Abstract
A gag-based molecular beacon assay utilizing real-time nucleic acid sequence-based amplification technology has been developed to differentiate between the two genetic subclusters of human immunodeficiency virus type 1 (HIV-1) subtype C (C and C′) circulating in Ethiopia. Of 41 samples, 36 could be classified as C or C′ by sequencing of the gag gene. All 36 isolates were correctly identified by the gag beacon test. Three isolates with genomes that were recombinant in gag were unambiguously typed as belonging to the C′ subcluster. Further analysis revealed that these contained the most sequence homology with a reference subcluster C′ sequence in the target region of the beacon and hence were correct for the analyzed region. For one sample, sequencing and gag molecular beacon results did not match, while another isolate could not be detected at all by the beacon assay. Overall, high levels of sensitivity and specificity were achieved for both beacons (90.5% sensitivity and 100% specificity for the C beacon and 100% sensitivity and 95.2% specificity for the C′ beacon). The availability of a diagnostic test which can quickly and reliably discriminate between C and C′ HIV-1 infections in Ethiopia is an important first step toward studying their respective biological characteristics. As the assay is specific to the Ethiopian HIV-1 subtype C epidemic, it will contribute to characterizing the circulating viruses in this population, thereby generating the information necessary for the development of a potential efficacious HIV-1 vaccine appropriate for the Ethiopian context.
The regional distribution of human immunodeficiency virus type 1 (HIV-1) subtypes in the world is well recognized, but undoubtedly the place with the most genetic diversity is Africa, sub-Saharan Africa in particular (16, 18). It is in this region that the impact of the pandemic is felt most acutely, and over 70% of HIV-1-infected individuals worldwide reside in it (29). Ethiopia, which is located in the horn of Africa, is one of the countries hit hardest by the HIV/AIDS pandemic. The most recent surveillance data available from the Joint United Nations Programme on HIV/AIDS (UNAIDS) indicate that it ranks as the fifth most affected country from sub-Saharan Africa and as the sixth in the global context in terms of total numbers of individuals carrying the virus (29). Various studies carried out at different times with Ethiopian HIV-1 isolates have indicated that the overwhelming majority belong to the subtype C group (2, 4, 5, 10, 15, 32). A more in-depth analysis has revealed the presence of two distinct subtype C genotypes (C and C′) based on a phylogenetic analysis of the C2V3 envelope gene (3). No association was found between the two groups of viruses and the geographic locations or risk groups into which their carriers were divided (3). The functional significance of the two subclusters of HIV-1 subtype C remains to be elucidated.
In the present paper, we describe the development of a rapid and highly specific test to differentiate between the two subclusters of HIV-1 subtype C circulating in the country. The assay can identify whether a studied isolate has a subcluster C (HIV C) or C′ (HIV C′) genetic composition based on the gag gene. The test, which is based on a real-time nucleic acid sequence-based amplification (NASBA) format, has a number of potential applications. Besides contributing to the surveillance and characterization of HIV-1 viruses which are already circulating in the population, the gag-based beacon assay could serve as a valuable tool for the fast identification of breakthrough infections at the time when HIV-1 vaccine trials get under way in Ethiopia.
MATERIALS AND METHODS
Sample selection.
Seventeen plasma samples from a cohort of HIV-1-positive individuals enrolled in the Ethio-Netherlands AIDS Research Project were randomly selected for analysis. The details of the cohort group have been described elsewhere (20, 21). In addition, 24 serum samples obtained from 1988 and 1996-1997 from two population groups in Ethiopia (commercial sex workers and blood donors) were included in the evaluation. The subtypes of all isolates were previously determined based on the gag region (unpublished data).
Sample preparation.
HIV-1 RNAs were isolated from 100 μl each of the plasma and serum samples by employing a standard silica-based method (6). After the elution of total nucleic acids in 50 μl of PCR-grade water, 10 μl was used in a reverse transcription reaction and the DNA was amplified by a nested PCR using two sets of oligonucleotides, as described earlier (7), generating a 729-bp fragment of the gag gene comprising p17 and a part of the p24 coding region. The presence and purity of the PCR products were monitored by 1% agarose gel electrophoresis visualized by ethidium bromide staining.
DNA sequencing and analysis.
The amplified DNAs were directly sequenced on an ABI 373A automated sequencer (Applied Biosystems, Foster City, Calif.), using a Thermo Sequenase fluorescence-labeled primer cycle-sequencing kit (Amersham International, Little Chalfont, England) according to the manufacturer's instructions. All nested primers used for DNA amplification were extended with the SP6 and T7 sequences, and the PCR products were sequenced with the dye-labeled primers SP6 (5′ GATTTAGGTGACATATAG 3′) and T7 (5′ TAATACGACTCACTATAGGG 3′). Alignment of the sequences was performed manually based on the alignment of the Los Alamos database reference sequences for subtyping. Phylogenetic analysis of the aligned sequences was performed by the neighbor-joining method of MEGA (Institute of Molecular Evolutionary Genetics, Pennsylvania State University) and was confirmed by the DNADIST, NEIGHBOR, and DRAWTREE options of the PHYLIP software package (University of Washington, Seattle [http://evolution.Genetics.Washington.edu/phylip.html]). The distance matrix was generated by Kimura's two-parameter estimation in the MEGA software, and the tree topology was confirmed by the maximum likelihood option. Bootstrap values of >70% were considered significant based on 100 replications (14). Sequences obtained from the Los Alamos database were included as reference sequences. The bootscanning method was used to detect and study the recombinant viruses, as implemented in the SIMPLOT program (22; http://sray.med.som.jhmi.edu/RaySoft/SimPlot). The analysis was performed by calculating the distances for a sliding window of 200 nucleotides (nt) of the test sequences, moving along the alignment of a panel of reference sequences by increments of 20 bp. One hundred replications were generated by the bootstrap method for each window, and the percent bootstrap values were plotted against the nucleotide positions of the sequence of the reference panel.
