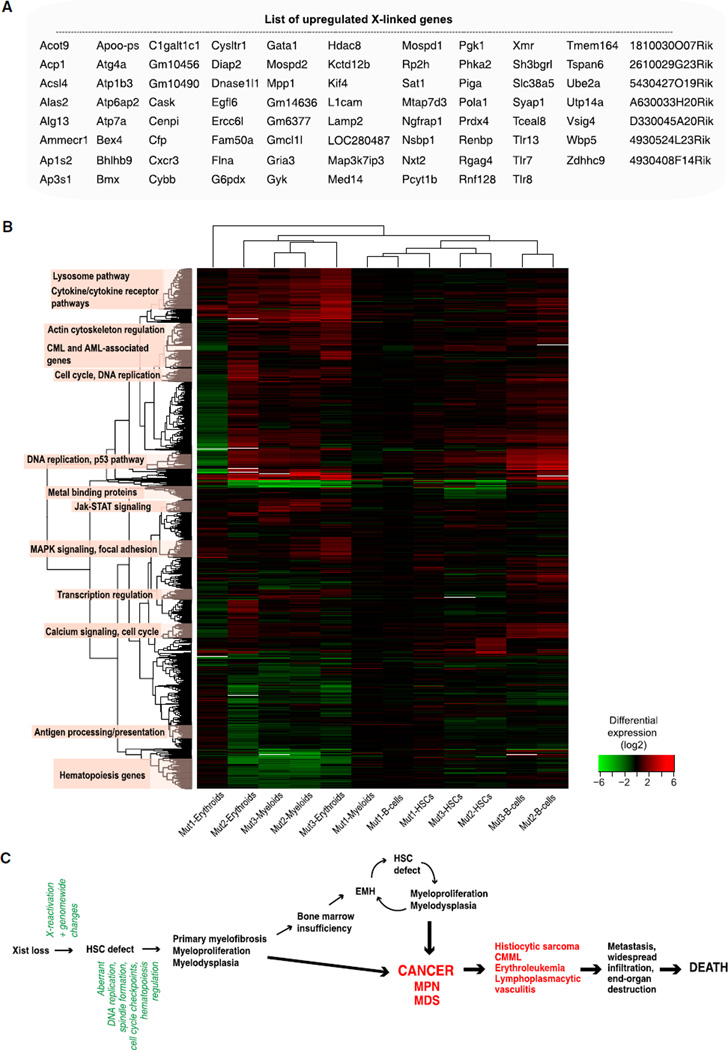
To reconstruct a genetic pathway toward cancer, we carried out expression profiling before disease (2 months old) and during overt MPN/MDS and CMML-like disease (27-1-1, XistΔ/+, 6 months; and 4-2-3, XistΔ/XistΔ, 12 months) and compared profiles of purified bone marrow HSC (LSK+CD34−), splenic B cells (B220+), myeloid cells (CD11b+), and erythroid cells (TER119+) against those of corresponding cell types from age-matched WT females. Significant changes were observed in all cell types before and during overt disease (Tables S1A–S1D). Compared to the autosomal average, the average X-linked gene expression was significantly higher (Figure 7A and Table S1A; enrichment of upregulated genes on X compared to autosomes; hypergeometric p ≈ 0.010). In the predisease state, upregulated X-linked genes included Tlr8, Gria8, Gyk, and Gm14636. During disease, upregulated X-linked genes in all cell types included Gata1, a crucial hematopoiesis factor frequently mutated in leukemia (Zheng and Blobel, 2010), and Kif4, a mitotic positioning motor protein aberrantly expressed in erythroleukemia (Mazumdar et al., 2011). Thus, deletion of Xist results in significant X reactivation, arguing that Xist is required not only for initiation but also for maintenance of XCI in vivo.
Figure 7. Gene Expression Profiling by Microarray Analysis Reveals X Reactivation and Genome-wide Changes.
(A) List of upregulated X-linked transcripts in blood cells of Xist mutants. See Figure S1A for details.
(B) Hierarchical clustering of transcripts that are differentially expressed between WT and mutant blood cells, as listed in Tables S1B,S1C, and S1D. Enriched functional categories are indicated on left. See also Figure S7 and Tables S1A–S1I.
(C) A model for the pathogenesis and progression of cancer resulting from Xist loss.
