Abstract
The aryl hydrocarbon receptor (AhR) is a ligand-dependent transcription factor that mediates the biological and toxicological effects of structurally diverse chemicals through its ability to bind specific DNA recognition sites (dioxin responsive elements (DREs)), and activate transcription of adjacent genes. While the DRE has a highly conserved consensus sequence, it has been suggested that the nucleotide specificity of AhR DNA binding may be ligand-dependent. The upstream regulatory regions of the murine Bax and human paraoxonase 1 (PON1) genes reportedly contain unique DRE-like sequences that respond to AhRs activated by some ligands but not others. Given the significant implications of this observation to understanding the diversity in AhR responses and that of other ligand-dependent nuclear receptors, a combination of DNA binding, nuclear translocation and gene expression analysis was used to investigate the molecular mechanisms underlying these ligand-selective responses. Although known AhR agonists stimulated AhR nuclear translocation, DRE binding and gene expression, the ligand-selective DRE-like DNA elements identified in the Bax and PON1 upstream regulatory regions failed to bind ligand-activated AhR or confer AhR-responsiveness upon a reporter gene. These results argue against the reported ligand-selectivity of AhR DNA binding and suggest DNA binding by ligand activated AhR involves DRE-containing DNA.
Keywords: Aryl hydrocarbon receptor, Bax, Paraoxonase 1, DNA Binding
INTRODUCTION
The AhR is a unique ligand-dependent transcription factor that can be bound and activated by structurally diverse chemicals to produce a dramatic pleiotropic spectrum of species- and tissue-specific toxic and biological effects [1-3]. These effects appear to result primarily from AhR-dependent alterations in gene expression, with metabolically stable ligands like 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD1, dioxin), producing a greater spectrum of toxic effects as a result of persistent alterations in gene expression and biological effects than AhR ligands like benzo(a)pyrene and 3-methylcholanthrene (3MC), that are metabolically labile and only transient AhR activators [4]. Mechanistically, AhR ligands diffuse into the cell wherein they bind to the cytosolic AhR protein complex to stimulate its nuclear translocation [5]. Once in the nucleus, the liganded AhR is released from its associated proteins by its specific interaction with the Ah receptor nuclear translocator (ARNT) protein and the resulting ligand:AhR:ARNT heterodimer gains the ability to bind to DNA with high affinity [6]. Binding of the ligand:AhR:ARNT complex to its specific DNA recognition sites (dioxin responsive elements (DREs)) leads to stimulation of transcription of adjacent genes and resulting biological effects [1, 7].
In addition to the classical AhR:ARNT:DRE-dependent mechanism of action, the AhR can produce biological effects through a multitude of other distinct mechanisms including acting as a nuclear coactivator or coregulator/dimerization partner for other transcription factors, altering intracellular calcium signaling and release, cross-talk with other cell signaling pathways, nuclear squelching effects and others [reviewed in 1, 8]. The structural diversity and differential binding of AhR ligands suggests the existence of ligands whose binding can selectively alter the structure and function of the AhR, leading to differential interactions with coactivator/coregulator proteins and alterations in the overall biological response. In fact, several selective AhR modulators (SAhRMs) have already been identified based on their ability to selectively produce some AhR-dependent responses and not others [9, 10]. Additionally, agonist-dependent changes in AhR protein structure have been observed [6] as has ligand-specific differences in AhR coactivator recruitment [11], suggestive of some ligand-dependent differences in AhR structure/function that can contribute to the diversity in AhR response. Two studies have proposed a novel mechanism, whereby differential ligand binding leads to alterations in AhR structure which changes the nucleotide specificity of AhR:ARNT DNA binding [12, 13] and this unique mechanism could contribute in very dramatic and unexpected ways to the functional diversity observed for of AhR ligands. Matikainen and co-workers [12] reported that the upstream regulatory region of the murine Bax gene contained two novel degenerate DRE-like sequences that were able to confer AhR-dependent responsiveness to a PAH metabolite (7,12-dimethylbenz[a]anthracene-3,4-dihydrodiol (DMBA-DHD)), but not to TCDD. They reported that insertion of a single nucleotide mutation 3’-ward of the invariant DRE core sequence (5’-GCGTG-3’) that restored the overall DRE consensus sequence also restored TCDD responsiveness. Gouédard and coworkers [13] identified a DRE-like sequence (4/5 consensus match with the invariant DRE core) present in the upstream region of the human paraoxonase 1 (PON1) gene that they reported was sufficient to allow the AhR ligands 3MC and quercetin, but not TCDD, to induce AhR-dependent gene expression.
Given the significance and major impact that ligand-selective changes in AhR DNA binding specificity can have in improving our understanding of the diversity and mechanisms of AhR toxic and biological effects, coupled with the lack of any subsequent studies actually confirming these novel ligand-selective effects, further studies are needed. Accordingly, here we describe studies examining the molecular mechanisms by which DMBA-DHD, 3MC, and quercetin are reported to selectively modulate the nucleotide specificity of AhR DNA binding to differentially activate gene expression.
MATERIALS AND METHODS
Materials
TCDD was provided by Dr. Stephen Safe (Texas A&M University). DMBA-DHD was obtained from the National Cancer Institute Chemical Carcinogen Reference Standards Repository and DMSO, quercetin, and 3MC were from Sigma-Aldrich (St. Louis, MO).
Oligonucleotides
Complementary DNA oligonucleotides containing a single copy of the murine DRE3 sequence (5’-CTAGCGATCAAGAGGCTCTTCTCACGCAACTCCGCCTGAGCTGA-3’), the Bax DRE-like sequence (5’-GATCCAGAAAGCCGGGCGTGGTGGCGCACGCCTTTAATCCCA-3’), mutated Bax DRE-like sequence (5’-GATCCAGAAAGCCGGGCGTGGTATCGCACGCCTTTAATCCCA-3’), or human PON1 DNA sequence (5’-GATCCGGAGGCTGCGGACCCGGCGGGGAGGGGTA-3’) were synthesized by Operon Biotechnologies (Huntsville, AL) with either BamHI and BglII or NheI and BglII compatible ends. The oligonucleotides were annealed and either radiolabeled using T4 polynucleotide kinase (New England Biolabs, Ipswich, MA) and [32-P]ATP (Perkin Elmer, Waltham, MA) for use as probes in electrophoretic mobility shift assays (EMSA) or phosphorylated and subcloned into a luciferase expression plasmid for transient transfections as described below.
Electrophoretic mobility shift assay (EMSA)
EMSA using guinea pig hepatic cytosol was performed as previously described [14] with minor modifications. Briefly, cytosol (8 mg protein/ml) was incubated with the indicated test compounds for 2 hours at 20°C followed by incubation with [32P]-labeled probes (~100,000 cpm, ~12 fmol/binding reaction). Samples were then subjected to non-denaturing polyacrylamide gel electrophoresis (PAGE) and protein/DNA complexes were visualized by autoradiography using a Molecular Dynamics PhosphorImager with ImageQuant software analysis or a FujiFilm FLA-9000 imager with Multi-Gauge software. In vitro expression of murine AhR and ARNT proteins and subsequent EMSA analysis was performed as described by Rushing and Denison [15] except that 5 μl aliquots of lysates containing AhR and ARNT were combined with 14.5 ul of HEDG buffer and 0.5 μl of test compounds in DMSO and allowed to incubate at 20°C for 3 hours. Ten microliters of this reaction was then combined with 15 μl of oligo buffer and allowed to incubate for 15 minutes, followed by the addition of the [32P]-labeled probes (as described above) and an additional 15 minute incubation. Loading buffer (4 ul) was added to each sample, and a 10 μl aliquot was loaded on a 4% non-denaturing polyacrylamide gel and protein/DNA complexes visualized as described above. EMSA analysis using nuclear proteins from hepa1c1c7 cells were performed as described by Denison et al. [16], except that poly(dI•dC) was reduced to 500 ng and the final DNA binding conditions were 25 mM Hepes, pH 7.5, 1 mM EDTA, 1 mM dithiothreitol, 10% (v/v) glycerol, 120 mM KCl with 3 μg of total protein. Preparation of nuclear proteins from HuH7 cells were as described by Denison et al. [16], except that 3 mM MgCl was added to both the initial HEPES wash buffer and the final extraction buffer. Final DNA binding conditions were modified to contain 250 ng poly(dI•dC) and 80 mM KCl with 3 μg of total protein.
