Abstract
Reactivation of latent porcine cytomegalovirus after coculture of peripheral blood mononuclear cells (PBMCs) from pigs with different genetic backgrounds was investigated. Nine of 10 allogeneic coculture pairs were PCR (DNA) positive, whereas 7 coculture pairs had porcine cytomegalovirus (PCMV) RNA, an indication of virus replication. The cell subpopulations harboring PCMV were monocytes and CD8+ T cells.
Cytomegaloviruses (CMVs) have as a common feature the capacity for long-term virus persistence after primary infection (21), resulting in latency. In contrast to other herpesviruses, such as herpes simplex virus, which persists in neurons, and Epstein-Barr virus, which persists in B lymphocytes, little is known about the localization of latent CMV. It is suggested that latent CMV can be found in multiple body sites, unlike other herpesviruses, which remain latent in highly restricted areas of the body (7). Monocytes, CD34+ hematopoietic progenitor cells, and endothelial cells are major sites of human CMV (HCMV) latency (4, 15). Viral DNA is present in mononuclear cells derived from peripheral blood or bone marrow of healthy seropositive individuals, but infectious virus is not present (19).
Xenotransplantation has become an important area of research due to the limited supply of human cells, tissues, and organs for clinical transplantation. An overriding issue that impacts the potential application of porcine cells and tissues for use in humans is xenozoonosis (i.e., transmission of an infection from the pig cells to the human). In view of the present interest in xenotransplantation, porcine CMV (PCMV) may present a risk to humans. Phase I and II trials are under way in the United States and Europe using porcine cells and tissues in extracorporeal liver assist systems for liver failure, pancreatic islets for diabetes, and neural tissue for treatment of Parkinson's patients (13). PCMV is of particular concern because, much like its human counterpart, it is characterized by high seroprevalency in the swine population, has diverse clinical manifestations, and establishes latent infection (1, 12). HCMV is the most frequent cause of infection in organ transplants and is associated with an immunosuppressive state and allograft rejection (2, 14). In addition, HCMV causes severe and life-threatening disease in newborn infants and immunocompromised individuals (10, 18).
We proposed a study to examine whether latent PCMV can be reactivated, after coculture of PBMCs, through allogeneic stimulation. The endpoints used were PCR for detection of PCMV DNA, reverse transcription-PCR (RT-PCR) for detection of PCMV mRNA, and virus isolation.
Peripheral blood mononuclear cells (PBMCs) from pigs from two different source herds (herds A and B) were used as a source of allogeneic cells. The cells from 10 pigs from a PCMV-infected herd (herd A) were cocultured in pairs with the PBMCs from 10 pigs of a high-health-status herd (herd B) or cultured alone for up to 35 days. PBMCs were isolated from blood samples as previously described (17). For allogeneic stimulation experiments, equal numbers of cells from two different pigs, each from a different source herd, at a concentration of 5 × 107 cells/ml were mixed before plating. The PBMCs of all 20 pigs were also individually cultured. Cultures were washed and then maintained in media composed of 50% spent medium-50% fresh medium, which was replenished every 3 days for up to 35 days poststimulation. PBMCs from the cocultured allogeneic cells were collected at 0, 7, 14, 21, 28, and 35 days of coculture for detection of PCMV gene products and/or for virus recovery. Cell samples were sonicated and plated onto PFT (9) and PTK-75 (8) cells at subconfluency for virus isolation. The cells were incubated for up to 21 days and observed daily for the presence of cytopathic effect (CPE). The cultures with CPE were further analyzed for the presence of PCMV DNA by PCR. For PCR analysis, cell samples were collected at the indicated time points by scraping cells from the flasks. DNA and RNA were prepared using the QIAGEN blood and cell culture DNA kit and the RNeasy kit, respectively, according to the manufacturer's instructions (QIAGEN, Inc., Valencia, Calif.). A specific primer pair of the PCMV DNA polymerase gene was used in the PCR and RT-PCRs: forward, 5′ CCCTGATCTTAAATGACGAGGACGTGAC 3′; and reverse, 5′ ACCGTCTGAGAGACTGAACTTCTCTGACAC 3′ (5). Thirty-five amplification cycles were carried out in a GeneAmp PCR system 9600 (Perkin-Elmer, Foster City, Calif.). As controls for PCR and RT-PCR, DNA and cDNA were prepared from uninfected and PCMV-infected PFT cells. Control samples for detection of accidental contamination contained all reagents except for DNA or cDNA. An additional control was the DNase treatment of the prepared RNA samples. The PCR and RT-PCR amplification products were run on a 1% agarose gel containing ethidium bromide.