Significant genome-wide changes occurred before (Figure S1B) and during (Figures S1C and S1D) disease. Commonalities and differences were also apparent between cell types (Figure S7 and Tables S1E–S1G). Comparing predisease and disease states, we observed a general increase in expression changes across and within cell types, probably reflecting disease progression. Notable autosomal deviations included Cenpl (centromere protein L); Erc6l (kinetochore assembly gene); Bmx (hematopoietic differentiation); Cxcr3 (leukocyte trafficking cytokine); Med14, Hdac8, and Hmgn5 (transcription and/or chromatin regulation); Pola1 (DNA replication); and Lamp2 (tumor metastasis). Using the DAVID tool (Huang et al., 2009), analysis of functional gene annotations among all differentially expressed genes revealed significant enrichment of genetic pathways involved in DNA replication, cell-cycle regulation, hematopoiesis, and primary immunodeficiency (Benjamini-Hochberg FDR ≤ 6.67 × 10−5).
To investigate further, we performed correlation analysis and hierarchical clustering of differentially expressed genes in Tables S1B–S1D and uncovered additional functional categories (Figure 7B). In addition to pathways involved in DNA replication, cell-cycle regulation, and hematopoiesis, we also identified those in Ca+2-dependent leukocyte signaling, Toll-like receptor signaling, MAPK kinase signaling, p53 regulation, and acute myeloid leukemia (AML) and/or chronic myeloid leukemia (CML) progression. When cross-referenced to the list of known oncogenes and tumor suppressors (Higgins et al., 2007), multiple upregulated oncogenes were identified (Table S1H), such as Clspn (cell-cycle arrest), Espl1 (chromosome segregation), Pparg (nuclear receptor signaling), Myb (megakaryocyte proliferation and maturation), and Csf1 (macrophage differentiation factor), and several tumor suppressor genes and hematopoiesis regulators were downregulated (Table S1I), such as Flt3 and c-Kit (Huang et al., 1998; Li et al., 2011). Several of these genes were previously implicated as single-gene mutations in MDS, MPN, and myeloid leukemias (e.g., Runx3, Kit, Foxo1, Flt3, Rap2a, p53, Ezh2, and Rras [Pellagatti et al., 2010; Bejar et al., 2011]; Aurka and Fos [Pang et al., 2011]; and Idh1, Tet2, and CebpA [Brecqueville et al., 2012; Muramatsu et al., 2012]). Taken together, our data indicate that MPN/MDS is initiated by an Xist deletion that leads to X reactivation, which in turn results in a series of autosomal changes causing DNA replication and mitotic anomalies, genome instability, and dysregulation of the hematopoiesis pathway (Figure 7C). Our data causally link the X chromosome to cancer.
DISCUSSION
Here we have demonstrated that Xist suppresses hematologic cancer. Deleting Xist, once thought to be essential only for dosage compensation, is sufficient to induce an aggressive, lethal blood cancer. The cancer is female specific and fully penetrant. Not a single mutant female–either heterozygous or homozygous–has escaped to date (n > 87). Our analysis suggests that upregulation of X-linked genes after deleting Xist leads to genome-wide changes in key developmental and homeostatic pathways, which in turn drive progression toward cancer (Figure 7C). A role for Xist RNA in suppressing cancer adds to the growing list of functions associated with long noncoding RNA (Huarte and Rinn, 2010; Lee, 2012).
The clinical presentation, histopathology, and cellular defects are most consistent with mixed MPN/MDS (Tefferi, 2011; Vainchenker et al., 2011). MPN and MDS have been proposed to be overlapping diseases along a spectrum; when there is significant overlap, they are categorized as mixed MPN/MDS. Significant components of the Xist mutant disease include primary myelofibrosis, HS, leukemia (CMML and an erythroleukemia-like disease), and lymphoplasmacytic vasculitis. Although the WHO does not currently recognize HS as a component of MPN/MDS, HS often occurs in the context of CMML and other monocytic, myeloid, and lymphocytic leukemias (e.g., [Doll et al., 1987; Laurencet et al., 1994; Feldman et al., 2004]). Thus, human HS may be linked to MPN/MDS. Furthermore, although a lymphoid leukemia is not a manifestation of the mouse disease, lymphoplasmacytic vasculitis is suggestive of an inflammatory or autoimmune disease, if not outright malignancy. The lymphoid lineage is therefore also clearly affected by the Xist deletion.
These multilineage defects or cancers coexist to varying degrees in each female mutant. Their relative dominance probably results from stochastic developmental differences that influence disease course. Like human MPN/MDS, the Xist-deficient disease has a variable course. In humans, death eventually results from leukemic progression and comorbid conditions (e.g., infection, bleeding [Tefferi, 2011; Vainchenker et al., 2011]). The mouse disease is also characterized by leukemic progression, widespread infiltration and/or invasion, end-organ failure, and a resulting constellation of comorbid conditions (Figure 7C).