Plasmids
The AhR and ARNT expression plasmids m AhR/pcDNA3 and mARNT/pcDNA3.1 have been previously described [15, 17]. To prepare the inducible luciferase expression vectors, complementary DNA oligonucleotides containing a single copy of the DRE3 sequence or Bax, mutant Bax, or PON1 DRE-like response elements (Figure 1A) were subcloned into the BglII site of the plasmid pGudLuc7.0 immediately upstream of the mouse mammary tumor virus (MMTV) promoter and firefly luciferase gene [18]. PCR analysis and DNA sequencing were used to confirm insertion of a single copy of the desired response element in the same orientation within each plasmid. The plasmids pPON1000-FL, p(XRE-PON)3-FL and p(mutXRE-PON)3-FL were kind gifts from Dr. Robert Barouki (Université René Descartes, Paris, France) [13].
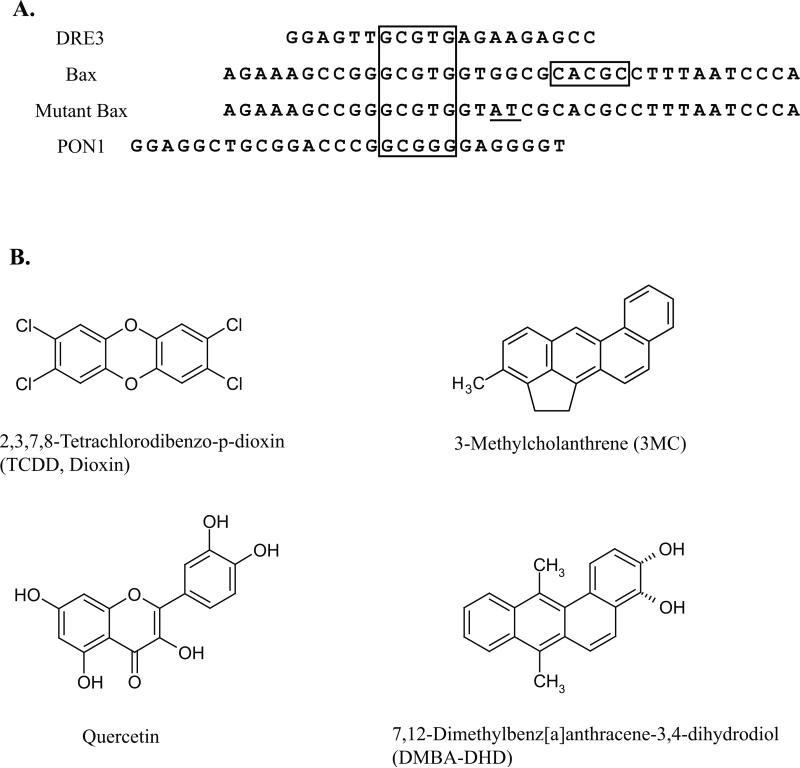
Figure 1.
DRE-like DNA elements (A) and chemicals (B) previously reported to induce AhR:ARNT-dependent reporter gene expression in a ligand-selective manner. Alignment of DRE-like sequences from the promoter regions of the murine Bax and human paraoxonase 1 (PON1) genes are shown along with the wild type DRE3. The DRE core nucleotides are boxed, and underlined bases represent mutations in the wild type Bax DRE-like response element that reportedly restore TCDD-responsiveness [12].
Cell culture, transfection, and luciferase measurement
Hepa1c1c7 and HuH7 cells were grown in alpha Minimum Essential Medium (MEM) (Gibco, Carlsbad, CA) or Dulbecco's Modified Eagle Medium (D-MEM) with high glucose (Gibco), respectively. Culture medium was supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals, Lawrenceville, GA) and the cells maintained at 37°C in an atmosphere of 5% CO2 and 85% humidity. For transient transfections, cells were plated at 100,000 cells per well in a 24-well plate and allowed to attach overnight, followed by transient transfection with 800 ng of the desired plasmid and 8 ng of pRL-TK (Promega, Madison, WI) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA) per the manufacturer's instructions. At 20-24 hours post-transfection, the cells were treated with the test compounds (final solvent concentration 0.1% in medium) and incubated for 4 hours. The medium was removed, the cells washed twice with phosphate-buffered saline and lysed with 100 μL of Promega passive lysis buffer. Luciferase activity in 10 μL of cell lysate was measured in an Orion microplate luminometer with the automatic injection of 50 μl of Luciferase Assay Reagent II (Dual-Luciferase Reporter Assay System, Promega), with a 2s delay and 10s firefly luciferase read integration parameters, followed by the addition of 50 μl of Stop and Glo reagent, a 2s delay, and 10s Renilla luciferase read integration. Firefly luciferase activity was then expressed relative to Renilla luciferase activity to normalize for transfection efficiency.
Nuclear translocation analysis
Ligand-dependent AhR nuclear translocation analysis was performed using recombinant mouse yAHAYc6 cells which contain a stably expressed recombinant chimeric AhR fused to yellow fluorescent protein fusion cells as previously described [19].
RESULTS
Examination of in vitro ligand-selective AhR:ARNT binding to DNA containing the Bax or PON1 DRE-like elements
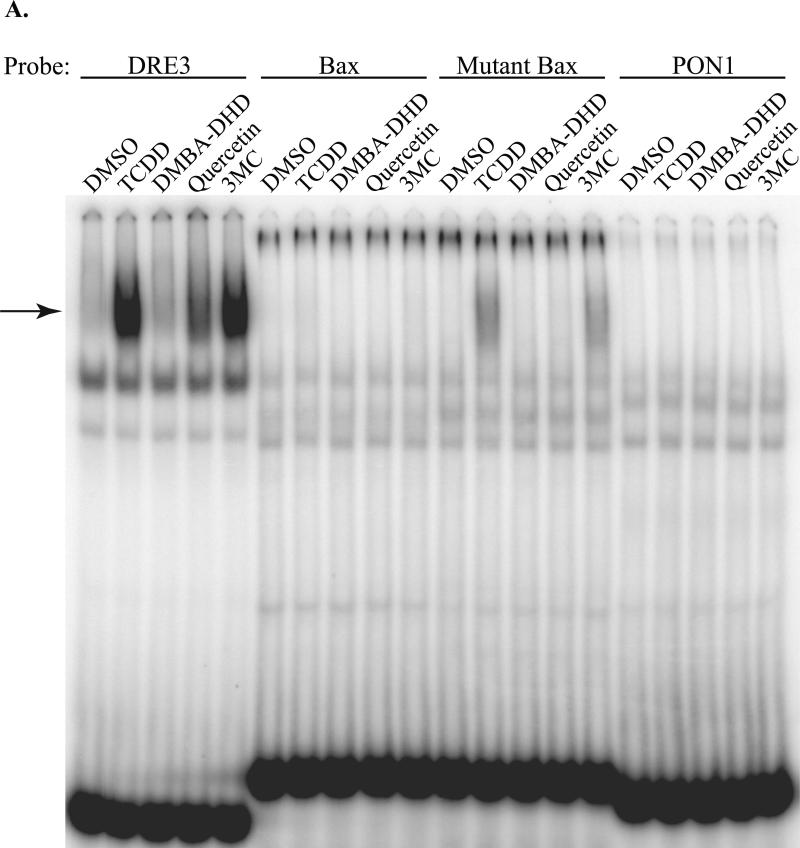
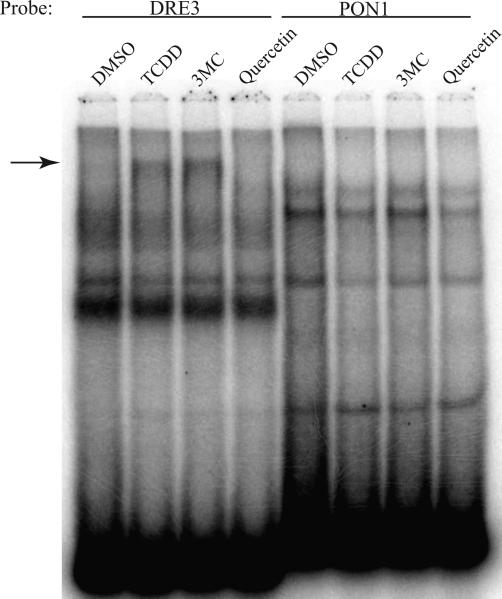
AhR-dependent expression of the murine Bax and human PON1 genes has been reported to occur in a ligand-selective manner mediated by novel DRE-like sequences (Figure 1A) present in their upstream regulatory regions [12, 13]. In order to confirm ligand-selective AhR:ARNT DNA binding to the Bax and PON1 DRE-like response elements, EMSA analysis was carried out using guinea pig hepatic cytosol as the source of AhR and ARNT. Guinea pig cytosolic AhR efficiently transforms into its high affinity DNA binding form in a ligand-dependent manner, producing a relatively large amount of inducible ligand:AhR:ARNT:DRE complex making it a good model system to examine AhR DNA binding [20, 21]. In initial experiments, we examined the ability of DMBA-DHD, 3MC, quercetin and TCDD (Figure 1B) to stimulate AhR binding to a DNA oligonucleotide containing a wild-type DRE (DRE3), the PON1 or Bax DRE-like sequence or the Bax DRE-like DNA element containing a mutation that nearly restores the full DRE consensus sequence (mutant Bax) [12]. As expected, incubation with the prototypical AhR ligands TCDD and 3MC stimulated AhR:ARNT:DRE3 complex formation (Figure 2A). Additionally, a small amount of AhR:ARNT:DRE3 complex was observed with cytosol incubated with the polyphenolic compound quercetin, similar to a previous study identifying it as an AhR agonist [22]; no ligand-induced AhR:ARNT:DRE3 complex was produced by DMBADHD. In contrast to the results obtained with the DRE3-containing oligonucleotide, no chemical-induced AhR:ARNT:DNA complex was observed with oligonucleotides containing the Bax and PON1 DRE-like sequences. In contrast, a small amount of TCDD- and 3MC-inducible protein-DNA complex formation was observed with an oligonucleotide containing the mutated Bax DRE-like sequence. While the substitutions inserted into the Bax DRE-like sequence appear to restore key nucleotides of the DRE consensus, other nucleotides in the mutant Bax sequence must negatively impact the binding of ligand:AhR:ARNT complexes. Given that significant species-specific differences in AhR:ARNT activation have been reported for a variety of ligands [reviewed in 8] we repeated our EMSA analysis using C57BL/6 mouse AhR and ARNT. Each protein was expressed in vitro, the proteins combined and incubated with the test ligands followed by the addition of [32P]-labeled oligonucleotide probes and detection of the protein-DNA complexes. The results (Figure 2B) reveal an identical pattern of chemical-induced protein-DNA complex formation with the different oligonucleotide probes as that observed with guinea pig AhR (Figure 2A). These results demonstrate an inability of DMBADHD, quercetin, 3MC or TCDD to stimulate binding of guinea pig or mouse AhR to the previously identified AhR-responsive Bax or PON1 DRE-like DNA elements.
