Abstract
Background
Tuberculosis (TB) is a major public health problem with high mortality and morbidity rates, especially in low-income countries. Disturbingly, the emergence of multidrug resistant (MDR) and extensively drug resistant (XDR) TB cases has worsened the situation, raising concerns of a future epidemic of virtually untreatable TB. Indeed, the rapid diagnosis of MDR TB is a critical issue for TB management. This study is an attempt to establish a rapid diagnosis of MDR TB by sequencing the target fragments of the rpoB gene which linked to resistance against rifampicin and the katG gene and inhA promoter region, which are associated with resistance to isoniazid.
Methods
For this purpose, 133 sputum samples of TB patients from Morocco were enrolled in this study. One hundred samples were collected from new cases, and the remaining 33 were from previously treated patients (drug relapse or failure, chronic cases) and did not respond to anti-TB drugs after a sufficient duration of treatment. All samples were subjected to rpoB, katG and pinhA mutation analysis by polymerase chain reaction and DNA sequencing.
Results
Molecular analysis showed that seven strains were isoniazid-monoresistant and 17 were rifampicin-monoresistant. MDR TB strains were identified in nine cases (6.8%). Among them, eight were traditionally diagnosed as critical cases, comprising four chronic and four drug-relapse cases. The last strain was isolated from a new case. The most recorded mutation in the rpoB gene was the substitution TCG > TTG at codon 531 (Ser531 Leu), accounting for 46.15%. Significantly, the only mutation found in the katG gene was at codon 315 (AGC to ACC) with a Ser315Thr amino acid change. Only one sample harbored mutation in the inhA promoter region and was a point mutation at the −15p position (C > T).
Conclusion
The polymerase chain reaction sequencing approach is an accurate and rapid method for detection of drug-resistant TB in clinical specimens, and could be of great interest in the management of TB in critical cases to adjust the treatment regimen and limit the emergence of MDR and XDR strains.
Keywords: Morocco, Mycobacterium tuberculosis, multidrug resistance, rpoB, katG, inhA promoter
Introduction
Tuberculosis (TB) remains one of the most prevalent infectious diseases worldwide, especially in developing countries, where the highest burden of TB is found.1 This is further exemplified by the high mortality and morbidity due to this disease. In this context, the emergence of multidrug resistant (MDR) TB (resistant to at least isoniazid and rifampicin) and extremely drug resistant TB (MDR plus additional resistance to a fluoroquinolone and any second-line injectable drug) cases has worsened the situation.2
According to reports from the World Health Organization (WHO), 250,000 TB patients were diagnosed in 2009,3 and of these, 30,000 (12%) were notified as MDR TB. The WHO estimated that approximately 650,000 of the 12 million prevalent TB cases in 2010 represented MDR TB.2 As for extensively drug resistant (XDR) TB, the first case was notified in South Africa in 2005, and by January 2010, 58 countries had reported at least one case of XDR TB.4
In Morocco, the incidence of TB has stagnated during recent years and was 81 per 100,000 in 2008 overall, but was significantly higher in several urban centers, or “hot spots”. Of the roughly 28,000 new TB cases nationally each year, 12% are retreatment cases (failure treatment, relapse or chronic cases).5 Among retreatment cases in Morocco, 12.2% are infected with MDR Mycobacterium tuberculosis (MTB) strains, whereas 0.4% of MDR TB are among new cases.
In Morocco, as in other resource-limited countries, WHO guidelines recommend the diagnosis of TB by smear microscopy in all new TB cases, and by smear microscopy, culture, and drug susceptibility testing in retreatment cases.6
Currently, drug susceptibility testing is mainly based on the proportion method, which is mainly performed on Lowenstein–Jensen medium.7 It can also be done using MGIT or BACTEC.8 Culture-based drug susceptibility testing is widely recognized as a reference method but requires several weeks and multiple methodologies to complete.7 Moreover, it is widely accepted that rapid drug susceptibility testing is critical in preventing emergence and expansion of drug resistance. Hence, there is an urgent need for new, rapid, and effective diagnostics to prevent the emergence and spread of MDR TB.