To determine which PBMC subpopulations harbored PCMV, porcine PBMCs were subjected to cell sorting using a fluorescence-activated cell analyzer (FACSCalibur; Becton Dickinson, Franklin Lakes, N.J.). The subpopulations of PBMCs were separated according to the following cell phenotypes for swine leukocyte antigens: CD4, CD8, and γδ for selected T-cell subpopulations; CD40 for B cells; and CD14 for monocytes/macrophages (6) using monoclonal fluorescein isothiocyanate-conjugated antibodies directed against the cell phenotype markers for swine leukocyte antigens. The specific cell types selected through the flow cytometry analysis were subjected to PCR analysis to determine which cell type(s) harbored the virus.
In this study, the possibility of reactivation of latent PCMV, after coculture of PBMCs, through allogeneic stimulation was investigated. The PBMCs of all 10 pigs from source herd A individually analyzed by PCR at days 0, 7, 14, 21, 28, and 35 were PCMV (DNA) positive. The PBMCs of all 10 pigs from source herd B individually analyzed by PCR at the indicated days 0 to 35 were PCMV (DNA) negative. Nine of the 10 pig-pig PBMC coculture pairs were PCR (DNA) positive at the same days of coculture. PCMV DNA polymerase RNA was first detected at day 14 poststimulation in 2 of 10 allogeneically stimulated cultures and at 21 days poststimulation in 7 of 10 allogeneically stimulated cultures (Table 1). The PBMCs of all 20 pigs individually analyzed by RT-PCR at the indicated days 0 to 35 postculture were RT-PCR negative. Infectious virus was demonstrated by isolation of PCMV from 5 out of 10 allogeneically stimulated cocultures at days 21 to 35 poststimulation (Table 2).
TABLE 1.
Detection of PCMV RNA following allogeneic stimulation
| Donor paira | Result at poststimulation day:
|
|||||
|---|---|---|---|---|---|---|
| 0 | 7 | 14 | 21 | 28 | 35 | |
| 1 + 2 | − | − | − | + | + | + |
| 3 + 4 | − | − | + | + | + | + |
| 5 + 6 | − | − | − | − | − | − |
| 7 + 8 | − | − | − | + | + | + |
| 9 + 10 | − | − | + | + | + | + |
| 11 + 12 | − | − | − | + | + | + |
| 13 + 14 | − | − | − | + | + | + |
| 15 + 16 | − | − | − | − | − | − |
| 17 + 18 | − | − | − | + | + | + |
| 19 + 20 | − | − | − | − | − | − |
Donor pigs with odd numbers came from source herd A, and donor pigs with even numbers came from source herd B.
TABLE 2.
Isolation of infectious virus from each donor pair
| Donor paira | Result at poststimulation dayb
|
|||||
|---|---|---|---|---|---|---|
| 0 | 7 | 14 | 21 | 28 | 35 | |
| 1 + 2 | − | − | − | − | − | − |
| 3 + 4 | − | − | − | + | + | + |
| 5 + 6 | − | − | − | − | − | − |
| 7 + 8 | − | − | − | − | − | − |
| 9 + 10 | − | − | − | + | + | + |
| 11 + 12 | − | − | − | + | + | + |
| 13 + 14 | − | − | − | − | − | − |
| 15 + 16 | − | − | − | − | + | + |
| 17 + 18 | − | − | − | − | − | − |
| 19 + 20 | − | − | − | − | + | + |
Donor pigs with odd numbers came from source herd A, and donor pigs with even numbers came from source herd b.