Our study argues that MPN/MDS arises from a primary, cell-autonomous defect in hematopoietic cells rather than bone marrow stroma. Which hematopoietic cell is the source of the disease phenotype cannot be definitively stated, but several lines of evidence point to HSC. First, Xist is conditionally deleted when HSCs first appear at E10.5 (Figure 1) and expression profiling reveals aberrant changes in this population even in predisease animals (Figures 7 and S7 and Table S1). Second, prolif-erative and dysplastic changes are present in all hematopoietic cell types (Figures 3 and 4). Third, malignancies occur in myelomonocytic, erythroid, and histiocytic lineages (Figures 3 and 4). Fourth, our analyses point to maturation defects of the HSC and age-dependent loss of long-term HSCs (Figure 6). Finally, reciprocal transplantation experiments between WT and mutant mice reveal recapitulation of MPN/MDS after transplantation of hematopoietic cells alone (Figures 5, 6, S3, and S4). These experiments also demonstrate that primary myelofibrosis and HS arise from hematopoietic cells.
The dramatic phenotypes probably reflect the extensive network of X-autosome interactions (Figures 7 and S7 and Table S1). Although the Xi is initially unaffected (Brown and Willard, 1994; Csankovszki et al., 1999), we have now shown that deleting Xist leads to failure of long-term Xi maintenance in vivo. We propose that carcinogenesis is driven by a series of changes occurring in the HSC and further accumulated in mature hematopoietic cells (Figure 7C). These changes are initiated by loss of Xist, which leads to progressive X reactivation, which in turn induces a cascade of unfavorable genome-wide changes that include dysregulation of genes involved in DNA replication, chromosome segregation, cell-cycle checkpoints, and hematopoiesis. A failure of HSC maturation and loss of long-term HSC in the marrow progressively shift hematopoiesis to extramedullary sites resulting in EMH. The defects are recapitulated and amplified at sites of EMH. Together, aberrant hematopoiesis at central and extramedullary sites result in MPN/MDS, with eventual progression to myelofibrosis, leukemia, HS, and death from comorbid conditions.
Although loss of the Barr body has been correlated with cancer for 60 years, here we have demonstrated direct causality in mice. Our study implies that human hematologic cancers may result from overdosage of X, either from XIST loss on Xi or from duplication of Xa. Interestingly, MDS is more common in women (Bennett and Orazi, 2009), with noted XIST deletions and X chromosome duplications occurring in MPN, MDS, and myeloid cancers (Dewald et al., 1989; Rack et al., 1994; Dierlamm et al., 1995; Paulsson et al., 2010). The association is not restricted to women, however, as extra X chromosomes are seen in ALL, AML, acute nonlymphoblastic leukemia (ANLL), adult T cell leukemia, CML, erythroleukemia, and non-Hodgkin lymphoma of both sexes (Sandberg, 1983); some 60% of childhood acute lymphoblastic leukemias (ALL) display extra X chromosomes and an extra X may be the only aneuploidy in some chronic myeloid leukemias (Heinonen et al., 1999; Yamamoto et al., 2002). The Xist-deficient mouse provides a unique opportunity to study mixed MPN/MDS, HS, and primary myelofi-brosis–one that can be approached from the perspective of long noncoding RNAs and the sex chromosome so frequently associated with human cancers.
EXPERIMENTAL PROCEDURES
Mice
Xist2lox/Xist2lox mice (129Sv/Jae strain) were a gift of R. Jaenisch (Csankovszki et al., 1999). To generate XistΔ/+ and XistΔ/Y mice, we crossed Xist2lox/Xist2lox females to Vav.Cre males [B6.Cg-Tg (Vav1-cre) A2Kio/J; Jackson Laboratory]. To generate homozygous mutants, we crossed Xist2lox/+;Vav-Cre females to Xist2lox/Y males or Xist2lox/+ females to Xist2lox/Y;Vav-Cre males. Mice were screened by PCR for Vav.Cre, XistWT, and Xist2lox alleles using the following primer sets: Vav.Cre (522-1: 5′-CTT CTC CAC ACC AGC TGT GGA-3′, 522-2: 5′-GAC AGG CAG GGC CTT CTC TGAA-3′; amplicon 580 bp) and Xist (Xint3F: 5′-GGC CAG TTT CTG ACA CCC TA-3′, Xint3R: 5′-CAC TGG CAA GGT GAA TAG CA-3′; XistWT 200 bp, Xist2lox 300 bp). Animal experiments were approved by the MGH Institutional Animal Care and Use Committee (IACUC). Dead and moribund animals (sacrificed per IACUC) were included in the kill curve.