Figure 2.
AhR:ARNT complexes do not bind to Bax or PON1 DRE-like DNA elements in vitro. Guinea pig hepatic cytosol (A) or in vitro synthesized mouse AhR and ARNT (B) were incubated with DMSO (2% (v/v)), TCDD (20 nM), DMBA-DHD (1μM), quercetin (50 μM) or 3MC (5 μM) for 2-3 hours at 20°C followed by incubation with [32P]-labeled oligonucleotide probes. Protein-DNA complexes were resolved by EMSA and visualized by PhosphorImager and ImageQuant software analysis. The arrow indicates the position of the inducible AhR:ARNT:DNA complex.
Nuclear ligand:AhR:ARNT complexes fail to bind to oligonucleotides containing Bax and PON1 DRE-like DNA sequences
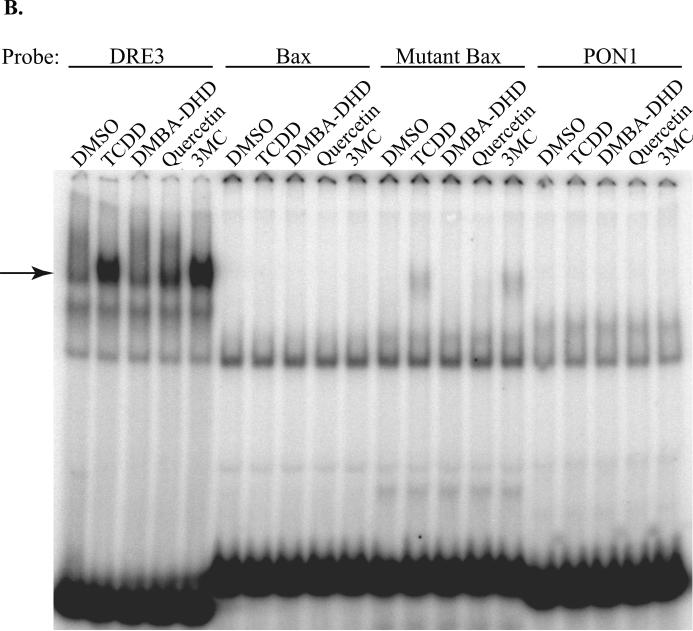
One possible explanation for the lack of inducible protein-DNA complexes with the Bax and PON1 DRE-like elements is that the DNA binding of DMBA-DHD-, quercetin- or 3MC-activated AhR:ARNT complexes requires an additional nuclear protein(s). To examine this possibility, mouse hepatoma (hepa1c1c7) cells were incubated with DMSO, TCDD or DMBADHD for 1 hour and EMSA analysis carried out with nuclear extracts and oligonucleotides containing DRE3, Bax or mutant Bax DNA sequences. Hepa1c1c7 cells were selected for these studies since these cells were derived from C57BL/6 mice, the same mouse strain utilized in the original Bax studies by Matikainen and coworkers [12]. While EMSA analysis revealed that nuclear extracts from TCDD treated cells resulted in an inducible protein-DRE3 complex, no comparable protein-DNA complex was observed with DMBA-DHD (Figure 3). Although no TCDD- or DMBA-DHD-induced protein-DNA complex was observed with the Bax or mutant Bax DNA oligonucleotides, a constitutive protein-DNA complex that migrated to the same position as the TCDD-induced protein-DNA complex was observed bound to the Bax DRE-like response elements (Figure 3). However, since these protein:Bax/mutant Bax DNA complexes were present using nuclear extracts from DMSO treated cells (and thus should contain no AhR) and they could not be competitively eliminated by unlabeled DRE-containing DNA (Supplemental Figure 1), these protein-DNA complexes do not represent DNA-bound AhR:ARNT complexes. Importantly, despite the use of nuclear protein extracts, we were unable to demonstrate the ability of DMBA-DHD to stimulate protein binding to the Bax DRE-like response element.
Figure 3.
Nuclear extracts from TCDD or DMBA-DHD treated mouse hepatoma cells do not exhibit ligand-inducible protein-DNA complexes with the Bax or mutant Bax DRE-like DNA element. Nuclear extracts were prepared from mouse hepatoma (hepa1c1c7) cells incubated with DMSO (0.1% (v/v)), TCDD (1 nM) or DMBA-DHD (1 μM) for 1 hour as described in Materials and Methods. Extracts (3 μg of protein) were incubated with the indicated [32P]-labeled DNA oligonucleotide probes and subjected to EMSA analysis to resolve the protein:DNA complexes. The arrow indicates the position of the induced AhR:ARNT:DNA complex.