NASBA primers and probes.
Real-time monitored NASBA (9) was used for the detection of HIV-1-derived RNAs and for genotype determination. Five microliters of RNA eluate obtained from the silica-based isolation method was added to the isothermal amplification reaction mixture, and the amplification was performed in a fluorimeter with a thermostat (Primagen, Amsterdam, The Netherlands). The oligonucleotide primers and probes and the conditions used were specifically adapted to recognize and amplify HIV-1 subtype C isolates from Ethiopia. The sequence of the 3′ antisense NASBA primer, elongated at its 5′ end with the T7 promoter recognition sequence (in italics), was AAT-TCT-AAT-ACG-ACT-CAC-TAT-AGG-GAG-AG-CTT-CTT-CTA-TTT-TGT-CTA-AGG-CT (nt 1108 to 1086 in strain HXB2 [GenBank accession number K03455]). The sequence of the 5′ sense primer was CAG-ACA-GGA-ACA-GAG-GAA-CT (nt 994 to 1013 in HXB2). The length of the amplicon was 115 nt. In developing the molecular beacons for the present study, we utilized the principle of incorporating additional mismatches as a way to enhance the specificity, as described earlier (8). The molecular beacons were targeted to an area within the amplified fragment of the gag gene (nt 1026 to 1045 in HXB2). A subtype C molecular beacon was designed to hybridize with isolates belonging to the C subcluster, while another molecular beacon (C′ molecular beacon) was designed to hybridize specifically with only those isolates belonging to the C′ subcluster. The nucleotides forming the typical stem structure of the beacons are represented in italics. The sequence of the subcluster C beacon was CGT-ACG-TAA-TAC-AGT-AGC-AAC-TCT-CTC-GTA-CG with the fluorophore FAM incorporated at its 5′ end, while the sequence of the subcluster C′ beacon was CGT-ACG-TAA-CAC-AGT-GTC-AAC-TCT-CTC-GTA-CG with the fluorophore ROX attached at its 5′ end. The 3′ ends of both molecular beacons were labeled with the dark quencher DABSYL. Real-time amplification detection reactions were performed at 41°C for 60 min, with a data point being generated every 30 s and measuring the fluorescence of both reporter labels. The fluorescence curves were standardized with the background set at 1.0. A sample was considered to react positively with the C molecular beacon of the monitored fluorophore (FAM) if the signal-to-noise ratio of the curve exceeded 1.3 and with the C′ beacon (ROX labeled) if this value exceeded 1.6.
RESULTS
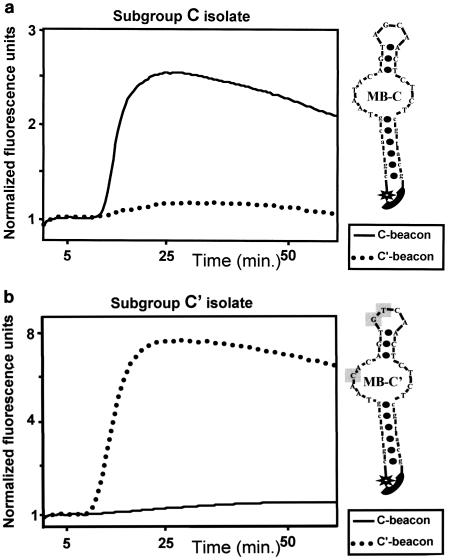
Two molecular beacons (Fig. 1a and b) were designed to distinguish between the two genetic subgroups of HIV-1 subtype C (C and C′) cocirculating in Ethiopia. They recognized the 3′ region of the p17 protein coding region in the gag gene, and both showed successful specific hybridization with their respective target sequences during NASBA amplification. Figure 1a demonstrates the amplification of a subgroup C virus and clearly shows the recognition of the amplicon by the beacon homologous to the C sequences but not by the one homologous to the C′ sequences. Likewise, the amplification of a subgroup C′ sequence and its recognition by the beacon homologous to the C′ sequences, but not by the one homologous to the C sequences, is shown in Fig. 1b. For each genotype, amplification led to a rise in fluorescence that was characteristic of the labeled fluorophores attached to their respective C or C′ sequence. The locus and sequence of the beacons and the amplification primers were chosen in a region of the gag gene near the junction of the p17 and p24 coding regions. This was done because in this region a sequence comparison showed significant homology between the two genotypes, so there would be no preferential amplification of either genotype. Nevertheless, in the beacon area there was a 2-nt variation between the two genotypes that was positioned in such a way that it would grant sufficient specificity of the beacons to the target area. Relative to the C beacon, an additional mismatch was incorporated into the C′ beacon design to enhance specificity. The results of the real-time NASBA amplification were compared to the characterization of the viruses by sequencing and subsequent phylogenetic analysis. Table 1 presents the sequencing results compared to those of the beacon test, illustrating a very good correlation. These results indicate that one mismatch from either the C or C′ beacon sequence in the samples tested did not adversely affect the capacity of the beacon to correctly predict the genotype.
