Abstract
Sixty-nine fecal samples from diarrheic puppies were examined by reverse transcription-PCR assays for the M and the S genes of canine coronaviruses (CCoVs). The isolates in 10 samples were recognized as CCoV type I, and the isolates in 6 samples were recognized as CCoV type II, while isolates of both genotypes were simultaneously detected in 53 samples.
Canine coronavirus (CCoV), a member of the family Coronaviridae, is an enveloped, positive-stranded RNA virus responsible for enteric disease in young puppies. CCoV has been detected in the feces of naturally infected puppies for up to 180 days (8, 10). Recently, by sequence analysis of the M genes of several CCoV isolates detected in infected puppies, a genetic drift to feline coronavirus (FCoV) was observed (7, 12). Subsequently, sequence analysis of the S gene revealed the presence of a new genotype of CCoV, tentatively designated CCoV type I on the basis of its genetic similarity to FCoV type I (13).
The results of a virological investigation by reverse transcription-PCR (RT-PCR) showing the simultaneous presence of both genotypes in the feces of diarrheic puppies are reported.
Sixty-nine fecal samples were collected from 6- to 12-week-old diarrheic puppies living in different regions of Italy. All samples had previously tested positive for CCoV by RT-PCR (5).
Feline cell whole fetus (fcwf-4) and A-72 dog cell lines were used for virus isolation, and an immunofluorescence assay (IFA) with a monoclonal antibody to CCoV was performed with each cell culture passage.
The fecal samples were tested for the S and M genes of CCoVs type I and type II by RT-PCR. In addition, RT-PCR assays were performed with the cryolysates of the third cell culture passages of the isolated strains. RNA extraction was performed according to the protocols of the manufacturer (Qiagen, GmbH, Germany). The sequences and the positions of all the primers are displayed in Table 1.
TABLE 1.
Sequences and positions of the primers used in the present study
| Primer | Gene | CCoV type | Sequence (5′ to 3′) | Sense | Position | Amplicon size (bp) |
|---|---|---|---|---|---|---|
| CCoV1aa | M | I | GTGCTTCCTCTTGAAGGTACA | + | 6900-6920c | 239 |
| CCoV2b | M | TCTGTTGAGTAATCACCAGCT | − | 7118-7138c | ||
| EL1F | S | I | CAAGTTGACCGTCTTATTACTGGTAG | + | 2611-2636d | 346 |
| EL1R | S | TCATATACGTACCATTATAGCTGAAGA | − | 2930-2956d | ||
| Can1F | M | II | TAACATTGCTCTCAGGGAATTTG | + | 6937-6959b | 202 |
| CCoV2b | M | TCTGTTGAGTAATCACCAGCT | − | 7118-7138b | ||
| S5 | S | II | TGCATTTGTGTCTCAGACTT | + | 3991-4010c | 694 |
| S6 | S | CCAAGGCCATTTTACATAAG | − | 4665-4684c |
The RNA was reverse transcribed with random hexamers by using murine leukemia virus reverse transcriptase (Applied Biosystems, Rome, Italy) and then amplified with AmpliTaq DNA polymerase (Applied Biosystems). For amplification of CCoV type I, the primer pairs CCoV1a-CCoV2, which amplify a fragment of the M gene (11), and EL1F-EL1R, the sequence for which was selected from a relatively conserved region of the Elmo/02 CCoV type I spike gene (13), were used.
For amplification of CCoV type II, primers Can1F and CCoV2 were chosen on the basis of the mismatches between the M gene of CCoV type II and CCoV type I strain 259/01 (12). Primers S5 and S6 were designed on the basis of comparative sequence analysis of the spike gene of the reference CCoV type II and FCoV type II strains.
The PCR products of four fecal samples (samples 4, 10, 13, and 19) positive for both genotypes were subjected to sequence analysis (Genome Express; Labo Grenoble, Meylan, France). The molecular analysis tools of the National Center for Biotechnology Information and the European Molecular Biology Laboratory were used for sequence comparison. Phylogenetic and molecular evolutionary analyses were performed with MEGA software, version 2.1 (3). Maximum-parsimony trees were processed by using a heuristic algorithm with bootstrapping of over 100 replicates.
The references and the nucleotide sequence European Molecular Biology Laboratory accession numbers for the M and S genes of the strains mentioned in this study are reported in Tables 2 and 3, respectively.
TABLE 2.
Accession numbers of the M genes of the strains mentioned in this study
| Strain | Virus and genotype | EMBL accession no. | Reference |
|---|---|---|---|
| Insavc | CCoV type II | D13096 | 2 |
| K 378 | CCoV type II | —a | |
| UCD1 | FCoV type I | AB086902 | 4 |
| Black | FCoV type I | AB086903 | 4 |
| 79-1146 | FCoV type II | X56496 | 15 |
| 79-1683 | FCoV type II | AB086904 | 4 |
| 259/01 | CCoV type I | AF502583 | 12 |
—, the strain was kindly supplied by L. E. Carmichael (J. Baker Institute, Ithaca, N.Y.), and the nucleotide sequence of the M gene was obtained by the authors.