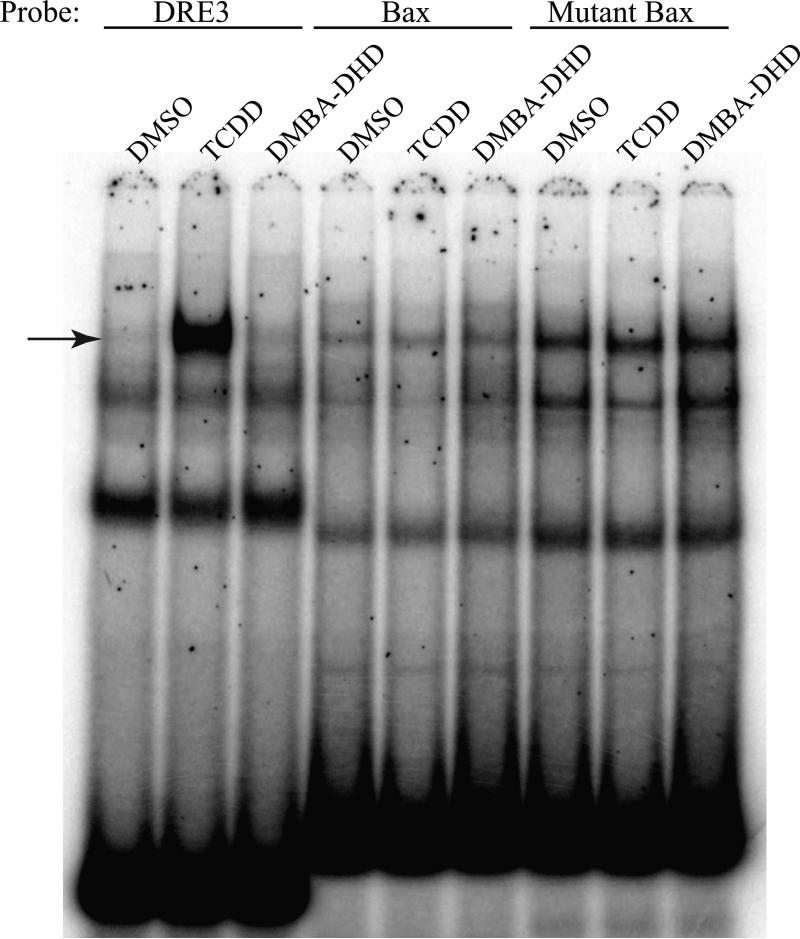
The study by Gouédard et al. [13] previously reported that a quercetin- and 3MC-dependent interaction of the AhR with the PON1 DRE-like DNA element could be observed using nuclear extracts from a human hepatoma (HuH7) cell line. Accordingly, to examine whether the lack of AhR binding to the PON1 DNA element we observed was species specific (i.e. occurring with human, but not mouse or guinea pig AhR) and/or required an additional human nuclear factor, we examined the ability of nuclear proteins from TCDD-, quercetin- and 3MC-treated HuH7 cells to bind an oligonucleotide containing the PON1 DRE-like DNA element using EMSA. While nuclear extracts from TCDD- and 3MC-treated HuH7 cells resulted in formation of inducible protein:DRE3 DNA complex formation at the expected relative migration position in the PAGE gel, albeit at relatively low levels (Figure 4), no quercetin-inducible protein-DRE3 DNA complex was observed. In contrast to the results of Gouédard et al. [13], no TCDD-, 3MC-or quercetin-inducible protein:PON1 DNA complex was detected with HuH7 extracts.
Figure 4.
Nuclear extracts from quercetin and 3MC exposed human HuH7 cells do not exhibit ligand-inducible protein-DNA complexes with the PON1 DRE-like element. Nuclear extracts were prepared from human hepatoma (HuH7) cells incubated with DMSO (0.1%), TCDD (10 nM), 3MC (5 μM) or quercetin (50 μM) for 1 hour. The extracts (3 μg of protein) were then incubated with [32P]-labeled DRE3 or PON1 oligonucleotide probes and subjected to EMSA to resolve the protein:DNA complexes. The arrow indicates the position of the induced protein:DNA complex.
DMBA-DHD is not an AhR agonist in intact cells and it does not stimulate AhR-dependent gene expression by the Bax DRE-like responsive element
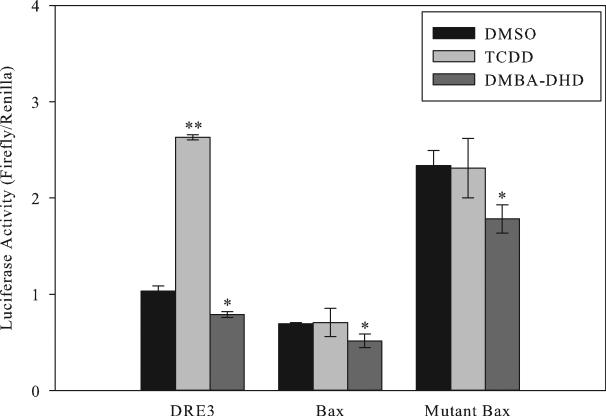
Despite our inability to demonstrate the ligand-dependent presence of the AhR:ARNT complex to DNA containing the Bax or PON1 DRE-like elements, the AhR and/or the AhR:ARNT complex could still be functioning as a transcriptional activator/cofactor to directly/indirectly stimulate gene expression through these DNA elements. To test this possibility, we first examined the functional activity of the Bax DRE-like DNA element. Mouse hepatoma cells were transiently transfected with a plasmid containing a single DRE3, Bax, or mutant Bax DRE-like element inserted immediately upstream of the MMTV promoter and firefly luciferase reporter gene, and the cells incubated for 4 hours with TCDD or DMBA-DHD after which luciferase activity was determined. While TCDD-stimulated luciferase gene expression from the reporter plasmid containing the DRE3 element, it failed to stimulate luciferase activity from plasmids containing the Bax or mutant Bax DNA elements (Figure 5). While DMBA-DHD did not increase luciferase activity above that of the DMSO control from plasmids containing the DRE3 or Bax or mutant Bax DRE-like element, it did produce a small, but significant, decrease in luciferase activity that may have resulted from the established toxicity of this chemical [23]. Interestingly, the presence of the mutant Bax DRE-like DNA sequence resulted in elevated basal luciferase activity that was greater than that of the wild-type Bax DRE-like sequence and may result from an interaction of a basal transcription factor(s) with the mutant Bax sequence. Taken together, our results demonstrate that the wild type Bax DNA element not only fails to bind ligand-activated AhR, but it does not mediate AhR-dependent gene expression by DMBA-DHD or other tested AhR agonists.
Figure 5.
TCDD and DMBA-DHD fail to stimulate luciferase reporter gene expression from the Bax or mutant Bax DRE-like response elements. Mouse hepatoma (hepa1c1c7) cells were transiently transfected with pGudLuc7.0 containing a single DRE3, Bax or mutant Bax DNA oligonucleotide present immediately upstream of the MMTV promoter and luciferase reporter gene. Cells were incubated with DMSO (0.1%), TCDD (1 nM), or DMBA-DHD (1 μM) for 4 hours after which luciferase activity was determined. Values represent the mean ± SD for triplicate determinations and the results are representative of three independent experiments. A significant change in luciferase activity compared to the DMSO control is indicated (*p < 0.05, **p < 0.001; Student's t-test).
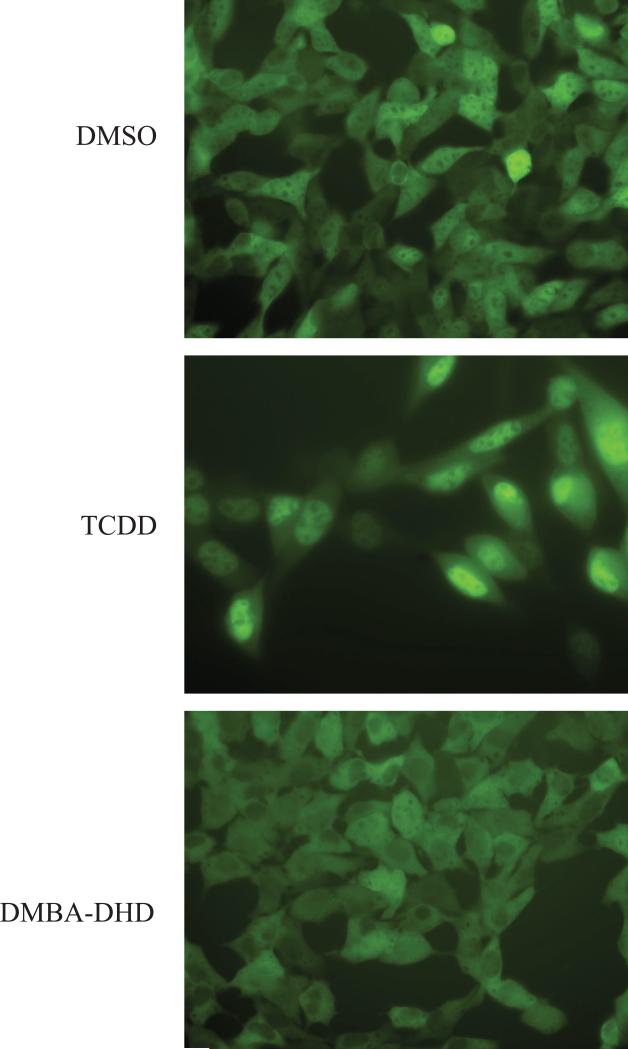
In contrast to our results, a previous study reported that DMBA-DHD was a competitive ligand for the cytosolic AhR from mouse hepatoma (hepa1c1c7) cells and that it could stimulate AhR translocation in the same cells based on western blot analysis of nuclear extracts from DMBADHD exposed cells [24]. However, since DMBA-DHD failed to stimulate AhR:ARNT DNA binding or transcriptional activity in the studies reported here, we questioned whether it was an AhR agonist. Accordingly, to determine whether DMBA-DHD had agonist activity, we examined its ability to stimulate AhR nuclear localization in intact cells. Recombinant mouse hepa1c1c7 cells containing a stably transfected yellow fluorescent protein (YFP)-tagged AhR (referred to as yAHAYc6 cells) were incubated with TCDD or DMBA-DHD for 1 hour and the cells visualized by fluorescent microscopy. While TCDD treatment resulted in almost complete AhR nuclear accumulation of YFP-AhR (Figure 6), DMBA-DHD had no apparent effect. Together the results of the DNA binding, nuclear translocation, and gene expression analysis reported here strongly suggest that DMBA-DHD is not an AhR agonist.
