Abstract
Efforts have been made to achieve full automation of molecular assays for quantitative detection of human immunodeficiency virus type 1 (HIV-1). In the present study, the Abbott LCx HIV RNA Quantitative assay was evaluated in conjunction with automated HIV-1 RNA extraction on the MagNA Pure LC instrument and compared to the conventional LCx HIV RNA Quantitative assay, which uses a manual nucleic acid extraction protocol. Accuracy, linearity, and interassay and intra-assay variations were determined. The performance of the assay in a routine clinical laboratory was tested with a total of 105 clinical specimens. When the accuracy of the LCx HIV RNA Quantitative assay with the automated sample preparation protocol was tested, all results were found to be within ±0.5 log unit of the expected results. Determination of linearity resulted in a quasilinear curve over 3.5 log units. For determination of interassay variation, coefficients of variation were found to be between 21 and 66% for the LCx HIV RNA Quantitative assay with the automated sample preparation protocol and between 10 and 69% for the LCx HIV RNA Quantitative assay with manual sample preparation. For determination of intra-assay variation, coefficients of variation were found to be between 7 and 25% for the LCx HIV RNA Quantitative assay with the automated sample preparation protocol and between 7 and 19% for the LCx HIV RNA Quantitative assay with manual sample preparation. When clinical samples were tested by the LCx HIV RNA Quantitative assay with the automated sample preparation protocol and the results were compared with those of the LCx HIV RNA Quantitative assay with manual sample preparation, 95% of all positive results were found to be within ±0.5 log unit. In conclusion, the assay with automated sample preparation proved to be suitable for use in the routine diagnostic laboratory and required significantly less hands-on time.
Molecular assays for quantitation of the human immunodeficiency virus type 1 (HIV-1) load in plasma have been shown to be important for the monitoring of HIV-1-infected patients and determination of the prognosis (14, 16, 22). Commercially available assays have been introduced and have been found to be useful for the routine diagnostic laboratory (3, 4, 6, 13, 20). The LCx HIV RNA Quantitative assay (Abbott Laboratories, Abbott Park, Ill.) for the quantitative detection of HIV-1 RNA in human plasma has recently been brought onto the market (9, 23). The nucleic acid extraction protocol included in this assay, however, is time-consuming and labor-intensive.
Automation of nucleic acid extraction has already been introduced into the routine diagnostic laboratory for the detection of pathogens in clinical specimens (10, 18). It has been reported that fully automated sample preparation on the MagNA Pure LC instrument (Roche Applied Science, Mannheim, Germany) meets the criteria of the routine diagnostic laboratory (11). It was found to be useful as a means of avoiding human error, increasing throughput, and decreasing the amount of hands-on time. The MagNA Pure LC instrument has also been shown to be suitable for the effective extraction of nucleic acids of different pathogens (5, 7, 15, 17, 21).
In the present study, we evaluated the performance of a new molecular assay for the quantitative detection of HIV-1 RNA in human plasma, which consisted of automated sample preparation on the MagNA Pure LC instrument and the LCx HIV RNA Quantitative assay. Accuracy was tested with a reference material; linearity was tested by use of a dilution series of samples with high titers. Both interassay and intra-assay variations were determined and compared with those of the standard version of the LCx HIV RNA quantitative assay, which included a manual sample preparation protocol. The performance of the new assay in the routine diagnostic laboratory was evaluated with routine clinical samples.
MATERIALS AND METHODS
Molecular assays.
The LCx HIV RNA Quantitative assay (Abbott) was performed according to the instructions of the manufacturer provided in the package insert. Sample preparation was done with either an automated or a manual sample preparation protocol. The LCx HIV RNA Quantitative assay uses an internal standard. The internal standard is a 163-bp RNA transcript, which is similar to the HIV-1 target except for a short, unique internal sequence. This transcript is added to the specimen during sample preparation and is simultaneously amplified by PCR in a competitive reaction.
Automated sample preparation protocol.