TABLE 3.
Accession numbers of the S genes of the strains mentioned in this study
Thirteen of the 69 fecal samples inoculated on A72 and fcwf-4 cells were CCoV positive by IFA at the first passage and showed the typical CCoV cytopathic effect at the second passage.
PCR amplicons of the expected sizes were obtained with primer pairs CCoV1a-CCoV2 (amplicon of 239 bp) and EL1F-EL1R (amplicon of 346 bp), which selectively recognized CCoV type I from 10 of 69 samples (14.5%). Otherwise, the PCR assays with primers Can1F-CCoV2 (amplicon of 202 bp) and S5-S6 (amplicon of 694 bp) detected CCoV type II in 6 of 69 samples (8.7%). CCoV type I and CCoV type II were simultaneously identified in 53 of 69 samples (76.8%). All 13 fecal samples from which the coronavirus strains were subsequently isolated in cell cultures belonged to the last group. Moreover, RT-PCR assays performed with the cryolysates of the third serial cell passages of the 13 CCoV strains were positive only for genotype II, showing that CCoV type I failed to grow in cell cultures.
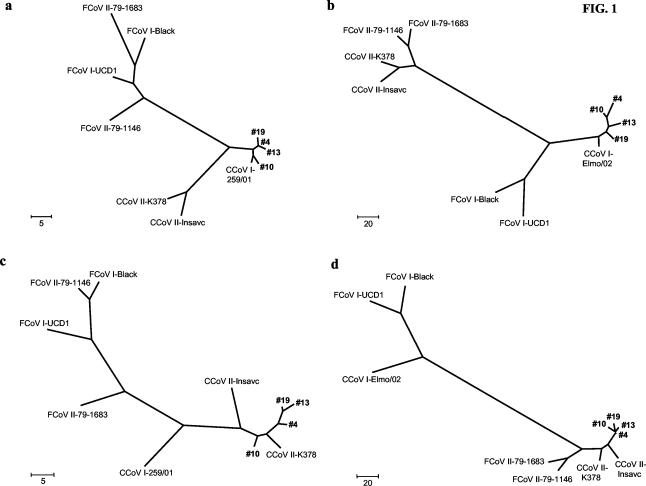
In the parsimony dendrograms based on the M- and S-gene fragments, the four CCoV type I strains examined were clearly clustered apart from the CCoV type II strains and segregated with the CCoV type I and FCoV type I reference strains (Fig. 1a and b).
FIG. 1.
Maximum-parsimony trees constructed with different genome fragments of CCoVs and FCoVs. (a) Fragment of the M gene obtained with primer pair CCoV1a-CCoV2 (239 bp); (b) fragment of the S gene obtained with primer pair EL1F-EL1R (346 bp); (c) fragment of the M gene obtained with primer pair Can1F-CCoV2 (202 bp); (d) fragment of the S gene obtained with primer pair S5-S6 (694 bp). The trees are unrooted and are drawn to scale. Bootstrap values are not shown. The numbers 4, 10, 13, and 19 indicate specimen designations of the PCR products from the fecal samples analyzed.
On the contrary, parsimony analysis based on the M- and S-gene fragments of the four CCoV type II strains examined revealed high degrees of homology with the CCoV type II and FCoV type II reference strains (Fig. 1c and d).
Serological investigations suggest that CCoV infection is widespread in pet and in kennel dogs (9, 14). In contrast, little evidence for CCoV-associated gastroenteritis in dogs has been collected, and only a few strains have been adapted to growth in vitro (6).
Therefore, the development of PCR assays for the detection of CCoV (1, 5) has provided important information on the diffusion and the epidemiology of CCoV infection.
Variations in the genomes of CCoV strains present in fecal samples of puppies with diarrhea have been reported (7), and a genetic drift to FCoV type II has been observed in the M genes of CCoV strains detected in the feces of two naturally infected puppies (10). Finally, an evident genetic divergence from the reference CCoV strains has been observed in the genomes of CCoVs identified in the feces of puppies, strongly indicating that a new genotype of CCoV is widespread in dogs (12, 13).
In the present study, analysis of the M and S genes from CCoV-positive fecal samples has confirmed the existence of a distinct genetic lineage of CCoV. Moreover, our results clearly show that CCoV infection in dogs is frequently characterized by the simultaneous presence of both CCoV type I and CCoV type II. Indeed, isolates of both genotypes were demonstrated in 53 of 69 samples. The significance of these data is still unclear.
Interestingly, viruses from only a few (13 of 69) samples PCR positive for CCoV have been adapted to growth in vitro. Moreover, the fecal samples positive for both virus genotypes yielded only CCoV type II in cell cultures. Failures to isolate CCoV type I in cell culture prevent authentic evaluations of the immunological characteristics of this new genotype of CCoV and, importantly, hinder the acquisition of key information on its pathogenic role in dogs.