Drug resistance in MTB develops through spontaneous mutations in target genes, followed by selection of these resistant bacteria upon exposure to anti-TB drugs, which could lead to a sequential accumulation of mutations in specific genes for MTB and consequently MDR TB cases.9
Mutations in the rifampicin resistance determining region (RRDR) of the rpoB gene, encoding the β-subunit of DNA-dependent RNA polymerase, leads to resistance to rifampicin, which is the key first-line anti-TB drug.10 Similarly, mutations in the katG gene and inhA promoter are largely responsible for resistance to isoniazid.11
Isoniazid resistance mutations are primarily found in the katG gene, encoding the catalase-peroxidase enzyme responsible for activating isoniazid,12 the inhA gene, which encodes the molecular target inhA of the activated drug, and the promoter region of the mabA-inhA operon, resulting in overexpression of inhA.13 Mutations in the katG gene and inhA promoter account for 50%–95% and 15%–34%, respectively, of isoniazid-resistant clinical isolates.13
During the past few years, molecular methods have been developed to identify drug resistance causing gene mutations.14,15 One of latest techniques is the GeneXpert MTB/RIF (Cepheid, Sunnyvale, CA, USA), which can detect mutations in the rpoB gene only; due to close association of rifampicin resistance and MDR TB, this technique has been used to detect MDR TB cases.4 The technique has been thoroughly evaluated16 and is used in many countries.17 It has a sensitivity and specificity of 90.4% and 98.4%, respectively.18
Moreover, the WHO, as part of the “Stop TB 2006–2015 Strategy”,19 strongly recommends the integration of molecular approaches, especially GeneXpert MTB/RIF as the initial diagnostic test in individuals suspected of having MDR TB or TB associated with human immunodeficiency virus for countries most affected by TB.4,19
The polymerase chain reaction (PCR) sequencing-based strategy,19 designed to detect mutations associated with drug resistance rapidly, is able to provide a same-day diagnosis from culture and even clinical samples with high sensitivity and specificity. This gold standard method can also detect new mutations that could be associated with drug resistance. Automated sequencing has been used by several groups in the clinical setting for detection of the most frequent mutations and has been found to confer excellent benefit for patient care.20–22
This study assessed the usefulness of the PCR sequencing approach for rpoB and katG genes and the inhA promoter region for rapid detection of MDR TB strains directly from sputum to guide treatment regimens for MDR TB. Further, this study would be of great interest when evaluating the efficiency with which the GeneXpert can identify MDR TB in Morocco.
Materials and methods
Sampling
The study was performed after gaining approval from the ethics committee of the Pasteur Institute in Casablanca, Morocco, and confidentiality of clinical and laboratory information on patients was preserved. Samples were collected at the Pasteur Institutes in Casablanca and Tangier. Casablanca is the biggest city and the economic capital of Morocco, which accounts for almost one fifth of the total cases of TB recorded in the country, according to the national anti-TB program.19 The Pasteur Institute in Casablanca receives samples from different Moroccan cities. Tangier is a big city in the north of Morocco and ranks fourth in the number of TB cases. The Pasteur Institute in Tangier receives samples from different cities in the northern region of the country.
A total of 133 Moroccan patients with confirmed pulmonary TB were included in this study. Three consecutive sputum samples were collected from each patient as recommended by the WHO23 and inoculated on Lowenstein–Jensen medium after decontamination by N-acetyl-l-cysteine.24 Direct microscopic examination was performed by the Ziehl–Neelsen method. Part of each decontaminated sputum sample was used to extract MTB DNA for molecular analysis.
Drug susceptibility testing
Cultures obtained on Lowenstein–Jensen medium were collected and tested for drug susceptibility to rifampicin and isoniazid. Drug susceptibility testing was performed using the proportional method with Lowenstein–Jensen medium. The critical drug concentrations were 0.2 μg/mL for isoniazid and 40 μg/mL for rifampicin. The critical proportion of resistant bacillus necessary to define a resistant strain is 1% for the two tested drugs.25
Bacterial lysis
Specimens were first thawed and centrifuged at 6,000 g for one minute. For each specimen, the supernatant was discarded and the pellet was treated by heat shock treatment as described by Aldous et al.26 The DNA thermolysate was used immediately for PCR amplification or stored at −20°C until use.