The presence of infectious virus was determined by the presence of CPE after cocultivation of sonicated PBMC allogeneic cultures with PFT and PTK-75 cells and was further confirmed by PCMV PCR.
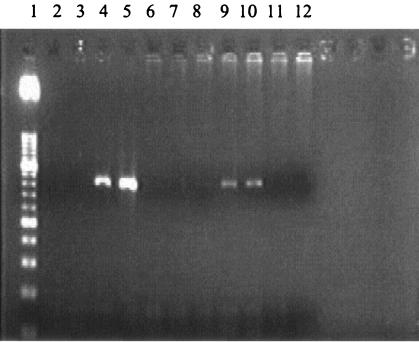
To identify the cell type(s) that harbors the virus, allogeneically stimulated cocultures were analyzed by flow cytometry for the expression of specific cell surface markers expressed on T and B cells and monocytes/macrophages. The PCMV DNA polymerase gene was detected in monocytes/macrophages as well as CD8+ cells (Fig. 1).
FIG. 1.
Presence of PCMV DNA in PBMC subpopulations at day 7 poststimulation. Lanes: 1, 50-bp DNA ladder; 2, PTK-75 cells as a negative control for PCMV Kanitz; 3, PFT cells as a negative control for PCMV Japan; 4, PCMV Kanitz as a positive control; 5, PCMV Japan as a positive control; 6, PCMV-negative CD40+ cells; 7, PCMV-negative γδ+ cells; 8, PCMV-negative CD4+ cells; 9, PCMV-positive CD8+ cells; 10, PCMV-positive CD14+ cells; and 11 and 12, PCMV-negative control with water instead of DNA.
The present study demonstrates that allogeneic stimulation resulted in reactivation of PCMV after coculture for up to 35 days. The experiments described in this study conclusively identify PBMCs as a cellular reservoir for PCMV and describe conditions allowing reactivation of latent virus from naturally infected cells. Since HCMV frequently reactivates in recipients of allogeneic blood transfusions or organ or bone marrow transplants, Soderberg-Naucler et al. (16) demonstrated HCMV could be reactivated from its latent state in allogeneically stimulated PBMCs obtained from naturally infected healthy blood donors. The endpoints used were PCR for detection of HCMV DNA, RT-PCR for detection of mRNA, and virus isolation. The detection of HCMV transcript through RT-PCR has been described as a way to differentiate between latency and active infection (3). In our study, a similar approach was used to differentiate between latency and active replication of PCMV by using primer sets based on the PCMV DNA polymerase sequence. The PCMV DNA polymerase gene is classified as a β2 (early) gene, and its expression requires prior expression of α (immediate-early) gene products (20), which leads to the assumption that viral DNA polymerase expression occurs only when the virus is actively replicating. Seven of 10 allogeneically stimulated cultures were RT-PCR positive for PCMV at 21 days poststimulation, and the individual PBMCs cultured alone were all RT-PCR negative. It therefore appears that, upon allogeneic stimulation, PCMV reactivation occurred.
Mueller et al. (11), using a pig-to-baboon xenotransplantation model, demonstrated that intense immunosuppression coupled with a xenogeneic immune response resulted in activation of PCMV and baboon CMV in both the baboon (the recipient) and in the porcine xenograft. While clinically evident disease was restricted to the native host species, the authors could detect low levels of PCMV in baboon tissue (and vice versa). All baboons used in this experiment (n = 6) demonstrated porcine genomic DNA in their tissues and baboon DNA in the porcine xenograft. The authors could not rule out whether this finding reflected infection, microchimerism, or uptake of DNA by resident phagocytic cells. These findings corroborate the present study, in which an allogeneic stimulation of pig PBMCs was enough to cause reactivation of PCMV.