Figure 6. DMBA-DHD fails to stimulate AhR nuclear localization in mouse hepatoma cells.
Recombinant mouse hepatoma (yAHAYc6) cells containing a stably transfected YFP-tagged AhR expression vector were treated with DMSO (0.1% (v/v)), TCDD (10 nM) or DMBA-DHD (1 μM) for 1 hour and localization of the YFP-AhR was visualized by fluorescent microscopy.
The PON1 DRE-like response element fails to confer 3MC or quercetin responsiveness upon a heterologous promoter and luciferase reporter gene
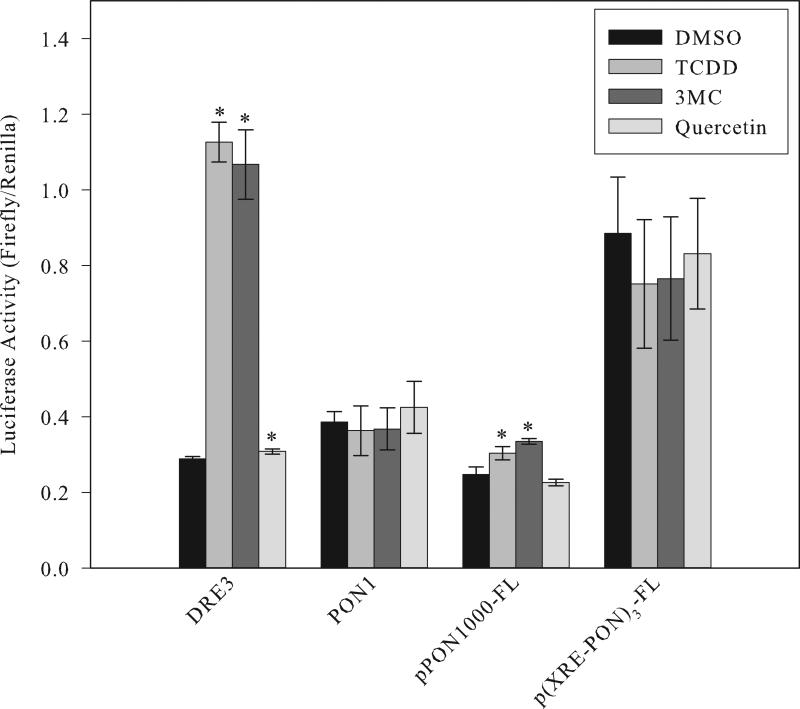
Studies were also carried out to examine the ability of the PON1 DRE-like sequence to confer 3MC- or quercetin-dependent responsiveness upon the luciferase reporter gene. HuH7 cells were transfected with a luciferase reporter plasmid containing a single DRE3 or PON1 DRE-like DNA element and luciferase activity determined after cells were incubated with DMSO, TCDD, 3MC, or quercetin for 4 hours. While TCDD and 3MC incubation greatly stimulated AhR- and DRE-dependent induction of luciferase activity, a very low, but significant, increase in luciferase activity was observed following incubation with quercetin (Figure 7). Similar to the Bax results, no chemical-dependent induction of luciferase activity was observed from the PON1-luciferase reporter plasmid transfected cells. These results are consistent with the DNA binding results that suggest that the AhR does not bind to the previously reported AhR-responsive PON1 DNA element.
Figure 7. TCDD, 3MC and quercetin fail to stimulate luciferase reporter gene expression from the PON1 DRE-like DNA element.
Human hepatoma (HuH7) cells were transiently transfected with pGudLuc7.0 containing a single DRE3 or PON1 DNA oligonucleotide present upstream of the luciferase reporter gene or with a plasmid containing either 1000 base pairs of the upstream regulatory region from the PON1 gene (pPON1000-FL) or three tandem copies of the wild type PON1 DRE-like sequence (p(XRE-PON)3-FL) upstream of the luciferase reporter gene [13]. Cells were incubated with DMSO (0.1% (v/v)), TCDD (10 nM), 3MC (5 μM) or quercetin (50 μM) for 4 hours followed by measurement of luciferase activity. Values represent the mean ± SD of triplicate determinations and are representative of three independent experiments. The asterisk indicates a significant (p < 0.05; Student's t-test) increase in luciferase activity compared to the DMSO control.
The previously published results of Gouédard et al. [13] reported that 3MC and quercetin stimulated AhR-dependent expression of the PON1 gene and this induction was mediated by a DNA element contained within 1000 base pairs of the immediate upstream regulatory region of the PON1 gene. Their subsequent deletion analysis led to the identification of the PON1 DRE-like element examined here as being responsible for the AhR-dependent activity. However, given our inability to demonstrate DNA binding of activated AhR to DNA containing this element or chemical-dependent activation of gene expression in the same cell line, additional examination of the AhR responsiveness of the original PON1 regulatory sequences was warranted. PON1 regulatory DNA-containing plasmids described in the original studies of Gouédard et al. [13] were obtained, and their responsiveness to TCDD, 3MC and quercetin in HuH7 cells were examined using similar experimental conditions. The specific luciferase reporter gene plasmids utilized in these studies contained either the 1000 base pairs of upstream regulatory sequences from the PON1 gene originally used to demonstrate AhR responsiveness (pPON1000-FL) or one containing three tandem copies of the PON1 DRE-like response element (p(XRE-PON)3-FL). These transfection studies revealed a very small, but significant induction of luciferase activity in pPON1000-FL transfected cells exposed to TCDD or 3MC, but no induction was observed with quercetin (Figure 7). The low levels of induction observed with TCDD and the somewhat greater activity with 3MC are consistent with the results of Gouédard et al. [13], however, while their studies reported even higher levels of induction with quercetin, we observed no induction. Additionally, we observed no ligand-dependent induction of reporter gene expression in cells transfected with p(XRE-PON1)3-FL, a plasmid that was also previously reported to be responsive to 3MC and quercetin [13]. This lack of induction is consistent with our PON1 DNA binding and transfection results. Taken together, while our results indicate that the upstream regulatory region of the PON1 gene can confer low levels of TCDD- and 3MC responsiveness upon a luciferase reporter gene, the AhR responsiveness observed in our studies appears not to be mediated by the PON1 DRE-like sequence previously reported by Gouédard and coworkers [13].
DISCUSSION
Nuclear receptors can exhibit significant diversity in the mechanisms by which they produce their spectrum of biological responses and many responses are directly related to nuclear factors (i.e. coactivators and corepressors) whose interactions with these receptors and their associated proteins are affected in a ligand-dependent manner. The binding and activation of nuclear receptors by various ligands can result in ligand-selective alterations in the overall structure of the receptor which can lead to changes in its functional activity as a result of differential binding to coactivator proteins. While ligand-dependent changes in coactivator binding by the AhR have been reported [11], novel ligand-selective alterations in AhR functional activity have also been reported, specifically that binding of the AhR by 3MC, quercetin or DMBA-DHD changes the nucleotide sequence to which the AhR can bind and stimulate gene expression [12, 13]. While nuclear hormone receptors can bind to response elements that exhibit a significant degree of nucleotide variation in the established consensus sequence [25, 26], no ligands have been identified that can alter the nucleotide specificity of DNA binding of these receptors. Accordingly, given this unique effect reported with some AhR ligands, we attempted to confirm and examine the molecular mechanisms by which binding to and activation of the AhR by DMBA-DHD, 3MC or quercetin changes the nucleotide specificity of AhR DNA binding using identical or very similar model systems to those described in the original studies identifying these novel AhR binding sites [12, 13]. In contrast to the published results, while TCDD, 3MC and quercetin were able to stimulate AhR binding to and transcriptional activation from the wild-type DRE, 3MC, quercetin or DMBA-DHD failed to stimulate AhR binding to or activation of transcription from the reported PON1 and Bax DRE-like DNA response elements. While the specific reasons for the differences in our results compared with those of Gouédard et al. [13] and Matikainen et al. [12] remain to be determined, our analyses indicate that these compounds do not change the nucleotide specificity of DNA binding of the AhR to allow it to bind, nor stimulate gene expression through, these unique DRE-like DNA elements. The results of our studies are consistent with a large amount of information on the well-established nucleotide specificity of DNA binding by other ligand-dependent nuclear receptors.