PCR for amplification of rpoB and katG genes and inhA promoter region
The katG and rpoB genes and the inhA promoter region were amplified by PCR using the corresponding primers (Table 1). Amplification reactions were performed in a total volume of 50 μL. The amplification mixture contained 0.5 mM of each primer, 2.5 mM of each dNTP (dATP, dCTP, dGTP and dTTP), 25 mM MgCl2, 1 unit of Hotstar Taq DNA polymerase (Invitrogen, Saint Aubin, France), and 5 μL of a DNA sample in 1 × Taq polymerase buffer. For the three target genes, the mixtures were denatured at 94°C for 7 minutes. Thirty-five cycles of PCR were then performed, with denaturation at 94°C for one minute, primer annealing for one minute at the corresponding melting temperature, and primer extension for one minute at 72°C. At the end of the final cycle, the mixture was incubated at 72°C for 7 minutes. For every reaction, a negative control in which DNA template was omitted from the amplification mixture and a positive control containing DNA from H37Rv strain was included. Amplicons were visualized after electrophoretic fractionation in 1.5% agarose gel in 1 × Tris/borate/ethylenediamine tetraacetic acid buffer and staining with ethidium bromide.
Table 1.
Primers for polymerase chain reaction amplification
Sequencing reaction
Direct sequencing of amplicons was performed using a Big Dye Terminator kit version 3.1 (Applied Biosystems, Foster City, CA, USA) that includes dideoxynucleotides labeled with four fluorochromes of different colors. For each PCR product, both strands were sequenced in independent reactions using the above-mentioned primers. The resulting chromatograms were manually edited to ensure sequence accuracy and added to the alignment component of Molecular Evolutionary Genetics Analysis software (MEGA) version 5 (Tempe, AZ, USA).
Results
A total of 133 clinically symptomatic TB patients were enrolled in this study and sent for MTB and MDR TB investigation. Among them, 100 new cases had active and chemonaïve TB or had received treatment for less than a month. The rest (n = 33) were previously treated patients (drug relapse or failure, chronic cases) nonresponsive to anti-TB treatment after a sufficient duration of treatment. Male gender was more common than female gender, with 27.48% being women and 72.52% being men, giving a sex ratio of 2.64. The median patient age was 38 (range 15–80) years.
It was found that 13.5% of the patients were rifampicin-monoresistant (18/133), 7.5% were isoniazid-monoresistant (10/133), and 8.3% were resistant to rifampicin and isoniazid (11/133). Moreover, nine MDR strains were isolated from previously treated patients, whereas only two MDR strains came from new TB cases.
Genotypic drug susceptibility testing of the 133 samples showed that seven were isoniazid-resistant (5.3%), 17 were rifampicin-resistant (12.8%), and nine had MDR TB (6.8%). Comparison of phenotypic and genotypic resistance showed that all strains harboring mutations in rpoB and katG genes and the inhA promoter were phenotypically resistant to rifampicin and/or isoniazid. All the mutations identified by partial sequencing of rpoB and katG genes and the inhA promoter region are listed in Table 2.
Table 2.