FIG. 1.
Amplification by real-time NASBA and differential beacon detection of two Ethiopian virus isolates. (a) Isolate F-0008 M grouped with genotype C by sequencing. (b) Isolate F-0337P grouped with genotype C′. The primers used for the amplifications were identical for both isolates, and a mixture of two different detection beacons was included in both reactions. The beacon specific to the C genotype was labeled with the fluorophore FAM and the beacon specific to the C′ genotype was labeled with the fluorophore ROX. Panel A shows the preferential fluorescence increase of the genotype C beacon, while panel B shows the preferential fluorescence increase of the genotype C′ beacon.
TABLE 1.
Nucleotide sequence in the area targeted by gag-based molecular beacons
| Sample | Beacon target sequence (positions 1026 to 1045 in HXB2)a |
gag genotype
|
|
|---|---|---|---|
| Sequencing | Molecular Beacon | ||
| Beacon_C | TAATACAGTAGCAACTCTCT | ||
| Beacon_C′ | ---C-----GT--------- | ||
| AA88055 | ---C-----G---------- | C′ | C′ |
| AM88051 | ---C-----G---------- | C′ | C′ |
| AM88052 | ---C---------------- | C′ | C′ |
| GO88019 | ---C-----G---------- | C′ | C′ |
| AA97202 | ---C-----G-----C---- | C′ | C′ |
| AM96148 | ---C-----G-----C---- | C′ | C′ |
| DD96066 | ---C-----G---------- | C′ | C′ |
| DE96035 | ---C-----G---------- | C′ | C′ |
| JM96102 | ---C-----G---------- | C′ | C′ |
| JM96125 | ---C-----G-----C---- | C′ | C′ |
| F-0266V | ---C-----G---------- | C′ | C′ |
| F-0337P | ---C-----G---------- | C′ | C′ |
| F-0587P | ---C-----G--------T- | C′ | C′ |
| F-0651B | ---C----GG---------- | C′ | C′ |
| W-7168U | ---C-----G---------- | C′ | C′ |
| W-7173A | ---C-----G---------- | C′ | C′ |
| W-7452G | ---C-----G------A--- | C′ | C′ |
| F-0770G | ---C-----G---------- | Recombinant | C′ |
| F-0078U | ---C-------T-------- | Recombinant | C′ |
| GO88052 | ---C-----G-----G---- | Recombinant | C′ |
| DD96078 | ---C---------------- | C | C′ |
| GO96009 | ---C---------------- | C | Negative |
| AA88022 | -------------------- | C | C |
| AS88328 | -------------------- | C | C |
| DD88379 | -------------------- | C | C |
| DD88477 | -------------------- | C | C |
| DE88174 | -------------------- | C | C |
| DE88404 | -------------------- | C | C |
| AA96204 | -------------------- | C | C |
| AM96145 | C------------------- | C | C |
| AM96146 | -------------------- | C | C |
| DE96043 | ---------------C---- | C | C |
| GO960 | ------------G------- | C | C |
| F-0008M | -------------------- | C | C |
| F-0091W | -------------------- | C | C |
| F-0144W | -------------------- | C | C |
| F-0177B | -------------------- | C | C |
| F-0701W | -------------------- | C | C |
| F-0360X | -------------------- | C | C |
| W-7823Q | -------------------- | C | C |
| F-0324J | Not available | C | C |
Dashes indicate conserved sequences.