DNA and RNA FISH
RNA and DNA FISH were performed using established protocols (Takizawa et al., 2008; Namekawa and Lee, 2011). Briefly, splenocytes were plated onto 1xPoly-L-Lysine-coated chamber slides in complete RPMI medium, incubated at 37°C for 2 hr, and fixed with 4% PFA in 1xPBS containing 10% acetic acid for 15 min at room temperature (RT). Cells were washed three times with 1xPBS at RT and kept in 70% EtOH at −20°C until use. To detect Xist RNA, we performed FISH using Alexa-fluor 488-dUTP-labeled Sx9 probe. We next performed sequential RNA and DNA FISH, whereby RNA FISH was performed first using Alexa-647-dUTP-labeled Sx9 probe, photographed, and then followed by DNA FISH using Alexa-fluor-488-dUTP-labeled Sx7 and Cy3-dUTP (Enzo Life Sciences)-labeled E9 probes. Fluorophore-labeled dUTPs were from Molecular Probes unless noted. Images were obtained with a Nikon Eclipse 90i microscope and a Hamamatsu CCD camera and analyzed using Volocity Software (Improvision, Perkin-Elmer).
Histopathology
Tissues were fixed in 10% buffered Formalin (Fisher Scientific) and bones were fixed and decalcified using Cal-Ex II (Fisher Scientific). Sections were stained with hematoxylin and eosin (H&E) stain, Prussian blue, or reticulin stains, as noted. For immunohistochemistry, B220 (550286; BD PharMingen), Ki67 (VP-RM04; Vector), CD3 (ab16669; Abcam), Ter119 (550565; BD PharMingen), MPO (sections from all organs, A0398; Dako; bone sections, RB-373; Neomarkers), Mac2 (CL8942AP; Cedarlane), S100 (Z0311; Dako), and F4/80 (MCA497GA; Serotec) antibodies were used. Images were obtained using a Nikon Eclipse90i microscope and a Q-imaging MicroPublisher RTV color camera and analyzed using Volocity (Improvision, Perkin-Elmer).
Bone Marrow and Peripheral Blood Cytology
For live bone marrow aspiration, mice were anesthetized using isoflurane and bone marrow cells aspirated using a 27 gauge needle from the femur through the patellar tendon. The cells were then prepared for FACS analysis (below). For bone marrow analysis at necropsy, cells were collected either by crushing or flushing tibias and femurs with 3 ml of 1xPBS/5% FBS solution into a 50 ml tube (Falcon) using a 22.5 gauge needle or by bone marrow brush preparations. Peripheral blood was collected by terminal cardiocentesis after CO2 administration or as survival bleeds using facial venipuncture into EDTA-treated tubes (Becton, Dickinson and Company). Fresh blood and bone marrow smears were stained with Wright-Giemsa (Fisher Scientific), Prussian blue, and PAS (Sigma-Aldrich). Automated complete blood counts were performed using the HemaTrue Veterinary Hematology Analyzer (Heska Corporation, Loveland, CO).
Transplantation Experiments
Splenocytes or bone marrow cells were used for transplantation. Spleen and bone (tibiae, femurs, iliac bones, humeri, and spine) were dissected, crushed in cold 1xPBS with 2% FBS, and passed through a 40 µm filter. Live mononuclear cells were isolated by gradient centrifugation using Ficoll-Paque Plus (GE Healthcare). Blood cells were collected by retro-orbital bleeds under anesthesia. The Ly-5.1 marker was used to distinguish donor versus recipient. For mutant-to-WT or WT-to-WT transplantation, 5×3 106 cells from mutant or WT littermate (Ly-5.2+) were transplanted into 6- to 12-week-old lethally irradiated (9.5 Gy) WT Ly-5.1+/Ly-5.2+ F1 hybrid recipients (129/SvJ x B6.SJL-PtprcaPepcb/BoyJ; Jackson Laboratories). For reverse transplantation (WT-to-mutant), 5 3 106 cells from Ly-5.1+ hybrids were transplanted into lethally irradiated mutant recipients. For competitive transplantation assays, 2 3 106 cells from mutant or WT littermates were mixed with 2 3 106 cells from B6.SJL-PtprcaPepcb/BoyJ (Ly-5.1+) WT competitors and transplanted into WT Ly-5.1 +/Ly-5.2+ hybrid recipients. Irradiated recipients were maintained on sterile water containing 0.5 g/l of enrofloxacin (Bayer, Shawnee Mission, KS) for 2 weeks after irradiation.