For fully automated sample preparation, isolation of HIV-1 RNA was done on the MagNA Pure LC instrument with the MagNA Pure LC Total Nucleic Acid Isolation kit—Large Volume (Roche). MagNA Pure LC software (version 3.01) was used. The input sample volume was 1,000 μl. For each sample, 3.5 μl of an HIV RNA quantitative internal standard was added to 1.35 ml of lysis reagent (including an additional volume because of the dead volume) prior to the start of the sample preparation procedure. Additionally, run controls, which are included in the LCx HIV RNA Quantitative Control kit, were prepared by addition of 20 μl of the control to 980 μl of lysis reagent for each control sample. An elution volume of 100 μl and a dilution volume of 0 μl were chosen. Other details, such as reagent volumes and the number of reaction tips needed for the run, were automatically calculated by the software. The MagNA Pure LC instrument performed all of the remaining steps of the procedure.
Manual sample preparation protocol.
Manual isolation of HIV RNA was done according to the instructions of the manufacturer of the LCx HIV RNA Quantitative Control kit provided in the package insert. The recommended specimen preparation procedure is based on the procedure for 1.0 ml from Qiagen GmbH (Hilden, Germany) and includes the LCx Vacuum System for the washing steps.
Reverse transcription, amplification, hybridization, and detection.
Following sample preparation and setup of the reaction mixture, reverse transcription and amplification of the target and the internal standard were simultaneously performed on the LCx Thermal Cycler (Abbott) in the same reaction tube. An additional cycle with a high-temperature denaturation and a low-temperature incubation allows the HIV-1- and the internal standard-specific detection probes, which are conjugated with different haptens (detection haptens), to hybridize to the amplification products. The tubes were then transferred unopened into an automated analyzer (LCx Analyzer; Abbott) for detection and quantitation with the microparticle enzyme immunoassay (MEIA) system. Two different conjugates (alkaline phosphatase and β-galactosidase) are used; the first one binds to the HIV-1-specific probes, and the second one binds to the internal standard-specific probes. The two substrates [7-β-galactosidase coumarin-4-acetic (2-hydroxyethylamine) and 4-methylumbelliferyl phosphate] were then added, and the resulting fluorescence was read by the MEIA optic system. A calibration was carried out with each different lot of HIV-1 RNA detected by the LCx assay by testing, in duplicate, six calibrators, in which the HIV-1 RNA and the internal standard concentrations were increased and decreased inversely from calibrator 1 through calibrator 6. The LCx software used the results to draw a curve, with which the results for the samples and the controls were subsequently calculated. The analyzer software was additionally used to carry out a second control analysis for possible inhibition of RNA amplification by checking the validity of the HIV-1/internal standard ratios for each sample and control. Three control levels (negative, low positive, and high positive) were tested in each run, and each run had to give valid results for the LCx instrument to express the HIV RNA levels in clinical samples.
Study design.
All experiments were done in the International Organization for Standardization-certified Molecular Diagnostics Laboratory, Institute of Hygiene, Medical University Graz.
In the first step, the accuracy of the LCx HIV RNA Quantitative assay with the automated sample preparation protocol was determined with original members and dilutions of commercially available HIV-1 RNA controls (NAC-HIV1E2 and NAC-HIV5E3; Acrometrix, Benicia, Calif.). Control members contained complete viral particles derived from infectious clones of HIV-1. The concentrations used were 5.0 × 104, 2.5 × 104, 1.0 × 103, 5.0 × 102, and 1.0 × 102 HIV-1 RNA copies/ml.
In the second step, the linearity of the LCx HIV RNA Quantitative assay with the automated sample preparation protocol was determined. Two routine clinical plasma samples which contained 7.9 × 105 and 5.0 × 105 HIV-1 RNA copies/ml, respectively, were used. Dilution series (0.5-log steps, i.e., 1:3.16 dilutions) of both serum samples were prepared by using HIV-1-negative human plasma. Each dilution was analyzed three times, and the mean HIV-1 RNA titer of each sample was determined.
In the third step, the interassay variation of the LCx HIV RNA Quantitative assay with the automated sample preparation protocol was determined and compared with that of the LCx HIV RNA Quantitative assay with the manual sample preparation protocol. Eight samples that contained different amounts of HIV-1 RNA ranging from 1.2 × 103 to 5.3 × 105 copies/ml were tested one time on each of 5 days.
In the fourth step, the intra-assay variations of the assays were compared. Four clinical routine samples were aliquoted, and each aliquot was analyzed five times in one run.