PCMV DNA was detected in monocytes/macrophages as well as CD8+ cells. Von Laer et al. (22) showed that HCMV DNA could be detected most frequently in monocytes, but they also found it in CD8+ lymphocytes. These reports corroborate our findings that porcine monocytes/macrophages and CD8+—the same cell types that harbor HCMV—can harbor PCMV.
Due to the ubiquitous nature of PCMV in pig populations and within various tissues and cells of pigs, transplanted pig tissue or cells will likely contain PCMV. The findings from this study suggest that allogeneic and by extension xenogeneic contact between the transplanted tissues or cells and the host cells will activate the PCMV. Whereas it is not clear if PCMV can infect human cells, the reactivation and thus productive replication of PCMV within the transplanted cells or tissues may destroy the transplanted tissues or cells.
Acknowledgments
This work was supported by funding from the University of Minnesota Faculty Research Development Program and Academic Health Center Faculty Development grant and was partially supported by Nextran, Inc., Princeton, N.J. M. I. M. C. Guedes was partially supported by the Brazilian government sponsoring agency “Coodernacao de Aperfeicoamento de Pessoal de Nivel Superior” (CAPES).
We thank Shidong Ma for technical assistance.
REFERENCES
- 1.Edington, N., W. Plowright, and R. G. Watt. 1976. Generalized porcine cytomegalic inclusion disease: distribution of cytomegalic cells and virus. J. Comp. Pathol. 86:191-202. [DOI] [PubMed] [Google Scholar]
- 2.Gorensek, M. J., W. D. Carey, D. Vogt, and M. Goormastic. 1990. A multivariate analysis of risk factors for cytomegalovirus infection in liver-transplant recipients. Gastroenterology 98:1326-1332. [DOI] [PubMed] [Google Scholar]
- 3.Gozlan, J., F. Caburet, C. Tancrede, and J. C. Petit. 1992. A reverse polymerase chain reaction method for detection of human cytomegalovirus late transcripts in cells infected in vitro. J. Virol. Methods 40:1-10. [DOI] [PubMed] [Google Scholar]
- 4.Hahn, G., R. Jores, and E. S. Mocarski. 1998. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc. Natl. Acad. Sci. USA 95:3937-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamel, A. L., L. Lin, C. Sachvie, E. Grudeski, and G. P. S. Nayar. 1999. PCR assay for detecting porcine cytomegalovirus. J. Clin. Microbiol. 37:3767-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haverson, K., A. Saalmuller, B. Alvarez, F. Alonso, M. Bailey, A. T. Bianchi, W. J. Boersma, Z. Chen, W. C. Davis, J. Domingues, H. Engelhardt, A. Ezquerra, L. S. Grosmaire, M. J. Hamilton, E. Hollemweguer, C. A. Huang, K. V. Khanna, G. Kuebart, G. Lackovic, J. A. Ledbetter, R. Lee, D. Llanes, J. K. Lunney, K. C. McCullough, T. Molitor, J. Nielsen, T. A. Niewold, M. D. Pescovitz, J. M. de la Lastra, Z. Rehakova, H. Salmon, W. M. Schnitzlein, J. Seeback, A. Simon, J. Sinkora, M. Sinkora, C. R. Stokes, A. Summerfield, L. Sver, E. Thacker, L. Valpotic, H. Yang, F. A. Zuckermann, and R. Zwart. 2001. Overview of the Third International Workshop on Swine Leukocyte Differentiation Antigens. Vet. Immunol. Immunopathol. 80:5-23. [DOI] [PubMed] [Google Scholar]
- 7.Ho, M. 1990. Epidemiology of cytomegalovirus infection. Rev. Infect. Dis. 12:S701-S710. [DOI] [PubMed] [Google Scholar]
- 8.Kanitz C. L., and M. E. Woodruff. 1976. Cell-cultured-indirect fluorescent antibody studies of porcine cytomegalovirus. Proceedings of the 9th International Pig Veterinary Society Meeting.