FACS Analyses
All antibodies were from BD PharMingen unless noted. The following monoclonal antibodies were used for HSC analysis: SCA-1 (D7; Biolegend), CD34 (RAM34; Ebioscience), CD135 (A2F10.1), CD150 (TC15-12F12.2; Biolegend), c-KIT (2B8), and CD48 (HM48-1). Biotinylated antibodies against CD11b (M1/70), Gr-1 (RB6-8C5), CD3ε (145-2C11), B220 (RA3-6B2), CD8a (53-6.7), CD4 (GK1.5), and TER-119 (TER-119) were used as lineage markers in combination with a Pacific Orange-Streptavidin conjugate (Invitrogen). Ly5.1 (A20; Biolegend) was used for donor cell tracing. For lineage-specific markers, the following were used: Erythroids (anti-Ter-119, TER-119 and anti-CD71, C2), T cells (anti-CD3, 500A2), B cells (anti-B220, RA3-6B2), and myeloids (anti-CD11b, M1/70, and anti-Gr-1, RB6-8C5). Biotin-labeled cocktails containing CD3, B220, Gr-1, Ter119, and CD11b antibodies (BD-Biotin mouse lineage panel, 559971) were used to differentiate LSK+ and LSK− cells in combination with SCA-1 (D7; Biolegend) and c-KIT (2B8) antibodies. SYTOX AADvanced Dead Cell Stain Kit was used to exclude dead cells. FACS data were acquired using a BD LSR II flow cytometer (BD Biosciences, San Jose, CA) and was analyzed with FlowJo version 8.8.6 for Mac.
Expression Profiling
B-lymphoid, myeloid, erythroid cells, and HSCs were isolated from predisease and diseased animals. For predisease cases, 38-1-2 and 38-1-1 (2 months old, XistΔ/+) and age-matched 37-1-1 (Xist2lox/+) were used. For leukemic cases, 27-1-1 (XistΔ/+, 6 months old, WBC 64 × 103 cells/µl) and 4-2-3 (XistΔ/XistΔ, 12 months old, 35.7 × 103 cells/µl) and age-matched control 27-1-3 were used. To isolate LSK+ HSCs, we depleted bone marrow cells of lineage-specific cells using Mouse Lineage mixture Biotin conjugate, MLM15 (Invitrogen), and MyOne Streptavidin T1 Dynabeads (Invitrogen). LSK+ CD34− HSCs were then sorted by using SCA-1 (D7)-PE-Cy7, c-KIT (2B8)-APC, and CD34-FITC (RAM34, eBioscience) antibodies. Between 3 × 103 and 6 × 103 HSCs were acquired. Splenocytes from the same animals were sorted for B cells (B220-APC and RA3-6B2), myeloids (CD11b–PE and M1/70), and erythroids (Ter119-APC and Ter-119). For each, 105–106 cells were obtained. Cells were sorted using BD Sorp Vantage SE DiVa cell sorter (BD Biosciences). All antibodies were from BD Biosciences unless noted. Total RNA was isolated using TRIzol Reagent (Invitrogen). RNA was processed using NuGEN Ovation Pico WTA System V2 paired with the Encore Biotin Module (NuGEN), and labeled cDNA probes were hybridized onto Affymetrix mouse Gene 1.0ST arrays (Affymetrix). Expression data were normalized by RMA (Irizarry et al., 2003). Data from diseased 27-1-1 and 4-2-3 were normalized to control 27-1-3; data from predisease 38-1-2 and 38-1-1 were normalized to control 37-1-1. Differentially expressed genes in Table S1 differed by two-fold or more between controls and mutants.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to members of the Lee lab for stimulating discussions. We thank R. Jaenisch forXist2lox/Xist2lox mice, G. Pihan and A. Sohani for hematopathology advice, H. Hock for advice on Vav-Cre mice, W. Press for maintenance of Xist2lox/Xist2lox mice colony, the MGH CCM-Clinical Pathology laboratory for assistance on hematological analysis, D. Dombkowski for FACS assistance, and A.J. Zall for survival bleed collections. This work was supported by the MGH ECOR Medical Discovery Fund (E.Y.), Clinician-Scientist Training Award from the Canadian Institutes of Health Research (F.E.M.), a National Institutes of Health grant (HL44851, D.T.S.), and the Howard Hughes Medical Institute, where J.T.L. is an Investigator.
Footnotes
ACCESSION NUMBERS
The GEO accession number for our microarray data set is GSE43961.
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.01.034.