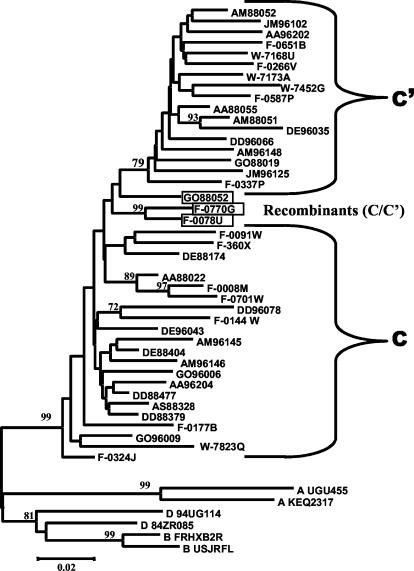
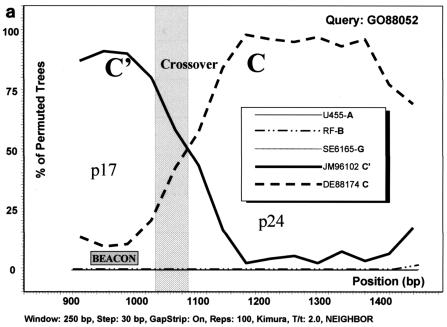
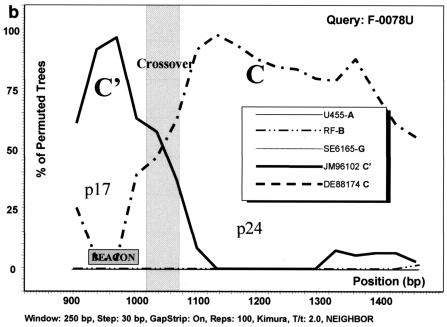
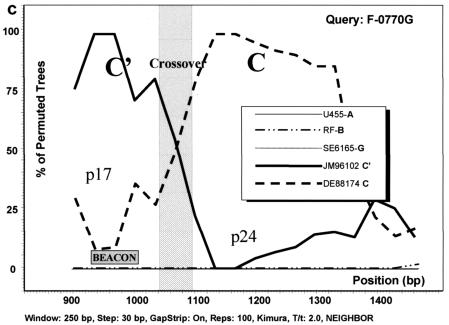
The phylogenetic analysis based on gag sequencing for all 41 samples studied revealed that most of the samples clustered with either the C (n = 21) or C′ (n = 17) genetic subgroup (Fig. 2). All bootstrap values of >70% are depicted, and the separation of the two genetic subclusters is evident. Three samples (GO88052, F-0078U, and F-0770G), although not clearly grouped with either genotype, reacted with the C′ molecular beacon in the NASBA assay (Table 1). All three were homologous to the C′ beacon at the target sequence. Under the assumption that these viruses were (C/C′) recombinants with a recombination breakpoint in the region of the gag gene that was amplified and sequenced, a sliding window bootscanning analysis was performed. The analysis revealed that these sequences were indeed C/C′ recombinants, and in all three cases, the area targeted by the beacon located in the p17 region of the gag gene clustered with significant bootstrap values with a reference C′ sequence (JM96102) (Fig. 3).
FIG. 2.
gag phylogenetic tree analysis of Ethiopian HIV-1 sequences by the neighbor-joining method of the DNADIST, NEIGHBOR, and DRAWTREE options of the PHYLIP software package. The two genotypes are indicated as C and C′. The sequences from the different towns are indicated by codes as follows: AA, Addis Ababa; AM, Arba Minch; AS, Assab; DD, Dire Dawa; DE, Dessie; JM, Jimma; and GO, Gondar. The first two digits following these codes indicate the year of sample collection, and the next three digits indicate the sample number. Subtype A (UG455 and KEQ2317), subtype D (94UG114 and 84ZR085), and subtype B (FRHxB2R and USJRFL) references were used. Numbers by the branches represent bootstrap values for 100 replications.
FIG. 3.
Analysis of C/C′ recombinant isolates by bootscanning, based on the neighbor-joining tree and Kimura 2 parameter methods with bootstrapping. The bootstrap values that supported the clustering of the sample sequences with the references are plotted. The chosen window size was 200 nt, moving in steps of 20 nt along the alignment, and the region of the gag gene studied was defined by nt 856 to 1587 of HXB2. (a) Isolate GO88052. (b) Isolate F-0078U. (c) Isolate F-0770G. The isolates UG455 (subtype A), RF (subtype B), and SE6165 (subtype G) together with DE88174 (Ethiopian subtype C) and JM96102 (Ethiopian subtype C′) were used as references. The plot analysis illustrates a crossover point between the p17 and p24 proteins in the gag gene, noted by shading, with the solid line identifying genotype C and the broken line identifying genotype C′. The shaded square shows the position of the beacon.
One sample (DD96078) showed a discrepancy between sequencing (shown to be of the C genotype) and molecular beacon prediction (predicted to be C′) of the genotype. Its sequence in the beacon region showed characteristics of C′ isolates by having a cytosine base at position 1029, identified as one of the signature motifs for identifying this genetic subcluster. However, it lacked the guanidine residue at position 1035 which is another distinguishing feature of C′ isolates, as shown in Table 1. Nevertheless, it was not possible to conclude that this particular mismatch will consistently lead to the same discrepancy, as another sample with the same sequence in the beacon region of its genome (AM88052) was correctly predicted to be C′ by the beacon, in agreement with the phylogenetic analysis of the sequence. For one other sample, there was not concurrence between sequencing and molecular beacon test results (GO96009). Although it belonged to the C group, as determined by sequencing, it was negative by the beacon assay. Its viral load, determined by a Nuclisens HIV-1 quantitative assay, was found to be below the quantification limit (80 copies/ml) (data not shown).
Of the 41 samples analyzed, there was one sample that tended to cluster separately from the main body of subtype C isolates (F-0324J), as shown in Fig. 2. A part of its sequence, including that at the position of the beacon, was not available, although sequencing of the remainder of the gag region amplified in the NASBA reaction showed that it belonged most closely to the C subcluster. The genotype by the gag beacon assay was also C. Table 1 summarizes the findings by the two methods for the 41 viral isolates studied.