There are, however, several factors that may contribute to the differences between our studies and those of Gouédard et al. [13] and Matikainen et al. [12]. First, the results of Gouédard et al. [13], demonstrate that 48 hours of exposure of HuH7 cells to 3MC and quercetin stimulates PON1 gene expression and this effect appears to be mediated by DNA contained within the first 1000 base pairs of the upstream regulatory region of the gene. However, our transfection results in the same cell line indicate that the relative level of induction by these compounds at 4 hours of incubation is extremely low, far below that mediated by a single wild-type DRE element (Figure 7), which suggests that gene induction may involve a mechanism distinct from the AhR, possibly an indirect effect or involving a metabolite of 3MC or quercetin. While in silico analysis of this 5’ upstream region using MatInspector (Genomatix) identified 229 putative transcription factor binding sites (38 of which were liver-specific), no DRE sequences were identified and it remains to be determined whether any of these putative transcription factor binding sites play any role in the TCDD/3MC induction observed in our studies or those of Gouédard et al. [13]. Another consideration is the nucleotide sequence of the DRE-like element that was proposed to regulate 3MC- and quercetin-dependent induction of PON1 gene expression. The PON1 DRE-like response element as reported by Gouédard et al. [13] is more similar to the DNA core consensus sequence for the transcription factor SP1 (5’-GGGCGGG-3’) than it is to the DNA core consensus sequence for the AhR:ARNT complex (i.e. 5’-GCGTG-3’ of the DRE). While DRE mutagenesis studies have identified that the AhR:ARNT complex can tolerate some variability in the first guanine of the DRE core sequence, substitution of the any of the remaining four bases completely eliminates AhR DNA binding, and this specificity was demonstrated for AhR from different species and with a variety of different ligands [20, 27]. Thus, it is somewhat surprising that the PON1 DRE-like sequence was reported to bind the AhR:ARNT complex when it contains a mutation (T to G) in a position of the DRE consensus sequence that has been unequivocally shown to eliminate AhR:ARNT binding. It is possible that the protein-DNA binding results of Gouédard et al. [13] represent the AhR bound to a different heterodimerization partner; however, we were unable to demonstrate inducible protein-DNA complex formation with nuclear extracts from the same cell line and chemical treatments. While quercetin has been well characterized as an inducer of PON1 mRNA levels and enzyme activity [28], a recent study has suggested that this effect occurs through a sterol regulatory element binding protein 2 (SREBP2)-dependent pathway, rather than through an AhR-dependent mechanism [29]. A subsequent report by Gouédard et al. [30] indicated that resveratrol, an AhR antagonist [31], could also stimulate AhR-dependent PON1 gene expression through the PON1 DRE-like element. The ability of an AhR antagonist (resveratrol) to produce the same mechanistically-based AhR-dependent induction response as an agonist (3MC and quercetin) is inconsistent with the current understanding of agonist versus antagonist action on the AhR and AhR signaling pathway and would be more consistent with an alternative mechanism of action for PON1 gene expression. Still, the results in this study do not preclude the possibility that PON1 may, in part, be regulated through an AhR-dependent mechanism, although species or strain specificity may play a role. Studies in rats have reported increases in PON1 gene expression or protein levels using classical AhR agonists (TCDD, 3MC, PCB 126) [32-34], while studies in C57Bl/6 mice failed to demonstrate a similar effect [35]. Although a clear mechanistic understanding for these biological effects is currently lacking, our results do not support AhR:ARNT binding to and transcriptional activity from the PON1 DRE-like response element in these induction responses.
In contrast to the PON1 DRE-like response element, the Bax DRE-like response element identified by Matikainen et al. [12] contains two DRE-like sequences both of which contain the invariant DRE core sequence of 5’-GCGTG-3’ (Figure 1A). However, previous studies have demonstrated that while this core sequence is necessary, it is not sufficient, and that nucleotides flanking the core sequence are critical for full AhR:ARNT binding affinity and transcriptional activation [36]. Neither of the Bax DRE-like sequences contain flanking nucleotides that completely match the defined wild type DRE3 consensus sequence, and while the AhR:ARNT complex is capable of binding to nonconsensus DREs [27], the variability in these flanking nucleotides is sufficient to prevent binding of ligand-activated AhR:ARNT complexes. Additionally, both Bax DRE-like sequences contain single nucleotide variations in their sequences that differ from those found in functional DREs (as defined by mutagenesis experiments) that would eliminate their ability to confer ligand-dependent AhR:ARNT transcriptional activation, despite only exhibiting moderate reductions in AhR:ARNT DNA binding [7, 27, 37]. Specifically, the responsible nucleotides are the guanine in the first base position immediately downstream of the DRE core, while in the second DRE, the responsible nucleotide is the cytosine in the third position downstream of the core (Figure 1A). Thus, it is not surprising that not only does the AhR:ARNT complex fail to bind to the Bax DRE-like response element, but these nucleotide variations would also be responsible for the lack of ligand- and AhR-dependent transcriptional activity of this DNA element in our transfection studies. In comparison, we also tested the mutant Bax DRE-like response element which contains a double mutation designed to make both sequences more “DRE like” [12]. While we observed that these mutations in the Bax DRE-like sequence restored the ability of TCDD- and 3MC-activated AhR:ARNT complex to bind to this DNA element, these changes were not sufficient to fully restore DNA binding to levels comparable to that of the wild type DRE3 (Figures 2A and 2B), suggesting that additional nucleotides negatively impact the binding of AhR:ARNT complexes to the mutant Bax DRE-like element. The inability of the mutant Bax DRE-like DNA element to confer ligand-dependent responsiveness upon the luciferase reporter gene also indicated that these mutations were not sufficient, within the context of the other nucleotide substitutions occurring in the sequence, to restore functional activity (at least to detectable levels) and this likely results from an insufficient AhR:ARNT DNA binding affinity.
Similar to the PON1 DRE-like response element, the inability of the AhR:ARNT complex to bind either the Bax or mutant Bax DRE-like response element does not necessarily exclude differential effects of DMBA-DHD and TCDD on Bax expression, although the role of AhR in this response is uncertain. For example, not only were we unable to demonstrate DMBA-DHD-stimulated AhR:ARNT DNA binding to the Bax DRE-like DNA element, but DMBA-DHD could not be confirmed as an AhR agonist working through the wild-type DRE. Although a previous study reported competitive ligand binding by DMBA-DHD to the murine AhR and its ability to stimulate AhR nuclear translocation, the ability of DMBA-DHD to stimulate AhR DNA binding and transcriptional activity were not examined [24]. The results presented here are consistent with our previous competitive ligand binding studies that demonstrated that the presence of a hydroxyl group at the 3 position on benzo(a)pyrene (equivalent to the hydroxyl group at the 4 position on DMBA-DHD) significantly reduced its binding to the AhR, suggesting that the hydroxyl groups on DMBA-DHD may similarly negatively impact on its ability to act as an AhR agonist [38]. Additionally, since the studies of Matikainen et al. [12] used over 2 kilobases of the murine Bax promoter and we utilized only a small 37 base pair piece to directly examine AhR:ARNT binding, the possibility exists that the differences in functionality observed between these two studies results from perturbations in other transcription factor(s) signaling pathways. Alternatively, the mouse oocytes used by Matikainen et al. [12] could contain unique factors and/or developmental targets susceptible to DMBA-DHD and/or TCDD that are not present in our mouse hepatoma cells, although both were derived from C57BL6 mice. Intriguingly, the endogenous ligand kynurenine was recently found to upregulate the Bax gene via an AhR-dependent pathway [39], which lend support for a role of the AhR in regulating Bax, although the exact mechanism(s) remains to be determined.