Acknowledgments
This study was supported by grants from the Ministry of Universities (2002) of Italy (Project Enteriti Virali del Cane) and CEGBA (Centro di Eccellenza di Genomica in Campo Biomedico e Agrario).
REFERENCES
- 1.Bandai, C., S. Ishiguro, N. Masuya, T. Hohdatsu, and M. Mochizuki. 1999. Canine coronavirus infections in Japan: virological and epidemiological aspects. J. Vet. Med. Sci. 61:731-736. [DOI] [PubMed] [Google Scholar]
- 1a.de Groot, R. J., J. Madurn, J. A. Lenstra, M. C. Horzinck, B. A. M. van der Ziejst, and W. J. M. Spaan. 1987. cDNA cloning and sequence analysis of the gene encoding the peplomer protein of feline infectious peritonitis virus. J. Gen. Virol. 68:2639-2646. [DOI] [PubMed] [Google Scholar]
- 2.Horsburgh, B. C., I. Brierley, and T. D. Brown. 1992. Analysis of a 9.6 kb sequence from the 3′ end of canine coronavirus genomic RNA. J. Gen. Virol. 73:2849-2862. [DOI] [PubMed] [Google Scholar]
- 3.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 4.Motokawa, K., T. Hohdatsu, H. Hashimoto, and H. Koyama. 1996. Comparison of the amino acid sequence and phylogenetic analysis of the peplomer, integral membrane and nucleocapsid proteins of feline canine and porcine coronaviruses. Microbiol. Immunol. 40:425-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pratelli, A., M. Tempesta, G. Greco, V. Martella, and C. Buonavoglia. 1999. Development of a nested PCR for the detection of canine coronavirus. J. Virol. Methods 80:11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pratelli, A., D. Buonavoglia, V. Martella, M. Tempesta, A. Lavazza, and C. Buonavoglia. 2000. Diagnosis of canine coronavirus infection using n-PCR. J. Virol. Methods 84:91-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pratelli, A., V. Martella, G. Elia, N. Decaro, A. Aliberti, D. Buonavoglia, M. Tempesta, and C. Buonavoglia. 2001. Variation of the sequence in the gene encoding for transmembrane protein M of canine coronavirus (CCV). Mol. Cell. Probes 15:229-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pratelli, A., V. Martella, G. Elia, M. Tempesta, F. Guarda, M. T. Capucchio, L. E. Carmichael, and C. Buonavoglia. 2001. Severe enteric disease in an animal shelter associated with dual infections by canine adenovirus type 1 and canine coronavirus. J. Vet. Med. B 48:385-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pratelli, A., G. Elia, V. Martella, A. Palmieri, F. Cirone, A. Tinelli, M. Corrente, and C. Buonavoglia. 2002. Prevalence of canine coronavirus antibodies in dogs in the south of Italy by an enzyme-linked immunosorbent assay. J. Virol. Methods 102:67-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pratelli, A., G. Elia, V. Martella, A. Tinelli, N. Decaro, F. Marsilio, D. Buonavoglia, M. Tempesta, and C. Buonavoglia. 2002. M gene evolution of canine coronavirus in naturally infected dogs. Vet. Rec. 151:758-761. [PubMed] [Google Scholar]
- 11.Pratelli, A., A. Tinelli, N. Decaro, M. Camero, G. Elia, A. Gentile, and C. Buonavoglia. 2002. PCR assay for the detection and the identification of atypical canine coronavirus in dogs. J. Virol. Methods 106:209-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pratelli, A., V. Martella, M. Pistello, G. Elia, N. Decaro, D. Buonavoglia, M. Camero, M. Tempesta, and C. Buonavoglia. 2003. Identification of coronaviruses in dogs that segregate separately from the canine coronavirus genotype. J. Virol. Methods 107:213-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pratelli, A., V. Martella, N. Decaro, A. Tinelli, M. Camero, F. Cirone, G. Elia, A. Cavalli, M. Corrente, G. Greco, D. Buonavoglia, M. Gentile, M. Tempesta, and C. Buonavoglia. 2003. Genetic diversity of a canine coronavirus detected in pups with diarrhoea in Italy. J. Virol. Methods 110:9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tennant, B. J., R. M. Gaskell, R. C. Jones, and C. J. Gaskell. 1993. Studies on the epizootiology of canine coronavirus. Vet. Rec. 132:7-11. [DOI] [PubMed] [Google Scholar]
- 15.Vennema, H., R. J. de Groot, D. A. Harbour, M. C. Horzinek, and W. J. Spaan. 1991. Primary structure of the membrane and nucleocapsid protein genes of feline infectious peritonitis virus and immunogenicity of recombinant vaccinia viruses in kittens. Virology 181:327-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wesseling, J. G., H. Vennema, G. Godeke, M. C. Horzinek, and P. J. M. Rottier. 1994. Nucleotide sequence and expression of the spike (S) gene of canine coronavirus and comparison with the S proteins of feline and porcine coronaviruses. J. Gen. Virol. 75:1789-1794. [DOI] [PubMed] [Google Scholar]