Frequency of mutations identified by sequencing in the katG and rpoB genes of multidrug-resistant Mycobacterium tuberculosis isolates
| Gene | Position | Type of mutation | Amino acid change | Frequency | Total |
|---|---|---|---|---|---|
| rpoB | 513 | Substitution of CAA → CCA | Glutamine → Proline | 1 (3.8%) | 26 |
| 513 | Substitution of GAA → CTA | Glutamine → Leucine | 1 (3.8%) | ||
| 516 | Substitution of GAC → GTC | Aspartate → Valine | 2 (7.7%) | ||
| 516 | Substitution of GAC → TAC | Aspartate → Tyrosine | 2 (7.7%) | ||
| 516 | Substitution of GAC → CAC | Aspartate → Histidine | 1 (3.8%) | ||
| 518 | Deletion of AAC | 2 (7.7%) | |||
| 520 | Deletion of CCG | 1 (3.8%) | |||
| 526 | Substitution of CAC → CGC | Histidine → Arginine | 1 (3.8%) | ||
| 526 | Substitution of CAC → AGC | Histidine → Serine | 1 (3.8%) | ||
| 527 | Substitution of AAG → CAG | Lysine → Glutamine | 1 (3.8%) | ||
| 531 | Substitution of TCG → TGG | Serine → Tryptophan | 1 (3.8%) | ||
| 531 | Substitution of TCG → TTG | Serine → Leucine | 12 (46%) | ||
| katG | 315 | Substitution of AGC → ACC | Serine → Threonine | 15 (93.75%) | 16 |
| inhA promoter | −15 | Substitution of C → T | 1 (6.25%) |
Our results show that the most recorded mutation in the RRDR of the rpoB gene was substitution of TCG > TTG at codon 531 (Ser531 Leu), accounting for 46.15% (Table 2). Other point mutations and deletions were found in a limited number of cases: Asp516Val, Asp516Tyr, 518AAC Δ was found in two cases each and GLn513Pro, Gln513Leu, Asp516His, His526Arg, His526Ser, Lys527Gln, Ser531Trp, and 520CCG Δ in one case each. Interestingly, no strain harbored more than one amino acid change.
Significantly, the only mutation found in the katG gene was at codon 315 (AGC to ACC) with a Ser315Thr amino acid change. Only one sample harbored mutation in the inhA promoter region and was a point mutation at the −15p position (C > T).
The distribution of genotypically resistant strains according to patient TB profiling is reported in Table 3. Strains monoresistant to isoniazid were found in both new cases (5%) and previously treated patients (6%). However, rifampicin-monoresistant strains prevailed in new cases (15%). As expected, the majority of MDR strains were isolated from previously treated patients (24.2%), especially chronic and drug relapse cases, whereas only one new case was infected with a genotypic MDR strain.
Table 3.
Distribution of MDR according to the tuberculosis profile of patients
| TB profile | n | Monoresistant strains
|
MDR strains | |
|---|---|---|---|---|
| Isoniazid | Rifampicin | |||
| New cases | ||||
| Smear positive | 72 | 5 | 12 | 1 |
| Smear negative | 28 | 0 | 3 | 0 |
| Previously treated patients | ||||
| Chronic | 7 | 0 | 0 | 4 |
| Drug relapse | 18 | 1 | 2 | 4 |
| Drug failure | 8 | 1 | 0 | 0 |
| Total | 133 | 7 | 17 | 9 |
Abbreviation: MDR, multidrug resistance.
Discussion
Worldwide, the success of national programs against TB relies on rapid diagnosis and good management of drug-resistant TB cases. Indeed, emergence of drug-resistant isolates of MTB poses a serious threat to global TB control. In Morocco, and according to TB control program recommendations, drug susceptibility testing should be performed for previously treated cases (relapse, failure, and chronic) and patients coinfected with TB and human immunodeficiency virus, where MDR TB cases are highly likely and there is a strong likelihood of acquiring further resistance. Therefore, the decision regarding treatment is crucial.19,28 Moreover, to improve the performance of the TB control program in Morocco, close evaluation of monitoring of treatment outcomes for relapse, failure, and default cases has been strongly recommended.29
MDR is a consequence of inappropriate chemotherapy, erratic drug supply, misuse of TB drugs, poor TB management, and lack of control. For these reasons, research efforts are directed towards finding ways to speed up the process of susceptibility testing, with limited cost and complexity.