As the field of AhR biology continues to expand, it is evident that the AhR functions in ways separate from its classically-defined mechanism as a ligand-activated transcription factor binding to DREs to stimulate gene transcription [1]. While some of these novel mechanisms involve the AhR partnering with proteins other than ARNT as well as it associating with non-DRE response elements through protein-protein interactions with other factors [40-42], these mechanisms are distinct from the AhR:ARNT-mediated mechanisms proposed to regulate the ligand-dependent activation of the Bax and PON1 genes. Specifically, we were unable to confirm differences in the nucleotide specificity of DNA binding by the AhR:ARNT complex when bound by ligands (DMBA-DHD, quercetin and 3MC) other than TCDD. These results are supported by a more systematic evaluation using a DNA selection and amplification approach and a series of structurally diverse AhR ligands [43] as well as our earlier studies that showed no significant ligand- or species-specific differences in the nucleotide specificity of AhR DNA binding [20]. We currently envision that if a compound is capable of stimulating AhR:ARNT DNA binding and gene activation through its classical mechanism of activation, than this heterodimer will bind to a sequence that contains the canonical DRE. As additional mechanisms of AhR action continue to be identified, ligand-specificity is expected to form the basis for many of these novel pathways. Indeed, studies on the selective AhR modulators (SAhRMs), ligand-specific effects on estrogen signaling, and differential recruitment of cofactors have already demonstrated that this is an important area of future research [11, 44, 45]. However, should ligand-selective differences in the nucleotide specificity of AhR:ARNT DNA binding be discovered as an additional pathway for AhR signaling, this would not only represent a major finding in AhR biology, but may also suggest alternative signaling pathways for other ligand-activated transcription factors. It would also have significant implications for our understanding of the molecular mechanisms by which the AhR modulates the toxic and biological effects of structurally diverse chemicals.
Supplementary Material
Highlights.
Ligand-specific differences in AhR:ARNT DNA binding and transcription were studied.
DRE-like sequences in the promoter regions of Bax and PON1 genes were analyzed.
The AhR:ARNT complex did not bind to either DRE-like sequence regardless of ligand.
AhR-dependent luciferase gene expression was not observed from either sequence.
Our results support the established role of the DRE in AhR:ARNT DNA binding.
ACKNOWLEDGEMENTS
We gratefully acknowledge Dr. Curtis Omiecinski (The Pennsylvania State University) for the HuH7 cells, Dr. Steven Safe for the TCDD, and Dr. Robert Barouki for the PON1 plasmids. Funding was provided by the National Institutes of Environmental Health Sciences (R01ES07685, P42ES04699 and training grant T32ES07059 (DED)), the California Agriculture Experiment Station and the American taxpayers.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Abbreviations: 3MC, 3-methylcholanthrene; AhR, aryl hydrocarbon receptor; ARNT, AhR nuclear translocator; DMBA-DHD, 7,12-dimethylbenz[a]anthracene-3,4-dihydrodiol; DMSO dimethyl sulfoxide; DRE, dioxin responsive element; EMSA, electrophoretic mobility shift assay; PAH, polycyclic aromatic hydrocarbon; PON1, paraoxonase 1; SAhRM, selective AhR modulator; TCDD, 2,3,7,8-tetracholordibenzo-p-dioxin
REFERENCES
- 1.Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B. Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor. Toxicological Sciences. 2011;124:1–22. doi: 10.1093/toxsci/kfr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annual Review of Pharmacology and Toxicology. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- 3.Furness SG, Whelan F. The pleiotropy of dioxin toxicity--xenobiotic misappropriation of the aryl hydrocarbon receptor's alternative physiological roles. Pharmacology and Therapeutics. 2009;124:336–353. doi: 10.1016/j.pharmthera.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 4.Poland A, Knutson JC. 2,3,7,8-Tetrachlorodibenzo-p-dioxin and related halogenated aromatic hydrocarbons: examination of the mechanism of toxicity. Annual Review of Pharmacology and Toxicology. 1982;22:517–554. doi: 10.1146/annurev.pa.22.040182.002505. [DOI] [PubMed] [Google Scholar]
- 5.Okey AB, Bondy GP, Mason ME, Kahl GF, Eisen HJ, Guenthner TM, Nebert DW. Regulatory gene product of the Ah locus. Characterization of the cytosolic inducer-receptor complex and evidence for its nuclear translocation. Journal of Biological Chemistry. 1979;254:11636–11648. [PubMed] [Google Scholar]
- 6.Soshilov A, Denison MS. Role of the Per/Arnt/Sim domains in ligand-dependent transformation of the aryl hydrocarbon receptor. Journal of Biological Chemistry. 2008;283:32995–33005. doi: 10.1074/jbc.M802414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denison MS, Elferink CF, Phelan D. The Ah receptor signal transduction pathway. In: Denison MS, Helferich WG, editors. Toxicant-Receptor Interactions in the Modulation of Signal Transduction and Gene Expression. Taylor and Francis; Philadelphia, PA: 1998a. pp. 3–33. [Google Scholar]
- 8.DeGroot D, He G, Fraccalvieri D, Bonati L, Pandini A, Denison MS. AHR Ligands: Promiscuity in binding and diversity in response. In: Pohjanvirta R, editor. The AH Receptor in Biology and Toxicology. John Wiley & Sons, Inc; Hoboken, NJ: 2011. pp. 63–79. [Google Scholar]
- 9.Patel RD, Murray IA, Flaveny CA, Kusnadi A, Perdew GH. Ah receptor represses acute-phase response gene expression without binding to its cognate response element. Laboratory Investigation. 2009;89:695–707. doi: 10.1038/labinvest.2009.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray IA, Krishnegowda G, DiNatale BC, Flaveny C, Chiaro C, Lin JM, Sharma AK, Amin S, Perdew GH. Development of a selective modulator of aryl hydrocarbon (Ah) receptor activity that exhibits anti-inflammatory properties. Chemical Research in Toxicology. 2010;23:955–966. doi: 10.1021/tx100045h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang S, Rowlands C, Safe S. Ligand-dependent interactions of the Ah receptor with coactivators in a mammalian two-hybrid assay. Toxicology and Applied Pharmacology. 2008;227:196–206. doi: 10.1016/j.taap.2007.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matikainen T, Perez GI, Jurisicova A, Pru JK, Schlezinger JJ, Ryu HY, Laine J, Sakai T, Korsmeyer SJ, Casper RF, Sherr DH, Tilly JL. Aromatic hydrocarbon receptor-driven Bax gene expression is required for premature ovarian failure caused by biohazardous environmental chemicals. Nature Genetics. 2001;28:355–360. doi: 10.1038/ng575. [DOI] [PubMed] [Google Scholar]
- 13.Gouedard C, Barouki R, Morel Y. Dietary polyphenols increase paraoxonase 1 gene expression by an aryl hydrocarbon receptor-dependent mechanism. Molecular and Cellular Biology. 2004;24:5209–5222. doi: 10.1128/MCB.24.12.5209-5222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denison MS, Rogers JM, Rushing SR, Jones CL, Tetangco SC, Heath-Pagliuso S. Analysis of the aryl hydrocarbon receptor (AhR) signal transduction pathway. In: Maines MD, Costa LG, Reed DJ, Sassa S, Sipes IG, editors. Current Protocols in Toxicology. John Wiley & Sons, Inc.; New York, NY: 2002. pp. 4.8.1–4.8.45. [DOI] [PubMed] [Google Scholar]
- 15.Rushing SR, Denison MS. The silencing mediator of retinoic acid and thyroid hormone receptors can interact with the aryl hydrocarbon (Ah) receptor but fails to repress Ah receptor-dependent gene expression. Archives of Biochemistry and Biophysics. 2002;403:189–201. doi: 10.1016/s0003-9861(02)00233-3. [DOI] [PubMed] [Google Scholar]
- 16.Denison MS, Fisher JM, Whitlock JP., Jr. Inducible, receptor-dependent protein-DNA interactions at a dioxin-responsive transcriptional enhancer. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:2528–2532. doi: 10.1073/pnas.85.8.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fukunaga BN, Hankinson O. Identification of a novel domain in the aryl hydrocarbon receptor required for DNA binding. Journal of Biological Chemistry. 1996;271:3743–3749. doi: 10.1074/jbc.271.7.3743. [DOI] [PubMed] [Google Scholar]