In the fifth step, the performance of the LCx HIV RNA Quantitative assay with the automated sample preparation protocol was evaluated in the routine diagnostic laboratory and compared with that of the LCx HIV RNA Quantitative assay with the manual sample preparation protocol. A total of 105 clinical plasma samples from patients with HIV-1 infection who were or who were not receiving antiretroviral therapy were investigated. The plasma samples contained HIV-1 subtypes B, E, AE, and AG. The HIV-1 subtypes were determined as described recently (12). Another aliquot of each of the samples had been tested earlier by the COBAS AMPLICOR HIV-1 MONITOR test (version 1.5) in the same routine diagnostic laboratory.
Statistical analysis.
SPSS (version 10.0) software (SPSS Inc., Chicago, Ill.) was used for computerized statistical analysis. A log transformation was used to adjust for the nonhomogeneity of the variance structure. Linearity was tested by comparison of the results of polynomial regression by use of the Mallows Cp statistic with a quadratic regression.
RESULTS
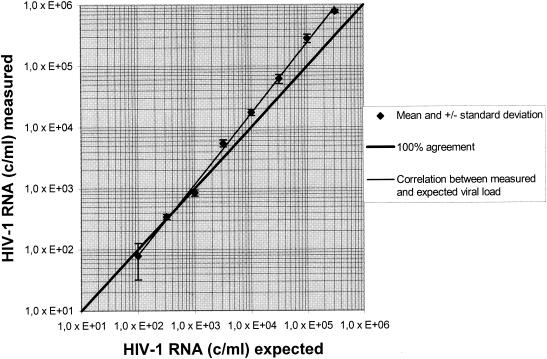
The accuracy of the LCx HIV RNA Quantitative assay with the automated sample preparation protocol was tested with HIV-1 RNA controls. All results were found to be within ±0.32 log unit of the expected results (Table 1). Linearity was tested with dilution series of two high-titer routine clinical samples. A linear curve was observed with dilutions containing up to 1.0 × 106 HIV-1 RNA copies/ml. Dilutions containing less than 1.0 × 102 HIV-1 RNA copies/ml were not detected (Fig. 1).
TABLE 1.
Accuracy of the automated sample preparation protocol by use of HIV-1 controls and their dilutions
| Vial no. | HIV RNA (no. of copies/ml)
|
Log expected results/actual results | |
|---|---|---|---|
| Expected resultsa | Actual results | ||
| 1 | 1.0 × 102 | 1.9 × 102 | −0.28 |
| 2 | 5.0 × 102 | 2.5 × 102 | 0.30 |
| 3 | 1.0 × 103 | 7.6 × 102 | 0.12 |
| 4 | 2.5 × 104 | 1.2 × 104 | 0.32 |
| 5 | 5.0 × 104 | 3.3 × 104 | 0.19 |
The values for viral stock material were determined by the Roche COBAS AMPLICOR HIV-1 MONITOR (version 1.5) assay, and viral loads were adjusted to the specified concentrations by dilution in human plasma.
FIG. 1.
Linearity of results for one of the high-titer routine clinical serum samples produced by automated sample preparation followed by the LCx HIV RNA Quantitative assay with a 0.5-log dilution (i.e., 1:3.16). c/ml, number of copies per milliliter.
For determination of interassay variation, mean plasma HIV-1 RNA titers ranged from 8.7 × 101 to 6.6 × 104 HIV-1 RNA copies/ml for the LCx HIV RNA Quantitative assay with the automated sample preparation protocol when they were tested one time on each of 5 days. The corresponding values for the LCx HIV RNA Quantitative assay with manual sample preparation were 1.1 × 102 and 5.3 × 104 HIV-1 RNA copies/ml. HIV-1 RNA in all replicates with the lowest concentration was detected by use of both extraction protocols. The coefficients of variation were found to be between 21 and 66% for the LCx HIV RNA Quantitative assay with the automated sample preparation protocol and between 10 and 69% for the LCx HIV RNA Quantitative assay with manual sample preparation (Table 2).
TABLE 2.