REFERENCES
- Adolfsson J, Månsson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Anguera MC, Sadreyev R, Zhang Z, Szanto A, Payer B, Sheridan SD, Kwok S, Haggarty SJ, Sur M, Alvarez J, et al. Molecular signatures of human induced pluripotent stem cells highlight sex differences and cancer genes. Cell Stem Cell. 2012;11:75–90. doi: 10.1016/j.stem.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Man-ero G, Kantarjian H, Raza A, Levine RL, Neuberg D, Ebert BL. Clinical effect of point mutations in myelodysplastic syndromes. N. Engl. J. Med. 2011;364:2496–2506. doi: 10.1056/NEJMoa1013343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett JM, Orazi A. Diagnostic criteria to distinguish hypocellular acute myeloid leukemia from hypocellular myelodysplastic syndromes and aplastic anemia: recommendations for a standardized approach. Haematologica. 2009;94:264–268. doi: 10.3324/haematol.13755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecqueville M, Rey J, Bertucci F, Coppin E, Finetti P, Carbuccia N, Cervera N, Gelsi-Boyer V, Arnoulet C, Gisserot O, et al. Mutation analysis of ASXL1, CBL, DNMT3A, IDH1, IDH2, JAK2, MPL, NF1, SF3B1, SUZ12, and TET2 in myeloproliferative neoplasms. Genes Chromosomes Cancer. 2012;51:743–755. doi: 10.1002/gcc.21960. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Willard HF. The human X-inactivation centre is not required for maintenance of X-chromosome inactivation. Nature. 1994;368:154–156. doi: 10.1038/368154a0. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafrenière RG, Xing Y, Lawrence J, Willard HF. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- Clemson CM, McNeil JA, Willard HF, Lawrence JB. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csankovszki G, Panning B, Bates B, Pehrson JR, Jaenisch R. Conditional deletion of Xist disrupts histone macroH2A localization but not maintenance of X inactivation. Nat. Genet. 1999;22:323–324. doi: 10.1038/11887. [DOI] [PubMed] [Google Scholar]
- de Boer J, Williams A, Skavdis G, Harker N, Coles M, Tolaini M, Norton T, Williams K, Roderick K, Potocnik AJ, Kioussis D. Trans-genic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 2003;33:314–325. doi: 10.1002/immu.200310005. [DOI] [PubMed] [Google Scholar]
- Dewald GW, Brecher M, Travis LB, Stupca PJ. Twenty-six patients with hematologic disorders and X chromosome abnormalities. Frequent idic(X)(q13) chromosomes and Xq13 anomalies associated with pathologic ringed sideroblasts. Cancer Genet. Cytogenet. 1989;42:173–185. doi: 10.1016/0165-4608(89)90085-x. [DOI] [PubMed] [Google Scholar]
- Dierlamm J, Michaux L, Criel A, Wlodarska I, Zeller W, Louwagie A, Michaux JL, Mecucci C, Van den Berghe H. Isodicentric (X)(q13) in haematological malignancies: presentation of five new cases, application of fluorescence in situ hybridization (FISH) and review of the literature. Br. J. Haematol. 1995;91:885–891. doi: 10.1111/j.1365-2141.1995.tb05405.x. [DOI] [PubMed] [Google Scholar]
- Doll DC, Grogan TM, Greenberg BR. Chronic myelomonocytic leukemia terminating as malignant histiocytosis. Hematol. Pathol. 1987;1:183–189. [PubMed] [Google Scholar]
- Feldman AL, Minniti C, Santi M, Downing JR, Raffeld M, Jaffe ES. Histiocytic sarcoma after acute lymphoblastic leukaemia: a common clonal origin. Lancet Oncol. 2004;5:248–250. doi: 10.1016/S1470-2045(04)01428-7. [DOI] [PubMed] [Google Scholar]
- Fentiman IS, Fourquet A, Hortobagyi GN. Male breast cancer. Lancet. 2006;367:595–604. doi: 10.1016/S0140-6736(06)68226-3. [DOI] [PubMed] [Google Scholar]
- Harrison DE. Competitive repopulation: a new assay for long-term stem cell functional capacity. Blood. 1980;55:77–81. [PubMed] [Google Scholar]