According to WHO recommendations and in order to implement GeneXpert for genotypic MDR analysis, this study was planned to undertake direct detection of MDR strains by a PCR sequencing-based approach. This standard method for genotyping drug susceptibility was limited to three specific genes, ie, rpoB, katG and inhA, which are the main ones responsible for resistance to the anti-TB drugs rifampicin and isoniazid, and are widely associated with MDR TB strains.9 Indeed, it is well documented that mutations in the rpoB gene, especially in RRDR, account for more than 95% of rifampicin resistance. Similarly, mutations in the katG gene at codon 315 exclusively confer high-level resistance to isoniazid in up to 75% of MDR TB strains.9–11
For both the katG and rpoB genes and the inhA promoter, the mutations or deletions found in this study have already been reported and are in concordance with previous published data.20,21 The most recorded mutation located in the RRDR of the rpoB gene is the Ser531 Leu (TCG → TTG) substitution, which has been widely reported.30,31 Interestingly, no double mutations were found in the samples analyzed. Our results clearly show that rifampicin monoresistance is relatively high. This resistance could be associated with other forms of isoniazid resistance, including other mutations in the katG gene or inhA promoter, or mutations in the ahpC gene, and other mechanisms of resistance. Moreover, this high frequency of rifampicin monoresistance could highlight that such resistance, as reported in previous studies, is not a rare event.32 On the other hand, only the Ser315Thr point mutation was found in the katG gene and is the most frequent mutation in this gene reported worldwide.11,31
It is widely accepted that genotypic drug susceptibility testing has a high sensitivity and specificity as compared with conventional culture-based drug susceptibility testing, but is still unable to detect all drug-resistant strains.11,33 Thus, the number of MDR strains detected by the PCR-based sequencing approach is underestimated. Of our 133 cases, nine were genotypically MDR, and the majority were isolated from previously treated patients. This is in agreement with reported data, and is of great interest for rapid detection of MDR strains, especially in critical cases, such as chronic and treatment failure or relapse patients, in which rapid diagnosis is mandatory for efficient chemotherapy.34
Moreover, eight MDR TB strains were isolated from 33 previously treated patients (21.1%). In 2004, 12.2% of previously treated patients were reported to be MDR.3 Despite the limited number of cases, there is evidence that MDR cases have increased in Morocco, thereby threatening the success of the TB control program.
In spite of its high cost and cumbersomeness, automated sequencing is an excellent and valid method for accurate and rapid detection of drug-resistant TB in clinical specimens and a valuable method for the management of suspected MDR TB cases.21 Significantly, one important advantage of sequence-based approaches is that the resulting data are virtually unambiguous, because the resistance-associated mutation is either present or absent.14
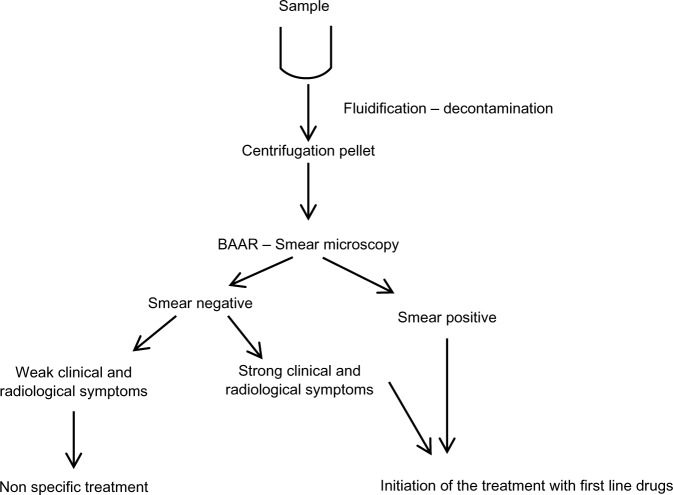
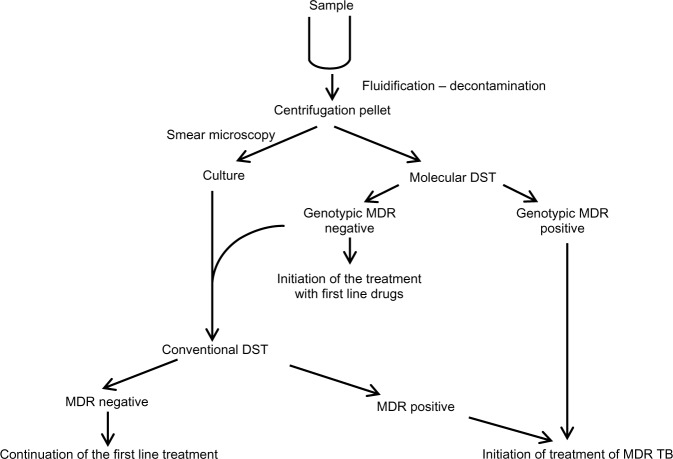
In Morocco, as in other countries with limited resources, introduction of the PCR-based sequencing approach to detect MDR cases should be of great benefit as a screening assay in the clinical setting for saving patients’ lives and preventing dissemination of TB and MDR TB in the community. Based on our results and those from several international studies,11,35–38 two algorithms associating conventional diagnosis, drug susceptibility testing, and genotypic drug susceptibility testing could be proposed (Figures 1 and 2) as an integrated model for reducing transmission of MDR strains in Morocco. Rapid detection of MDR strains can play a critical role in limiting the emergence of virtually untreatable XDR TB strains. In fact, the complications of drug resistance in MDR TB can lead to XDR TB.