- 9.Kawamura, H., T. Tajima, T. Hironao, T. Kajikawa, and T. Kotani. 1992. Replication of porcine cytomegalovirus in the 19-PFT cell line. J. Vet. Med. Sci. 54:1209-1211. [DOI] [PubMed] [Google Scholar]
- 10.Leach, C. T., J. D. Cherry, P. A. English, K. Hennessey, J. V. Giorgi, B. R. Visscher, J. P. Dudley, and R. Detels. 1993. The relationship between T-cell levels and CMV infection in asymptomatic HIV-1 antibody-positive homosexual men. J. Acquir. Immune Defic. Syndr. 6:407-413. [PubMed] [Google Scholar]
- 11.Mueller, N. J., R. N. Barth, S. Yamamoto, H. Kitamura, C. Patience, K. Yamada, D. K. C. Cooper, D. H. Sachs, A. Kaur, and J. A. Fishman. 2002. Activation of cytomegalovirus in pig-to-primate organ xenotransplantation. J. Virol. 76:4734-4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narita, M., M. Shimizu, H. Kawamura, M. Haritani, and M. Moriwaki. 1985. Pathologic changes in pigs with prednisolone-induced recrudescence of herpesvirus infection. Am. J. Vet. Res. 46:1506-1510. [PubMed] [Google Scholar]
- 13.Patterson, A. P. 1998. Current clinical trials in xenotransplantation, p. 18-26. In Developing U.S. public health policy in xenotransplantation. U.S. Department of Health and Human Services, Rockville, Md.
- 14.Pollard, A. B. 1988. Cytomegalovirus infections in renal, heart, heart-lung and liver transplantation. Pediatr. Infect. Dis. J. 7:97-102. [PubMed] [Google Scholar]
- 15.Sinclair, J. H., and J. G. Sissons. 1996. Latent and persistent infections of monocytes and macrophages. Intervirology 39:293-301. [DOI] [PubMed] [Google Scholar]
- 16.Soderberg-Naucler, C., K. N. Fish, and J. A. Nelson. 1997. Reactivation of latent human cytomegalovirus by allogenic stimulation of blood cells from healthy donors. Cell 91:119-126. [DOI] [PubMed] [Google Scholar]
- 17.Solano-Aguilar, G. I., K. G. Vengroski, E. Beshah, and J. K. Lunney. 2000. Isolation and purification of lymphocyte subsets from gut-associated lymphoid tissue in neonatal swine. J. Immunol. Methods 241:185-199. [DOI] [PubMed] [Google Scholar]
- 18.Stagno, S., R. F. Pass, G. Cloud, W. J. Britt, R. E. Henderson, P. D. Walton, D. A. Veren, F. Page, and C. A. Alford. 1986. Primary cytomegalovirus infection in pregnancy. Incidence, transmission to fetus, and clinical outcome. JAMA 256:1904-1908. [PubMed] [Google Scholar]
- 19.Stanier, P., D. L. Taylor, A. D. Kitchen, N. Wales, Y. Tryhorn, and A. S. Tyms. 1989. Persistence of cytomegalovirus in mononuclear cells in peripheral blood from blood donors. Br. Med. J. 299:897-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stinski, M. F. 1978. Sequence of protein synthesis in cells infected by human cytomegalovirus: early and late virus-induced polypeptides. J. Virol. 26:686-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor-Wiedeman, J., L. K. Borysiewicz, J. G. Sissons, and J. H. Sinclair. 1991. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J. Gen. Virol. 72:2059-2064. [DOI] [PubMed] [Google Scholar]
- 22.Von Laer, D., A. Serr, U. Meyer-Konig, G. Kirste, F. T. Hufert, and O. Haller. 1995. Human cytomegalovirus immediate early and late transcripts are expressed in all major leukocyte populations in vivo. J. Infect. Dis. 172:365-370. [DOI] [PubMed] [Google Scholar]