Interassay variation of results obtained by the automated versus the manual sample extraction protocol
| Sample no. | Mean no. of HIV-1 RNA copies/ml detected by the following protocol:
|
SD
|
Coefficient of variation (%)
|
|||
|---|---|---|---|---|---|---|
| Automateda | Manualb | Automated | Manual | Automated | Manual | |
| 1 | 8.7 × 101 | 1.1 × 102 | 5.8 × 101 | 8.0 × 101 | 66 | 69 |
| 2 | 4.3 × 103 | 6.2 × 103 | 2.3 × 103 | 2.2 × 103 | 53 | 36 |
| 3 | 8.0 × 103 | 1.3 × 104 | 2.9 × 103 | 5.9 × 103 | 36 | 46 |
| 4 | 9.8 × 103 | 5.7 × 103 | 2.8 × 103 | 2.4 × 103 | 28 | 41 |
| 5 | 1.9 × 104 | 1.5 × 104 | 6.2 × 103 | 3.7 × 103 | 32 | 24 |
| 6 | 2.0 × 104 | 2.0 × 104 | 8.9 × 103 | 1.9 × 103 | 44 | 10 |
| 7 | 2.8 × 104 | 2.9 × 104 | 8.1 × 103 | 7.9 × 103 | 29 | 27 |
| 8 | 6.6 × 104 | 5.3 × 104 | 1.4 × 103 | 2.6 × 104 | 21 | 48 |
Automated sample preparation followed by the LCx HIV RNA Quantitative assay.
Manual sample preparation followed by the LCx HIV RNA Quantitative assay.
Determination of intra-assay variation was tested with four routine clinical samples by analyzing them five times. The coefficients of variation were found to be between 7 and 25% for the LCx HIV RNA Quantitative assay with the automated sample preparation protocol and between 7 and 19% for the LCx HIV RNA Quantitative assay with manual sample preparation (Table 3).
TABLE 3.
Intra-assay variation of results obtained by the automated versus the manual sample preparation protocola
| Sample no. | Mean no. of HIV-1 RNA copies/ml detected by the following protocol:
|
SD
|
Coefficient of variation (%)
|
|||
|---|---|---|---|---|---|---|
| Automatedb | Manualc | Automated | Manual | Automated | Manual | |
| 1 | 9.7 × 102 | 9.3 × 102 | 2.1 × 102 | 1.7 × 102 | 22 | 18 |
| 2 | 7.3 × 103 | 5.1 × 103 | 1.8 × 103 | 3.6 × 102 | 25 | 7 |
| 3 | 3.5 × 104 | 1.6 × 104 | 8.4 × 103 | 2.6 × 103 | 24 | 17 |
| 4 | 5.1 × 104 | 3.6 × 104 | 3.6 × 103 | 6.8 × 103 | 7 | 19 |
Data are from five replicates of one run.
Automated sample preparation followed by the LCx HIV RNA Quantitative assay.
Manual sample preparation followed by the LCx HIV RNA Quantitative assay.
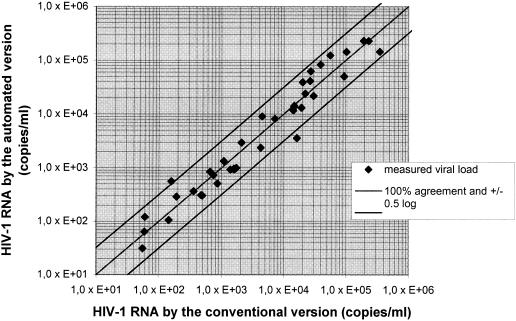
When 105 clinical samples were tested by the LCx HIV RNA Quantitative assay with the automated sample preparation protocol, all samples with positive results (n = 39) were also found to be positive by the LCx HIV RNA Quantitative assay with manual sample preparation, and the results for 37 samples (95%) with positive results were found to be within ±0.5 log unit (Fig. 2). One sample was negative by the LCx HIV RNA Quantitative assay with the automated sample preparation protocol but was positive (3.5 × 103 HIV-1 RNA copies/ml) by the LCx HIV RNA Quantitative assay with manual sample preparation. This sample had earlier tested negative by the COBAS AMPLICOR HIV-1 MONITOR test. Of 66 negative samples, the results for 4 samples were found to be invalid due to a low internal standard rate by the LCx HIV RNA Quantitative assay with the automated sample preparation protocol, and the result for another sample was found to be invalid due to a low internal standard rate by the LCx HIV RNA Quantitative assay with manual sample preparation. On repetition of the assay, the results for two samples with invalid results by the LCx HIV RNA Quantitative assay with automated sample preparation were still found to be invalid due to a low internal standard rate.
FIG. 2.