Figure 1.
Management of suspected and new cases.
Abbreviation: BAAR, bacillus acido-alcohol resistant.
Figure 2.
Place of genotypic DST in the global management of TB for specific cases: previously treated patients (chronic, relapse, failure, default), co-infection with HIV, patients with close contact to MDR TB cases.
Abbreviations: MDR, multidrug resistance; DST, drug susceptibility testing; TB, tuberculosis; HIV, Human immunodeficiency virus.
In conclusion, implementation of molecular approaches for direct diagnosis of MDR TB, as a part of routine analysis in the laboratories of health care institutions, will be of great benefit in adapting treatment regimens, limiting dissemination of MDR TB strains, and limiting the emergence of XDR MTB strains for better management of TB in Morocco.
Acknowledgments
The authors would like to thank TC Victor and H Mardassi for their critical review of the manuscript and relevant comments. This study was funded in part by the International Atomic Energy Agency under the RAF6040 project and Regional Office for the Eastern Mediterranean/World Health organization under the RPC/RAB&GH 10/11-03 project.
Footnotes
Disclosure
The authors have no conflicts of interest in this work.
References
- 1.Rodrigues LC, Smith PG. Tuberculosis in developing countries and methods for its control. Trans R Soc Trop Med Hyg. 1990;84(5):739–744. doi: 10.1016/0035-9203(90)90172-b. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization; Global Tuberculosis Control 2011 Geneva, Switzerland: World Health Organization; 2011Available from: http://whqlibdoc.who.int/publications/2011/9789241564380_eng.pdfAccessed August 23, 2013 [Google Scholar]
- 3.World Health Organization Global tuberculosis control: epidemiology, strategy, financing WHO/HTM/TB/ Geneva, Switzerland: World Health Organization; 2011Available from: http://whqlibdoc.who.int/publications/2011/9789241564380_eng.pdfAccessed August 23, 2013 [Google Scholar]
- 4.World Health Organization Rapid implementation of the Xpert MTB/RIF diagnostic test (technical and operational ‘how-to’ practical considerations): WHO Library Cataloguing-in-Publication Data Geneva, Switzerland: World Health Organization; 2011Available from: http://whqlibdoc.who.int/publications/2011/9789241501569_eng.pdfAccessed August 23, 2013 [Google Scholar]
- 5.World Health Organization Anti-tuberculosis drug resistance in the world Geneva, Switzerland: World Health Organization; 2008Available from: http://www.who.int/tb/publications/mdr_surveillance/en/Accessed August 23, 2013 [Google Scholar]
- 6.World Health Organization Interim recommendations for the surveillance of drug resistance in tuberculosis Geneva, Switzerland: World Health Organization; 2007Available from: http://whqlibdoc.who.int/hq/2007/WHO_HTM_TB_2007.385_eng.pdfAccessed August 23, 2013 [Google Scholar]
- 7.Reller LB, Weinstein MP, Woods GL. Susceptibility testing for mycobacteria. Clin Infect Dis. 2000;31(5):1209–1215. doi: 10.1086/317441. [DOI] [PubMed] [Google Scholar]
- 8.Huang T-S, Lee SS-J, Tu H-Z, et al. Use of MGIT 960 for rapid quantitative measurement of the susceptibility of Mycobacterium tuberculosis complex to ciprofloxacin and ethionamide. J Antimicrob Chemother. 2004;53(4):600–603. doi: 10.1093/jac/dkh120. [DOI] [PubMed] [Google Scholar]
- 9.Van Rie A, Warren R, Mshanga I, et al. Analysis for a limited number of gene codons can predict drug resistance of Mycobacterium tuberculosis in a high-incidence community. J Clin Microbiol. 2001;39(2):636–641. doi: 10.1128/JCM.39.2.636-641.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramaswamy S, Musser J. Molecular genetic basis of antimicrobial agent resistance in Mycobacterium tuberculosis: 1998 update. Tuber Lung Dis. 1998;79(1):3–29. doi: 10.1054/tuld.1998.0002. [DOI] [PubMed] [Google Scholar]