The sensitivity and specificity characteristics of the beacons were evaluated for the panel of 41 isolates included in the present study. For the analysis, each beacon was considered separately, and the results are shown in Tables 2 and 3. Accordingly, sensitivities of 90.5 and 100% were obtained for the C and C′ beacons, respectively. The specificity values for the C and C′ beacons were 100 and 95.2%, respectively.
TABLE 2.
Sensitivity and specificity of subcluster C gag molecular beacona
| Result with subcluster C beacon | No. of samples with indicated result by sequencing
|
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 19 | 0 | 19 |
| Negative | 2 | 20 | 22 |
| Total | 21 | 20 | 41 |
Sensitivity of subcluster C beacon = 19/(19 + 2) × 100 = 90.5%. Specificity of subcluster C beacon = 20/(20 + 0) × 100 = 100%.
TABLE 3.
Sensitivity and specificity of subcluster C′ gag molecular beacona
| Result with subcluster C′ beacon | No. of samples with indicated result by sequencing
|
||
|---|---|---|---|
| Positive | Negative | Total | |
| Positive | 20 | 1 | 21 |
| Negative | 0 | 20 | 20 |
| Total | 20 | 21 | 41 |
Sensitivity of subcluster C′ beacon = 20/(20 + 0) × 100 = 100%. Specificity of subcluster C′ beacon = 20/(20 + 1) × 100 = 95.2%.
DISCUSSION
The use of molecular diagnostics in medicine has become widespread, particularly during the last decade (27). NASBA technology, which began in the mid-1990s, has been extensively used as a diagnostic tool for the detection of HIV RNA for both qualitative and quantitative screening of HIV-infected populations (11, 26, 30, 31). The most recent versions of diverse NASBA tests have incorporated molecular beacons into the assay format, enabling real-time detection of RNA amplification (28). The present report focuses on the development of a qualitative NASBA assay that utilizes molecular beacons specific for HIV-1 subtypes C and C′, which are found in Ethiopia. The rationale for developing such a test is twofold. Firstly, it will contribute to efforts at identifying and conducting surveillance of HIV-1 viruses that are transmitted in the population. Secondly, it will provide an appropriate tool for accurately distinguishing between the two subclusters in order to evaluate whether there are biological differences which characterize them. The intrasubtype clustering of sequences has also been reported for India, where like in Ethiopia, HIV-1 clade C dominates the epidemic (23). However, also in this case, clustering is based on sequence data, and due to the lack of an easily performed subtyping assay, epidemiological data related to intrasubtypes are missing.
The relative ease and reduced cost of performing the C/C′ gag molecular beacon NASBA assay compared to sequencing, considered the “gold standard” for genotyping, make the test a viable and less costly alternative for screening large numbers of samples, especially given the high rates of specificity. The results reported here show a marked improvement over other tests that have been evaluated in Ethiopia to determine HIV-1 genotypes, namely a V3 peptide enzyme-linked immunosorbent assay and a heteroduplex mobility assay (15). Although not directly comparable to the study reported in the present paper, the previous study aimed to compare the two methods by evaluating their capacities to distinguish between different HIV-1 genotypes, without any distinction of intrasubtype (C/C′) clustering. The sensitivities of these two methods in recognizing subtype C isolates were 82 and 88%, respectively (15). In particular, the V3 peptide enzyme-linked immunosorbent assay showed a high rate of serum cross-reactivity, especially between subtypes A and C, rendering it less useful as a diagnostic tool for subtyping purposes. Other investigators have also shown the limitations of using V3 serotyping to determine subtypes (25). The present results show the genotype for gag only, but together with a NASBA-molecular beacon test which targets the V3 region (currently under development), it may be possible to identify the genotype based on the env gene as well. This will allow more possibilities for conducting more facilitated epidemiological surveillance of the C/C′ HIV-1 epidemic in Ethiopia, including the identification of recombinants.
The sensitivity and specificity of the gag C and C′ beacons have been determined without reference to other subtypes in the present case, which considering the situation in Ethiopia, is not an anomaly. All reports to date show an overwhelming predominance of HIV-1 subtype C in that country. Subtype A viruses have been reported only in very few instances (2, 3, 24, 32) and more recently in a single individual from a cohort of HIV-1-infected women presenting with sexually transmitted diseases in Addis Ababa in 2001 (M. Adal, personal communication). In Ethiopia, except for two reported cases of subtype D viruses (3, 15), no other subtypes have been documented. These data fit well with observations from neighboring countries and others in the region, in which along with HIV-1 subtype C, other subtypes, mainly subtypes A and D, also cocirculate (12, 13, 17, 19). These subtypes are the ones which are most likely to appear in Ethiopia, given the proximity of the areas in which they predominate.