Correlation between viral load measurements for 39 HIV-1-positive plasma samples obtained by the LCx HIV RNA Quantitative assay with the automated sample preparation protocol and the LCx HIV RNA Quantitative assay with manual sample preparation.
During the whole study, the results for the low- and high-positive run controls were in the range required, and those for the negative run controls tested were negative. Because of the different capacities of the MagNA Pure LC instrument (32 samples) and the LCx Analyzer (24 samples), an additional set of run controls was added in every third run.
The LCx HIV RNA Quantitative assay with the automated sample preparation protocol proved to be quick and non-labor-intensive. The automated HIV-1 RNA extraction with the MagNA Pure LC instrument was completed within 120 min for 24 samples. This included 15 min for setup of the MagNA Pure LC instrument. In contrast, the manual extraction of 24 samples was completed within 180 min and involved 150 min of hands-on time. For both methods, the extracted samples had to be added manually to the PCR tubes containing the prefilled master mixture. The time needed for addition of the samples and the activation reagent to the PCR tube, amplification on the LCx Thermocycler, and detection on the LCx Analyzer was 270 min, which involved a total of 30 min for manual steps. The total time to the retrieval of results was 6.5 h with the automated sample preparation protocol and 7.5 h with the manual sample preparation protocol.
DISCUSSION
The LCx HIV RNA Quantitative assay has been described to be useful for routine quantitation of HIV-1 RNA in plasma (1, 2, 9, 19). This assay, however, uses a manual RNA extraction protocol, which is laborious and time-consuming. Automated nucleic acid extraction devices have recently been introduced and have been found to be useful for extraction of viral DNAs or RNAs in the routine diagnostic laboratory (8, 11, 18). In the present study, the usefulness of the LCx HIV RNA Quantitative assay with an automated sample preparation protocol was examined, and the results were compared to those obtained by the conventional LCx HIV RNA Quantitative assay with manual sample preparation.
The MagNA Pure LC instrument allows the use of sample volumes between 200 and 1,000 μl. To optimize HIV-1 RNA recovery, the large-volume version of the nucleic acid isolation kit, which allows the introduction of 1,000 μl, was chosen. Prior to the start of the fully automated extraction procedure, an adequate amount of internal standard was manually introduced into the lysis reagent. Because amplification may fail because of interference from PCR inhibitors, an internal standard must be incorporated into every molecular assay used in the routine diagnostic laboratory.
The LCx HIV RNA Quantitative assay with the automated sample preparation protocol showed good accuracy and sufficient linearity. With increasing HIV-1 RNA concentrations, however, the results were found to be slightly higher than expected. Similar to our results, it has recently been reported (9) that the conventional version of the LCx HIV RNA Quantitative assay (with manual sample preparation) proved to be linear between 5.0 × 101 copies/ml and 1.0 × 106 copies/ml with a sample volume of 1,000 μl. In this study, the interassay and intra-assay variations of both assays were found to be similar. These results are comparable to those reported for the conventional LCx HIV RNA Quantitative assay with a sample volume of 1,000 μl and other molecular assays based on PCR amplification (8, 9, 18).
When the LCx HIV RNA Quantitative assay with the automated sample preparation protocol was applied for routine use, no differences in the results due to the presence of different HIV-1 subtypes could be observed by either assay. The only discrepant result (for a sample positive by the assay with the manual extraction protocol but with RNA below the detection limit by both the new assay with the automated extraction protocol and the COBAS AMPLICOR HIV-1 MONITOR assay, version 1.5) might be explained by contamination of the sample during manual sample preparation, which may further support automation efforts. The results for four samples were found to be invalid due to a low internal standard rate by the LCx HIV RNA Quantitative assay with the automated sample preparation protocol. Invalid results may have been due to inhibition or loss of the internal standard during extraction. In this study, none of the samples were contaminated with heparin or found to be hyperlipemic. Because the internal standard is a short (163-bp) RNA transcript, a decreased level of recovery as a result of the use of the automated sample preparation protocol with the MagNA Pure instrument appears to be more likely.