- 11.Müller B, Streicher EM, Hoek KGP, et al. inhA promoter mutations: a gateway to extensively drug-resistant tuberculosis in South Africa? Int J Tuberc Lung Dis. 2011;15(3):344–351. [PubMed] [Google Scholar]
- 12.Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358(6387):591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 13.Mayers D, Karakousis P. Mechanisms of action and resistance of antimycobacterial agents. In: Mayers DL, editor. Antimicrobial Drug Resistance: New York. NY: Humana Press; 2009. [Google Scholar]
- 14.Neonakis IK, Gitti Z, Krambovitis E, Spandidos DA. Molecular diagnostic tools in mycobacteriology. J Microbiol Methods. 2008;75(1):1–11. doi: 10.1016/j.mimet.2008.05.023. [DOI] [PubMed] [Google Scholar]
- 15.García de Viedma D. Rapid detection of resistance in Mycobacterium tuberculosis: a review discussing molecular approaches. Clin Microbiol Infect. 2003;9(5):349–359. doi: 10.1046/j.1469-0691.2003.00695.x. [DOI] [PubMed] [Google Scholar]
- 16.Marlowe EM, Novak-Weekley SM, Cumpio J, et al. Evaluation of the Cepheid Xpert MTB/RIF assay for direct detection of Mycobacterium tuberculosis complex in respiratory specimens. J Clin Microbiol. 2011;49(4):1621–1623. doi: 10.1128/JCM.02214-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barnard M, Warren R, Van Pittius NG, et al. GenoType MTBDRsl line probe assay shortens time to diagnosis of XDR-TB in a high-throughput diagnostic laboratory. Am J Respir Crit Care Med. 2012;186(12):1298–1305. doi: 10.1164/rccm.201205-0960OC. [DOI] [PubMed] [Google Scholar]
- 18.Chang K, Lu W, Wang J, et al. Rapid and effective diagnosis of tuberculosis and rifampicin resistance with Xpert MTB/RIF assay: a meta-analysis. J Infect. 2012;64(6):580–588. doi: 10.1016/j.jinf.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 19.PNLAT [Tuberculosis Guide for use by health care professionals] Guide de la lutte antituberculeuse à l’usage des professionnel de la santé. 3rd ed. Rabat, Morocco: SMR; 2011. Direction de l’épidémiologie et de lutte contre les maladies. DELM. [Google Scholar]
- 20.Kourout M, Chaoui I, Sabouni R, et al. Molecular characterisation of rifampicin-resistant Mycobacterium tuberculosis strains from Morocco. Int J Tuberc Lung Dis. 2009;13(11):1440–1442. [PubMed] [Google Scholar]
- 21.Abdelaal A, El-Ghaffar H, Zaghloul M, El mashad N, Badran E, Fathy A. Genotypic detection of rifampicin and isoniazid resistant Mycobacterium tuberculosis strains by DNA sequencing: a randomized trial. Ann Clin Microbiol Antimicrob. 2009;8(1):4. doi: 10.1186/1476-0711-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Choi J-H, Lee KW, Kang H-R, et al. Clinical efficacy of direct DNA sequencing analysis on sputum specimens for early detection of drug-resistant mycobacterium tuberculosis in a clinical setting. Chest. 2010;137(2):393–400. doi: 10.1378/chest.09-0150. [DOI] [PubMed] [Google Scholar]
- 23.World Health Organization Guidelines for speciation within the Mycobacterium tuberculosis complex. Emerging and other communicable diseases, surveillance and control Geneva, Switzerland: World Health Organization; 1996Available from: http://whqlibdoc.who.int/hq/1996/who_emc_zoo_96.4.pdfAccessed August 23, 2013 [Google Scholar]