The predominance of subtype C HIV-1 in Ethiopia which has been previously reported is confirmed by the present findings. Based on the gag gene, most of the samples included in this study clustered into two distinct groups, with the C′ group forming a tighter cluster, as has been observed previously for the env gene (2, 3). Another study has shown that a number of Ethiopian HIV-1 isolates have recombinant (C/C′) genomes, with the env and gag genes being from the C′ and C subclusters, respectively (1). Three of the 41 isolates included in the present study (GO88052, F-0078U, and F-0770G) were identified to be recombinants in gag by sequencing. They were observed to have similar crossover points of about 60 nt between p17 and p24, either indicative of a “hot spot” for recombination within gag or else due to a founder effect. The gag beacon assay identified all three samples as belonging to the C′ subcluster because the region targeted by the beacon fell strictly in one part of the gag p17 gene where their sequences most closely resembled typical C′ isolates (Fig. 3). Because the hybridization sequence of the molecular beacon is only 20 nt, the possibility for identifying intragene recombinant viruses will be limited. In order to do so, a larger fragment of the genome would need to be targeted, which is not achievable by the beacon assay. Additionally, certain isolates identified as subtype C or C′ in gag by sequencing but having sequence variations in the area where the beacons hybridize (containing two or more mismatches with the beacon sequence) may be undetectable, despite substantial viral loads. Although such samples were not encountered in the present sample set, there have been observed cases of beacon negativity with other samples with known high viral loads (data not shown). Therefore, the interpretation of negative results in the gag beacon assay should not be taken as conclusive on its own but should be viewed in the context of clinical parameters as well. The negative sample GO96009 included in the present study had a low viral load which was below the lower limit of quantification of standard viral load assays. This may have contributed to the sample being undetected by the gag molecular beacon assay.
For one sample that yielded discrepant results (DD96078), the reason for the discrepancy could not be inferred from its sequence. The possibility of a double infection cannot be ruled out. If two viruses cocirculate in the same individual and one of them is present in a significantly smaller quantity, the chance of detection of that virus by PCR and sequencing is small. For this study, population (not clonal) sequencing was performed, so even if minor variants were present, it was still possible to miss the detection of a virus that was circulating in very low numbers with a sequence sufficiently distinct from the population consensus sequence represented by the majority in the population pool. In the sample mentioned above, although the phylogeny as determined by sequencing was clearly C, it is still possible that sequences from the C′ virus may have been selectively amplified in the NASBA reaction. One application of beacon technology in the future could be the investigation of dual virus infections.
Taken together, with the constraints of the assay included, the results presented here indicate that for the Ethiopian setting there is now a technology available which is capable of providing subtype information for gag, with nearly the same level of accuracy as sequencing. The method is able to distinguish between the two genotypes of subtype C (intrasubtype differentiation) better than previous technologies have been able to distinguish between different subtypes of HIV-1. The introduction of such a diagnostic tool opens up possibilities for more facilitated epidemiological surveillance of the HIV-1 C/C′ epidemic in that country. It remains to be elucidated if the genotypic differences translate into functional differences in the two groups, and one clinical parameter of interest is an investigation of possible pathogenic differences between them. This will be easier to carry out on a large scale given the availability of an assay which can quickly and reliably distinguish between the two subclusters. The technology may also be relevant for studying the response variation to antiretroviral therapy in persons infected with one or the other genetic subcluster of subtype C HIV-1 virus. In addition, it will help to elucidate which subcluster is more prone to developing resistance to specific antiretroviral drugs. Taking this a step further, the development of this type of technology could be expanded with the view to detect specific mutations which confer drug resistance in vivo. Furthermore, the NASBA-molecular beacon assay described in this paper has the potential for use as a diagnostic tool for identifying breakthrough infections during HIV vaccine trials, which may take place in Ethiopia in the future. In that event, the assay will yield valuable information by indicating which of the two genetic subclusters of subtype C HIV-1 is more likely to undergo vaccine escape.
In summary, the development of an assay to differentiate between the C and C′ genetic subclusters of HIV-1 subtype C is an important first step for studying their respective characteristics, be they similar or different. The C/C′ gag-based NASBA-molecular beacon assay is highly specific and compares favorably with sequencing results. It is also less labor-intensive, and results can be available within 2 h or less. These characteristics of the assay make it an ideal tool for conducting epidemiological surveys on a large scale. It also has a potential role in assessing the efficacy of intervention strategies such as vaccine trials. More specifically, since the assay is tailor-made for the Ethiopian HIV-1 epidemic, it will contribute to characterizing circulating viruses with the information necessary for the development of a potential efficacious HIV vaccine of local relevance.
Acknowledgments
This study is part of the Ethio-Netherlands AIDS Research Project (ENARP), a collaborative effort of the Ethiopian Health and Nutrition Research Institute (EHNRI), the Amsterdam Municipal Health Service (GG/GD), the Central Laboratory of The Netherlands Red Cross Blood Transfusion Service (CLB), and the Academic Medical Center of the University of Amsterdam (AMC). ENARP is financially supported by The Netherlands Ministry of Foreign Affairs and the Ethiopian Ministry of Health (MOH) as a bilateral project.
REFERENCES
- 1.Abebe, A. 2000. HIV-1 subtype C in Ethiopia: genotypic and phenotypic variation. Ph.D. thesis. University of Amsterdam, Amsterdam, The Netherlands.