- 18.Rogers JM, Denison MS. Recombinant cell bioassays for endocrine disruptors: development of a stably transfected human ovarian cell line for the detection of estrogenic and anti-estrogenic chemicals. In Vitro & Molecular Toxicology. 2000;13:67–82. [PubMed] [Google Scholar]
- 19.Zhao B, Degroot DE, Hayashi A, He G, Denison MS. CH223191 is a ligand-selective antagonist of the Ah (Dioxin) receptor. Toxicological Sciences. 2010;117:393–403. doi: 10.1093/toxsci/kfq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bank PA, Yao EF, Phelps CL, Harper PA, Denison MS. Species-specific binding of transformed Ah receptor to a dioxin responsive transcriptional enhancer. European Journal of Pharmacology. 1992;228:85–94. doi: 10.1016/0926-6917(92)90016-6. [DOI] [PubMed] [Google Scholar]
- 21.Denison MS, Phelps CL, DeHoog J, Kim HJ, Bank PA, Yao EF, Harper PA. Species Variation in Ah Receptor Transformation and DNA Binding. In: Gallo MA, Scheuplein RJ, Van Der Heijden KA, editors. Banbury Report No. 35: Biological Basis of Risk Assessment of Dioxins and Related Compounds. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1991. pp. 337–347. [Google Scholar]
- 22.Ciolino HP, Daschner PJ, Yeh GC. Dietary flavonols quercetin and kaempferol are ligands of the aryl hydrocarbon receptor that affect CYP1A1 transcription differentially. Biochemical Journal. 1999;340(Pt 3):715–722. [PMC free article] [PubMed] [Google Scholar]
- 23.Modi BG, Neustadter J, Binda E, Lewis J, Filler RB, Roberts SJ, Kwong BY, Reddy S, Overton JD, Galan A, Tigelaar R, Cai L, Fu P, Shlomchik M, Kaplan DH, Hayday A, Girardi M. Langerhans cells facilitate epithelial DNA damage and squamous cell carcinoma. Science. 2012;335:104–108. doi: 10.1126/science.1211600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mann KK, Matulka RA, Hahn ME, Trombino AF, Lawrence BP, Kerkvliet NI, Sherr DH. The role of polycyclic aromatic hydrocarbon metabolism in dimethylbenz[a]anthracene-induced pre-B lymphocyte apoptosis. Toxicology and Applied Pharmacology. 1999;161:10–22. doi: 10.1006/taap.1999.8778. [DOI] [PubMed] [Google Scholar]
- 25.Gruber CJ, Gruber DM, Gruber IM, Wieser F, Huber JC. Anatomy of the estrogen response element. Trends in Endocrinology and Metabolism. 2004;15:73–78. doi: 10.1016/j.tem.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Mason CE, Shu FJ, Wang C, Session RM, Kallen RG, Sidell N, Yu T, Liu MH, Cheung E, Kallen CB. Location analysis for the estrogen receptor-alpha reveals binding to diverse ERE sequences and widespread binding within repetitive DNA elements. Nucleic Acids Research. 2010;38:2355–2368. doi: 10.1093/nar/gkp1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao EF, Denison MS. DNA sequence determinants for binding of transformed Ah receptor to a dioxin-responsive enhancer. Biochemistry. 1992;31:5060–5067. doi: 10.1021/bi00136a019. [DOI] [PubMed] [Google Scholar]
- 28.Costa LG, Giordano G, Furlong CE. Pharmacological and dietary modulators of paraoxonase 1 (PON1) activity and expression: the hunt goes on. Biochemical Pharmacology. 2011;81:337–344. doi: 10.1016/j.bcp.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garige M, Gong M, Varatharajalu R, Lakshman MR. Quercetin up-regulates paraoxonase 1 gene expression via sterol regulatory element binding protein 2 that translocates from the endoplasmic reticulum to the nucleus where it specifically interacts with sterol responsive element-like sequence in paraoxonase 1 promoter in HuH7 liver cells. Metabolism: Clinical and Experimental. 2010;59:1372–1378. doi: 10.1016/j.metabol.2009.12.025. [DOI] [PubMed] [Google Scholar]
- 30.Gouedard C, Barouki R, Morel Y. Induction of the paraoxonase-1 gene expression by resveratrol. Arteriosclerosis, Thrombosis, and Vascular Biology. 2004;24:2378–2383. doi: 10.1161/01.ATV.0000146530.24736.ce. [DOI] [PubMed] [Google Scholar]
- 31.Casper RF, Quesne M, Rogers IM, Shirota T, Jolivet A, Milgrom E, Savouret JF. Resveratrol has antagonist activity on the aryl hydrocarbon receptor: implications for prevention of dioxin toxicity. Molecular Pharmacology. 1999;56:784–790. [PubMed] [Google Scholar]
- 32.Shen H, Robertson LW, Ludewig G. Regulation of paraoxonase 1 (PON1) in PCB 126-exposed male Sprague Dawley rats. Toxicology Letters. 2012;209:291–298. doi: 10.1016/j.toxlet.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boverhof DR, Burgoon LD, Tashiro C, Sharratt B, Chittim B, Harkema JR, Mendrick DL, Zacharewski TR. Comparative toxicogenomic analysis of the hepatotoxic effects of TCDD in Sprague Dawley rats and C57BL/6 mice. Toxicological Sciences. 2006;94:398–416. doi: 10.1093/toxsci/kfl100. [DOI] [PubMed] [Google Scholar]
- 34.Rodrigo L, Hernandez AF, Lopez-Caballero JJ, Gil F, Pla A. Immunohistochemical evidence for the expression and induction of paraoxonase in rat liver, kidney, lung and brain tissue. Implications for its physiological role. Chemico-Biological Interactions. 2001;137:123–137. doi: 10.1016/s0009-2797(01)00225-3. [DOI] [PubMed] [Google Scholar]
- 35.Cheng X, Klaassen CD. Hormonal and chemical regulation of paraoxonases in mice. Journal of Pharmacology and Experimental Therapeutics. 2012;342:688–695. doi: 10.1124/jpet.112.194803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denison MS, Fisher JM, Whitlock JP., Jr. The DNA recognition site for the dioxin-Ah receptor complex. Nucleotide sequence and functional analysis. Journal of Biological Chemistry. 1988;263:17221–17224. [PubMed] [Google Scholar]
- 37.Shen ES, Whitlock JP., Jr. Protein-DNA interactions at a dioxin-responsive enhancer. Mutational analysis of the DNA-binding site for the liganded Ah receptor. Journal of Biological Chemistry. 1992;267:6815–6819. [PubMed] [Google Scholar]
- 38.Denison MS, Wilkinson CF. Identification of the Ah receptor in selected mammalian species and induction of aryl hydrocarbon hydroxylase. European Journal of Biochemistry. 1985;147:429–435. doi: 10.1111/j.1432-1033.1985.tb08767.x. [DOI] [PubMed] [Google Scholar]
- 39.Opitz CA, Litzenburger UM, Sahm F, Ott M, Tritschler I, Trump S, Schumacher T, Jestaedt L, Schrenk D, Weller M, Jugold M, Guillemin GJ, Miller CL, Lutz C, Radlwimmer B, Lehmann I, von Deimling A, Wick W, Platten M. An endogenous tumour-promoting ligand of the human aryl hydrocarbon receptor. Nature. 2011;478:197–203. doi: 10.1038/nature10491. [DOI] [PubMed] [Google Scholar]
- 40.Vogel CF, Sciullo E, Li W, Wong P, Lazennec G, Matsumura F. RelB, a new partner of aryl hydrocarbon receptor-mediated transcription. Molecular Endocrinology. 2007;21:2941–2955. doi: 10.1210/me.2007-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang G, Elferink CJ. A novel nonconsensus xenobiotic response element capable of mediating aryl hydrocarbon receptor-dependent gene expression. Molecular Pharmacology. 2012;81:338–347. doi: 10.1124/mol.111.075952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oesch-Bartlomowicz B, Huelster A, Wiss O, Antoniou-Lipfert P, Dietrich C, Arand M, Weiss C, Bockamp E, Oesch F. Aryl hydrocarbon receptor activation by cAMP vs. dioxin: divergent signaling pathways. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9218–9223. doi: 10.1073/pnas.0503488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.DeGroot DE, Denison MS. Does the binding of structurally diverse chemicals to the Ah receptor alter its nucleotide specificity of DNA binding? Organohalogen Compounds. 2011;74:1033–1036. [Google Scholar]
- 44.Murray IA, Flaveny CA, Chiaro CR, Sharma AK, Tanos RS, Schroeder JC, Amin SG, Bisson WH, Kolluri SK, Perdew GH. Suppression of cytokine-mediated complement factor gene expression through selective activation of the Ah receptor with 3′,4′-dimethoxy-alpha-naphthoflavone. Molecular Pharmacology. 2011;79:508–519. doi: 10.1124/mol.110.069369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okino ST, Pookot D, Basak S, Dahiya R. Toxic and chemopreventive ligands preferentially activate distinct aryl hydrocarbon receptor pathways: implications for cancer prevention. Cancer Prevention Research (Philadelphia, Pa.) 2009;2:251–256. doi: 10.1158/1940-6207.CAPR-08-0146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.