- 24.Ratnam S, Stead FA, Howes M. Simplified acetylcysteine-alkali digestion-decontamination procedure for isolation of mycobacteria from clinical specimens. J Clin Microbiol. 1987;25(8):1428–1432. doi: 10.1128/jcm.25.8.1428-1432.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.David H, Levy Frebault V, Thorel M. Laboratory Methods for Clinical Mycobacteriology. Paris, France: Institut Pasteur Paris; 1987. [Google Scholar]
- 26.Aldous W, Pounder J, Cloud JL, Woods G. Comparison of six methods of extracting M. tuberculosis DNA from processed sputum for testing by quantitative real-time PCR. J Clin Microbiol. 2005;43:2471–2473. doi: 10.1128/JCM.43.5.2471-2473.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cingolani A, Antinori A, Sanguinetti M, et al. Application of molecular methods for detection and transmission analysis of Mycobacterium tuberculosis drug resistance in patients attending a reference hospital in Italy. J Infect Dis. 1999;179(4):1025–1029. doi: 10.1086/314677. [DOI] [PubMed] [Google Scholar]
- 28.Quy HTW, Lan NTN, Borgdorff MW, et al. Drug resistance among failure and relapse cases of tuberculosis: is the standard re-treatment regimen adequate? Int J Tuberc Lung Dis. 2003;7(7):631–636. [PubMed] [Google Scholar]
- 29.Ottmani SE, Zignol M, Bencheikh N, et al. Results of cohort analysis by category of tuberculosis retreatment cases in Morocco from 1996 to 2003. Int J Tuberc Lung Dis. 2006;10(12):1367–1372. [PubMed] [Google Scholar]
- 30.Telenti A, Imboden P, Marchesi F, et al. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341(8846):647–651. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 31.Aziz MA, Wright A, Laszlo A, et al. Epidemiology of antituberculosis drug resistance (the Global Project on Anti-tuberculosis Drug Resistance Surveillance): an updated analysis. Lancet. 2006;368(9553):2142–2154. doi: 10.1016/S0140-6736(06)69863-2. [DOI] [PubMed] [Google Scholar]
- 32.Mukinda FK, Theron D, van der Spuy GD, et al. Rise in rifampicin-monoresistant tuberculosis in Western Cape, South Africa. Int J Tuberc Lung Dis. 2012;16(2):196–202. doi: 10.5588/ijtld.11.0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barnard M, Albert H, Coetzee G, O’Brien R, Bosman ME. Rapid molecular screening for multidrug-resistant tuberculosis in a high-volume public health laboratory in South Africa. Am J Respir Crit Care Med. 2008;177(7):787–792. doi: 10.1164/rccm.200709-1436OC. [DOI] [PubMed] [Google Scholar]
- 34.Miotto P, Saleri N, Dembele M, et al. Molecular detection of rifampin and isoniazid resistance to guide chronic TB patient management in Burkina Faso. BMC Infect Dis. 2009;9(1):142. doi: 10.1186/1471-2334-9-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balabanova Y, Drobniewski F, Nikolayevskyy V, et al. An integrated approach to rapid diagnosis of tuberculosis and multidrug resistance using liquid culture and molecular methods in Russia. PLoS One. 2009;4(9):e7129. doi: 10.1371/journal.pone.0007129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tukvadze N, Kempker RR, Kalandadze I, et al. Use of a molecular diagnostic test in AFB smear positive tuberculosis suspects greatly reduces time to detection of multidrug resistant tuberculosis. PLoS One. 2012;7(2):e31563. doi: 10.1371/journal.pone.0031563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson ML. Recent advances in the laboratory detection of Mycobacterium tuberculosis complex and drug resistance. Clin Infect Dis. 2011;52(11):1350–1355. doi: 10.1093/cid/cir146. [DOI] [PubMed] [Google Scholar]
- 38.Rachow A, Zumla A, Heinrich N, et al. Rapid and accurate detection of Mycobacterium tuberculosis in sputum samples by Cepheid Xpert MTB/RIF assay. A clinical validation study. PLoS One. 2011;6(6):e20458. doi: 10.1371/journal.pone.0020458. [DOI] [PMC free article] [PubMed] [Google Scholar]