- 2.Abebe, A., C. L. Kuiken, J. Goudsmit, M. Valk, T. Messele, T. Sahlu, H. Yeneneh, A. Fontanet, F. De Wolf, and T. F. R. de Wit. 1997. HIV type 1 subtype C in Addis Ababa, Ethiopia. AIDS Res. Hum. Retrovir. 13:1071-1075. [DOI] [PubMed] [Google Scholar]
- 3.Abebe, A., G. Pollakis, A. L. Fontanet, B. Fisseha, B. Tegbaru, A. Kliphuis, G. Tesfaye, H. Negassa, M. Cornelissen, J. Goudsmit, and T. F. R. de Wit. 2000. Identification of a genetic sub-cluster of HIV-1 subtype C (C′) widespread in Ethiopia. AIDS Res. Hum. Retrovir. 16:1909-1914. [DOI] [PubMed] [Google Scholar]
- 4.Ayehunie, S., B. Johansson, A. Sonnerborg, D. W. Zewdie, S. Britton, and O. Strannegard. 1993. Sequence analysis of selected regions of the env (V3 loop and gp41) and gag (p7) reading frames of Ethiopian human immunodeficiency virus type 1 strains. Arch. Virol. 128:229-239. [DOI] [PubMed] [Google Scholar]
- 5.Ayehunie, S., B. Johansson, M. Salminen, P. Leinikki, A. Sonnerborg, D. W. Zewdie, S. Britton, and O. Strannegard. 1991. HIV-1 in Ethiopia: phylogenetic divergence from other HIV-1 strains. Virus Genes 5:359-366. [DOI] [PubMed] [Google Scholar]
- 6.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. Van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cornelissen, M., G. Kampinga, F. Zorgdrager, J. Goudsmit, and the UNAIDS Network for HIV Isolation and Characterization. 1996. Human immunodeficiency virus type 1 subtypes defined by env show high frequency of recombinant gag genes. J. Virol. 70:8209-8212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Baar, M. P., E. C. Timmermans, M. Bakker, E. de Rooij, B. van Gemen, and J. Goudsmit. 2001. One-tube real-time isothermal amplification assay to identify and distinguish HIV-1 subtypes A, B, and C and CRFs AE and AG. J. Clin. Microbiol. 39:1895-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Baar, M. P., M. W. van Dooren, E. de Rooij, M. Bakker, B. van Gemen, J. Goudsmit, and A. de Ronde. 2001. Single rapid real-time monitored isothermal RNA amplification assay for quantification of human immunodeficiency virus type 1 isolates from groups M, N, and O. J. Clin. Microbiol. 39:1378-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Wit, T. F. R., A. Tsegaye, D. Wolday, B. Hailu, M. Aklilu, E. Sanders, M. Hagos, A. Kliphuis, G. Pollakis, A. Krol, R. Geskus, F. Miedema, J. Goudsmit, R. Coutinho, and A. L. Fontanet. 2002. Primary HIV-1 subtype C infection in Ethiopia. J. Acqir. Immun. Defic. Syndr. 30:463-470. [DOI] [PubMed] [Google Scholar]
- 11.Guatelli, J. C., K. M. Whitfield, D. Y. Kwoh, K. J. Barringer, D. D. Richman, and T. R. Gingeras. 1990. Isothermal in vitro amplification of nucleic acids by multi-enzyme reaction modeled after retroviral replication. Proc. Natl. Acad. Sci. USA 87:1874-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris, M. E., D. Serwadda, N. Sewankambo, B. Kim, G. Kigozi, N. Kiwanuka, J. B. Phillips, F. Wabwire, M. Meehen, T. Lutalo, J. R. Lane, R. Merling, R. Gray, M. Wawer, D. L. Birx, M. L. Robb, and F. E. McCutchan. 2002. Among 46 near full length HIV type 1 genome sequences from Rakai District, Uganda, subtype D and AD recombinants predominate. AIDS Res. Hum. Retrovir. 18:1281-1290. [DOI] [PubMed] [Google Scholar]
- 13.Hierholzer, M., R. R. Graham, I. El Khidir, S. Tasker, M. Darwish, G. D. Chapman, A. H. Fagbami, A. Soliman, D. L. Birx, F. E. McCutchan, and J. K. Carr. 2002. HIV type 1 strains from east and west Africa are intermixed in Sudan. AIDS Res. Hum. Retrovir. 18:1163-1166. [DOI] [PubMed] [Google Scholar]
- 14.Hillis, D. M., and J. J. Bull. 1993. An empirical test for bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst. Biol. 42:182-192. [Google Scholar]
- 15.Hussein, M., A. Abebe, G. Pollakis, M. Brouwer, B. Petros, A. L. Fontanet, and T. F. R. de Wit. 2000. HIV-1 subtype C in commercial sex workers in Addis Ababa, Ethiopia. J. Acquir. Immun. Defic. Syndr. 23:120-127. [DOI] [PubMed] [Google Scholar]
- 16.Janssens, W., A. Buve, and J. N. Nkengasong. 1997. The puzzle of HIV-1 subtypes in Africa. AIDS 11:705-712. [DOI] [PubMed] [Google Scholar]