The new LCx HIV RNA Quantitative assay with automated sample preparation showed good overall functionality and user-friendliness. The automated sample preparation protocol took 34% less total time than the manual one and required 90% less hands-on time. It must, however, be taken into consideration that aliquots of the extracted samples must still be pipetted manually into the tubes containing the master mixtures. Nevertheless, because of the significantly lower number of manipulations required, the probability that false-positive results will be obtained because of contamination may be less. For optimal work flow, 32 samples should be analyzed on the MagNA Pure instrument. Only 24 samples can be processed on the LCx Analyzer, which requires an additional set of run controls in every third sample preparation run.
In conclusion, the Abbott LCx HIV RNA Quantitative assay with automated nucleic acid purification on the MagNA Pure LC instrument proved to be a step forward in meeting the requirements of the routine diagnostic laboratory. Compared to the conventional assay, this assay provides significant reductions in hands-on work time and labor intensity.
Acknowledgments
This work was supported in part by a grant from the Austrian-Hungarian Scientific and Education Cooperation Action Program and Abbott GmbH Diagnostika.
We thank Henriette Wachtberger for technical assistance and Andrea Holensteiner and John Robinson for stimulating discussions.
REFERENCES
- 1.Abravaya, K., C. Esping, R. Hoenle, J. Gorzowski, R. Perry, P. Kroeger, J. Robinson, and R. Flanders. 2000. Performance of a multiple qualitative PCR LCx assay for detection of human immunodeficiency virus type 1 (HIV-1) group M subtypes, group O, and HIV-2. J. Clin. Microbiol. 38:716-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berger, I., H. F. Rabenau, A. Stief, H. Troonen, and H. W. Doerr. 2001. Evaluation of the new LCx HIV RNA quantitative assay: comparison with the Cobas Amplicor HIV Monitor assay. Med. Microbiol. Immunol. 190:129-134. [DOI] [PubMed] [Google Scholar]
- 3.de Baar, M. P., M. W. van Dooren, E. de Rooij, M. Bakker, B. van Gemen, J. Goudsmit, and A. de Ronde. 2001. Single rapid real-time monitored isothermal RNA amplification assay for quantification of human immunodeficiency virus type 1 isolates from groups M, N, and O. J. Clin. Microbiol. 39:1378-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erali, M., and D. Hillyard. 1999. Evaluation of the ultrasensitive Roche Amplicor HIV-1 Monitor assay for the quantification of human immunodeficiency virus type 1 RNA. J. Clin. Microbiol. 37:792-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Espy, M. J., P. N. Rys, A. D. Wold, J. R. Uhl, L. M. Sloan, G. D. Jenkins, D. M. Ilstrup, F. R. Cockerill III, R. Patel, J. E. Rosenblatt, and T. F. Smith. 2001. Detection of herpes simplex virus DNA in genital and dermal specimens by LightCycler PCR after extraction using the IsoQuick, MagNA Pure, and BioRobot 9604 methods. J. Clin. Microbiol. 39:2233-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fischer, M., W. Huber, A. Kallivroussis, P. Ott, M. Opravil, R. Lüthy, R. Weber, and R. W. Cone. 1999. Highly sensitive methods for quantitation of human immunodeficiency virus type 1 RNA from plasma, cells, and tissue. J. Clin. Microbiol. 37:1260-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grisold, A. J., E. Leitner, G. Mühlbauer, E. Marth, and H. H. Kessler. 2002. Detection of methicillin-resistant Staphylococcus aureus and simultaneous confirmation by automated nucleic acid extraction and real-time PCR. J. Clin. Microbiol. 40:2392-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hölzl, G., M. Stöcher, V. Leb, H. Stekel, and J. Berg. 2003. Entirely automated quantification of human immunodeficiency virus type 1 (HIV-1) RNA in plasma by using the Ultrasensitive COBAS AMPLICOR HIV-1 Monitor Test and RNA purification on the MagNA Pure LC instrument. J. Clin. Microbiol. 41:1248-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johanson, J., K. Abravaya, W. Caminiti, D. Erickson, R. Flanders, G. Leckie, E. Marshall, C. Mullen, Y. Ohhashi, R. Perry, J. Ricci, J. Salituro, A. Smith, N. Tang, M. Vi, and J. Robinson. 2001. A new ultrasensitive assay for quantitation of HIV-1 RNA in plasma. J. Virol. Methods 95:81-92. [DOI] [PubMed] [Google Scholar]