- Heinonen K, Mahlamäki E, Riikonen P, Meltoranta RL, Rahiala J, Perkkiö M. Acquired X-chromosome aneuploidy in children with acute lymphoblastic leukemia. Med. Pediatr. Oncol. 1999;32:360–365. doi: 10.1002/(sici)1096-911x(199905)32:5<360::aid-mpo9>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Higgins ME, Claremont M, Major JE, Sander C, Lash AE. CancerGenes: a gene selection resource for cancer genome projects. Nucleic Acids Res. 2007;35(Database issue):D721–D726. doi: 10.1093/nar/gkl811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Jean D, Luca M, Tainsky MA, Bar-Eli M. Loss of AP-2 results in downregulation of c-KIT and enhancement of melanoma tumorigenicity and metastasis. EMBO J. 1998;17:4358–4369. doi: 10.1093/emboj/17.15.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Sherman BT, Lempicki RA. Systematic and integra-tive analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer? Hum. Mol. Genet. 2010;19(R2):R152–R161. doi: 10.1093/hmg/ddq353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive Xnucleation center. Cell. 2011;146:119–133. doi: 10.1016/j.cell.2011.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S, Purushothaman S, Bowen RB, Starmer J, Magnuson T. Evidence of Xist RNA-independent initiation of mouse imprinted X-chromosome inactivation. Nature. 2009;460:647–651. doi: 10.1038/nature08161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami T, Okamoto K, Sugihara H, Hattori T, Reeve AE, Ogawa O, Okada Y. The roles of supernumerical X chromosomes and XIST expression in testicular germ cell tumors. J. Urol. 2003;169:1546–1552. doi: 10.1097/01.ju.0000044927.23323.5a. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Laurencet FM, Chapuis B, Roux-Lombard P, Dayer JM, Beris P. Malignant histiocytosis in the leukaemic stage: a new entity (M5c–AML) in the FAB classification? Leukemia. 1994;8:502–506. [PubMed] [Google Scholar]
- Lee JT. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat. Rev. Mol. Cell Biol. 2011;12:815–826. doi: 10.1038/nrm3231. [DOI] [PubMed] [Google Scholar]
- Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–1439. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- Li L, Bailey E, Greenblatt S, Huso D, Small D. Loss of the wild-type allele contributes to myeloid expansion and disease aggressiveness in FLT3/ITD knockin mice. Blood. 2011;118:4935–4945. doi: 10.1182/blood-2011-01-328096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao DJ, Du QQ, Yu BW, Grignon D, Sarkar FH. Novel perspective: focusing on the X chromosome in reproductive cancers. Cancer Invest. 2003;21:641–658. doi: 10.1081/cnv-120022385. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermato-genesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- Mazumdar M, Sung MH, Misteli T. Chromatin maintenance by a molecular motor protein. Nucleus. 2011;2:591–600. doi: 10.4161/nucl.2.6.18044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhoubad S, Bock C, de Boer AS, Kiskinis E, Meissner A, Eggan K. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell. 2012;10:595–609. doi: 10.1016/j.stem.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KL, Barr ML. The sex chromatin in benign tumours and related conditions in man. Br. J. Cancer. 1955;9:246–252. doi: 10.1038/bjc.1955.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller AM, Medvinsky A, Strouboulis J, Grosveld F, Dzierzak E. Development of hematopoietic stem cell activity in the mouse embryo. Immunity. 1994;1:291–301. doi: 10.1016/1074-7613(94)90081-7. [DOI] [PubMed] [Google Scholar]
- Muramatsu H, Makishima H, Maciejewski JP. Chronic myelomonocytic leukemia and atypical chronic myeloid leukemia: novel pathoge-netic lesions. Semin. Oncol. 2012;39:67–73. doi: 10.1053/j.seminoncol.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa SH, Lee JT. Detection of nascent RNA, single-copy DNA and protein localization by immunoFISH in mouse germ cells and preimplantation embryos. Nat. Protoc. 2011;6:270–284. doi: 10.1038/nprot.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namekawa SH, Payer B, Huynh KD, Jaenisch R, Lee JT. Two-step imprinted X inactivation: repeat versus genic silencing in the mouse. Mol. Cell. Biol. 2010;30:3187–3205. doi: 10.1128/MCB.00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S, Nakauchi H, Nagayoshi K, Nishikawa S, Miura Y, Suda T. In vivo and in vitro stem cell function of c-kit- and Sca-1-positive murine hematopoietic cells. Blood. 1992;80:3044–3050. [PubMed] [Google Scholar]
- Pageau GJ, Hall LL, Ganesan S, Livingston DM, Lawrence JB. The disappearing Barr body in breast and ovarian cancers. Nat. Rev. Cancer. 2007;7:628–633. doi: 10.1038/nrc2172. [DOI] [PubMed] [Google Scholar]