- 17.Koulinska, I. N., G. Msamanga, D. Mwakagile, M. Essex, and B. Renjifo. 2002. Common genetic arrangements among human immunodeficiency virus type 1 A and D recombinant genomes vertically transmitted in Tanzania. AIDS Res. Hum. Retrovir. 18:947-956. [DOI] [PubMed] [Google Scholar]
- 18.Louwagie, J., W. Janssens, J. Mascola, L. Heyndrickx, P. Hegerich, G. van der Groen, F. E. McCutchan, and D. S. Burke. 1995. Genetic diversity of the envelope glycoprotein from human immunodeficiency virus type 1 isolates of African origin. J. Virol. 69:263-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poss, M., J. Gosink, E. Thomas, J. K. Kreiss, J. Ndinya-Achola, K. Mandaliya, J. Bwayo, and J. Overbaugh. 1997. Phylogenetic evaluation of Kenyan HIV type 1 isolates. AIDS Res. Hum. Retrovir. 13:493-499. [DOI] [PubMed] [Google Scholar]
- 20.Sahlu, T., A. Fontanet, T. F. R. de Wit, T. Messele, R. Doorly, H. Yeneneh, P. Bindels, and R. Coutinho. 1998. Identification of a site for a cohort study on natural history of HIV infection in Ethiopia. J. Acquir. Immun. Defic. Syndr. Hum. Retrovir. 17:149-155. [DOI] [PubMed] [Google Scholar]
- 21.Sahlu, T., E. Kassa, T. Agonafer, A. Tsegaye, T. F. R. de Wit, H. Gebre-Mariam, R. Doorly, I. Spijkerman, H. Yeneneh, R. Coutinho, and A. L. Fontanet. 1999. Sexual behaviours, perception of risk of HIV infection, and factors associated with attending HIV post-test counseling in Ethiopia. AIDS 13:1263-1271. [DOI] [PubMed] [Google Scholar]
- 22.Salminen, M. O., J. K. Carr, D. S. Burke, and F. E. McCutchan. 1995. Identification of breakpoints in intergenotypic recombinants of HIV type 1 by bootscanning. AIDS Res. Hum. Retrovir. 11:1423-1425. [DOI] [PubMed] [Google Scholar]
- 23.Shankarappa, R., R. Chatterjee, G. H. Learn, D. Neogi, M. Ding, P. Roy, A. Ghosh, L. Kingsley, L. Harrison, J. I. Mullins, and P. Gupta. 2001. Human immunodeficiency virus type 1 env sequences from Calcutta in eastern India: identification of features that distinguish subtype C sequences in India from other subtype C sequences. J. Virol. 75:10479-10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sherefa, K., B. Johansson, M. Salminen, and A. Sonnerborg. 1998. Full-length sequence of human immunodeficiency virus type 1 subtype A, recombined with subtype C in the env V3 domain. AIDS Res. Hum. Retrovir. 14:289-292. [DOI] [PubMed] [Google Scholar]
- 25.Sherefa, K., M. Sallberg, B. Johansson, M. Salminen, and A. Sonnerborg. 1997. Subtyping of human immunodeficiency virus type 1 strains by using antibodies specific for the third variable domain (V3) of gp120: results may be affected by divergent V3 sequences. J. Clin. Microbiol. 35:2419-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simonds, R. J., T. M. Brown, D. M. Thea, S. L. Orloff, R. W. Steketee, F. K. Lee, P. E. Palumbo, and M. L. Kalish for the Perinatal AIDS Collaborative Transmission Study. 1998. Sensitivity and specificity of a qualitative RNA detection assay to diagnose HIV infection in young infants. AIDS 12:1545-1549. [DOI] [PubMed] [Google Scholar]
- 27.Tang, Y. W., G. W. Procop, and D. H. Persing. 1997. Molecular diagnostics of infectious diseases. Clin. Chem. 43:2021-2038. [PubMed] [Google Scholar]
- 28.Tyagi, S., and F. R. Kramer. 1996. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14:303-308. [DOI] [PubMed] [Google Scholar]
- 29.UNAIDS. 2002. Report on the global HIV/AIDS epidemic. UNAIDS/02.26E. UNAIDS, Geneva, Switzerland.
- 30.Van Gemen, B., T. Kievits, P. Nara, H. G. Huisman, S. Jurriaans, J. Goudsmit, and P. Lens. 1993. Qualitative and quantitative detection of HIV-1 RNA by nucleic acid sequence-based amplification. AIDS 7:S107-S110. [DOI] [PubMed] [Google Scholar]
- 31.Van Gemen, B., T. Kievits, R. Schukkink, D. van Strijp, L. T. Malek, R. Sooknanan, H. G. Huisman, and P. Lens. 1993. Quantification of HIV-1 RNA in plasma using NASBA during HIV-1 primary infection. J. Virol. Methods 43:177-187. [DOI] [PubMed] [Google Scholar]
- 32.Zewde, A., S. Bahiru, E. Sanders, T. Tilahun, A. Beyene, M. Alebachew, A. Schaap, D. Wolday, and T. F. R. de Wit. 2002. HIV-1 seroprevalence and subtypes in police recruits from Afar regional state, Ethiopia. Ethiop. Med. J. 40(Suppl. 1):1-10. [PubMed] [Google Scholar]