- 10.Jungkind, D. 2001. Automation of laboratory testing for infectious diseases using the polymerase chain reaction—our past, our present, our future. J. Clin. Virol. 20:1-6. [DOI] [PubMed] [Google Scholar]
- 11.Kessler, H. H., G. Mühlbauer, E. Stelzl, E. Daghofer, B. I. Santner, and E. Marth. 2001. Fully automated nucleic acid extraction: MagNA Pure LC. Clin. Chem. 47:1124-1126. [PubMed] [Google Scholar]
- 12.Kessler, H. H., D. Deuretzbacher, E. Stelzl, E. Daghofer, B. I. Santner, and E. Marth. 2001. Determination of human immunodeficiency virus type 1 subtypes by a rapid method useful for the routine diagnostic laboratory. Clin. Diagn. Lab. Immunol. 8:1018-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poljak, M., K. Seme, and S. Koren. 1997. Evaluation of automated COBAS AMPLICOR hepatitis C virus PCR system. J. Clin. Microbiol. 35:2983-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Powderly, W. G., M. S. Saag, S. Chapman, G. Yu, B. Quart, and N. J. Clendeninn. 1999. Predictors of optimal virological response to potent antiretroviral therapy. AIDS 13:1873-1880. [DOI] [PubMed] [Google Scholar]
- 15.Rabenau, H. F., A. M. Clarici, G. Muhlbauer, A. Berger, A. Vince, Z. Muller, E. Daghofer, B. I. Santner, E. Marth, and H. H. Kessler. 2002. Rapid detection of enterovirus infection by automated RNA extraction and real-time fluorescence PCR. J. Clin. Virol. 25:155-164. [DOI] [PubMed] [Google Scholar]
- 16.Raboud, J. M., J. S. G. Montaner, B. Conway, S. Rae, P. Reiss, S. Vella, D. Cooper, J. Lange, M. Harris, M. A. Wainberg, P. Robinson, M. Myers, and D. Hall. 1998. Suppression of plasma viral load below 20 copies/ml is required to achieve a long-term response to therapy. AIDS 12:1619-1624. [DOI] [PubMed] [Google Scholar]
- 17.Raggam, R. B., E. Leitner, G. Mühlbauer, J. Berg, M. Stocher, A. Grisold, E. Marth, and H. H. Kessler. 2002. Qualitative detection of Legionella species in bronchoalveolar lavages and induced sputa by automated DNA extraction and real-time polymerase chain reaction. Med. Microbiol. Immunol. 191:119-125. [DOI] [PubMed] [Google Scholar]
- 18.Stelzl, E., A. Kormann-Klement, J. Haas, E. Daghofer, B. I. Santner, E. Marth, and H. H. Kessler. 2002. Evaluation of an automated sample preparation protocol for quantitative detection of hepatitis C virus RNA. J. Clin. Microbiol. 40:1447-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swanson, P., B. J. Harris, V. Holzmayer, S. G. Devare, G. Schochetman, and J. Hackett, Jr. 2000. Quantification of HIV-1 group M (subtypes A-G) and group O by the LCx HIV RNA quantitative assay. J. Virol. Methods 89:97-108. [DOI] [PubMed] [Google Scholar]
- 20.Swanson, P., V. Soriano, S. G. Devare, and J. Hackett, Jr. 2001. Comparative performance of three viral load assays on human immunodeficiency virus type 1 (HIV-1) isolates. J. Clin. Microbiol. 39:862-870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Doornum, G., J. J. Guldemeester, A. D. Osterhaus, and H. G. Niesters. 2003. Diagnosing herpesvirus infections by real-time amplification and rapid culture. J. Clin. Microbiol. 41:576-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Praag, R. M., F. W. Wit, S. Jurriaans, F. de Wolf, J. M. Prins, and J. M. Lange. 2002. Improved long-term suppression of HIV-1 replication with triple-class multidrug regimen compared with standard of antiretroviral therapy. AIDS 16:719-725. [DOI] [PubMed] [Google Scholar]
- 23.Zanchetta, N., G. Nardi, L. Tocalli, L. Drago, C. Bossi, F. R. Pulvirenti, C. Galli, and M. R. Gismondo. 2000. Evaluation of the Abbott LCx HIV-1 RNA Quantitative, a new assay for quantitative determination of human immunodeficiency virus type 1 RNA. J. Clin. Microbiol. 38:3882-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]