- Pang WW, Price EA, Sahoo D, Beerman I, Maloney WJ, Rossi DJ, Schrier SL, Weissman IL. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc. Natl. Acad. Sci. USA. 2011;108:20012–20017. doi: 10.1073/pnas.1116110108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsson K, Haferlach C, Fonatsch C, Hagemeijer A, Andersen MK, Slovak ML, Johansson B MDS Foundation. The idic(X)(q13) in myeloid malignancies: breakpoint clustering in segmental duplications and association with TET2 mutations. Hum. Mol. Genet. 2010;19:1507–1514. doi: 10.1093/hmg/ddq024. [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu. Rev. Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Pellagatti A, Cazzola M, Giagounidis A, Perry J, Malcovati L, Della Porta MG, Jädersten M, Killick S, Verma A, Norbury CJ, et al. Deregulated gene expression pathways in myelodysplastic syndrome hematopoietic stem cells. Leukemia. 2010;24:756–764. doi: 10.1038/leu.2010.31. [DOI] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. Requirement for Xist in X chromosome inactivation. Nature. 1996;379:131–137. doi: 10.1038/379131a0. [DOI] [PubMed] [Google Scholar]
- Percy DH, Barthold SW. Mouse. Ames, IA: Blackwell; 2007. [Google Scholar]
- Rack KA, Chelly J, Gibbons RJ, Rider S, Benjamin D, Lafreniére RG, Oscier D, Hendriks RW, Craig IW, Willard HF, et al. Absence of the XIST gene from late-replicating isodicentric X chromosomes in leukaemia. Hum. Mol. Genet. 1994;3:1053–1059. doi: 10.1093/hmg/3.7.1053. [DOI] [PubMed] [Google Scholar]
- Sandberg AA, editor. Cytogenetics of the mammalian X-chromosome, Part B: X Chromosome Anomalies and Their Clinical Manifestations. New York: Alan R Liss, Inc.; 1983. “The Xchromosomein human neoplasia, including sex chromatin and congenital conditions with X chromosome anomalies.”; pp. 459–498. [Google Scholar]
- Savarese F, Flahndorfer K, Jaenisch R, Busslinger M, Wutz A. Hematopoietic precursor cells transiently reestablish permissiveness for X inactivation. Mol. Cell. Biol. 2006;26:7167–7177. doi: 10.1128/MCB.00810-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt A, Guichard J, Massé JM, Debili N, Cramer EM. Of mice and men: comparison of the ultrastructure of megakaryocytes and platelets. Exp. Hematol. 2001;29:1295–1302. doi: 10.1016/s0301-472x(01)00733-0. [DOI] [PubMed] [Google Scholar]
- Shen Y, Matsuno Y, Fouse SD, Rao N, Root S, Xu R, Pellegrini M, Riggs AD, Fan G. X-inactivation in female human embryonic stem cells is in a nonrandom pattern and prone to epigenetic alterations. Proc. Natl. Acad. Sci. USA. 2008;105:4709–4714. doi: 10.1073/pnas.0712018105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva SS, Rowntree RK, Mekhoubad S, Lee JT. X-chromo-some inactivation and epigenetic fluidity in human embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2008;105:4820–4825. doi: 10.1073/pnas.0712136105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttie AW. Histopathology of the spleen. Toxicol. Pathol. 2006;34:466–503. doi: 10.1080/01926230600867750. [DOI] [PubMed] [Google Scholar]
- Takizawa T, Gudla PR, Guo L, Lockett S, Misteli T. Allele-specific nuclear positioning of the monoallelically expressed astrocyte marker GFAP. Genes Dev. 2008;22:489–498. doi: 10.1101/gad.1634608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tefferi A. Primary myelofibrosis: 2012 update on diagnosis, risk stratification, and management. Am. J. Hematol. 2011;86:1017–1026. doi: 10.1002/ajh.22210. [DOI] [PubMed] [Google Scholar]
- Vainchenker W, Delhommeau F, Constantinescu SN, Bernard OA. New mutations and pathogenesis of myeloproliferative neoplasms. Blood. 2011;118:1723–1735. doi: 10.1182/blood-2011-02-292102. [DOI] [PubMed] [Google Scholar]
- Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 2011;12:542–553. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell. 2000;5:695–705. doi: 10.1016/s1097-2765(00)80248-8. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Nagata K, Kida A, Hamaguchi H. Acquired gain of an X chromosome as the sole abnormality in the blast crisis of chronic neutrophilic leukemia. Cancer Genet. Cytogenet. 2002;134:84–87. doi: 10.1016/s0165-4608(01)00603-3. [DOI] [PubMed] [Google Scholar]
- Zhang LF, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng R, Blobel GA. GATA Transcription Factors and Cancer. Genes Cancer. 2010;1:1178–1188. doi: 10.1177/1947601911404223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.