Abstract
Background
The estimated prevalence of HCV infection in Argentina is around 2%. However, higher rates of infection have been described in population studies of small urban and rural communities. The aim of this work was to compare the origin and diversification of HCV-1b in samples from two different epidemiological scenarios: Buenos Aires, a large cosmopolitan city, and O'Brien, a small rural town with a high prevalence of HCV infection.
Patients and Methods
The E1/E2 and NS5B regions of the viral genome from 83 patients infected with HCV-1b were sequenced. Phylogenetic analysis and Bayesian Coalescent methods were used to study the origin and diversification of HCV-1b in both patient populations.
Results
Samples from Buenos Aires showed a polyphyletic behavior with a tMRCA around 1887–1900 and a time of spread of infection approximately 60 years ago. In contrast, samples from ÓBrien showed a monophyletic behavior with a tMRCA around 1950–1960 and a time of spread of infection more recent than in Buenos Aires, around 20–30 years ago.
Conclusion
Phylogenetic and coalescence analysis revealed a different behavior in the epidemiological histories of Buenos Aires and ÓBrien. HCV infection in Buenos Aires shows a polyphyletic behavior and an exponential growth in two phases, whereas that in O'Brien shows a monophyletic cluster and an exponential growth in one single step with a more recent tMRCA. The polyphyletic origin and the probability of encountering susceptible individuals in a large cosmopolitan city like Buenos Aires are in agreement with a longer period of expansion. In contrast, in less populated areas such as O'Brien, the chances of HCV transmission are strongly restricted. Furthermore, the monophyletic character and the most recent time of emergence suggest that different HCV-1b ancestors (variants) that were in expansion in Buenos Aires had the opportunity to colonize and expand in O’Brien.
Introduction
Hepatitis C virus (HCV) infection is a major cause of chronic hepatitis, liver cirrhosis and hepatocellular carcinoma [1], [2]. About 150 million people worldwide are chronically infected with HCV, and more than 350 000 people die every year from HCV-related liver diseases. Taxonomically, HCV is classified into six major genotypes (1 to 6) and into many subtypes based on phylogenetic analysis [3], [4]. Genotype distribution differs by epidemiological characteristics and geographical areas [5]. Before the adoption of anti-HCV control measures in blood banks (∼1990), HCV was mainly transmitted via blood transfusion. Today, needle sharing among injecting drug users is the most common form of HCV transmission [6], [7]. Therefore, the distribution of certain subtypes such as HCV-1a, 1b and 3a is largely related to risk factors such as blood transfusions, intravenous drug use or inadequately sterilized medical equipment [5], [8], [9].
The prevalence of HCV infection varies among different geographic regions. Kershenobich et al. has recently published a review describing the status of HCV infection in six countries of Latin America, including Argentina [10]. These authors observed that HCV prevalence ranges from 1.0 to 2.3% and that genotype 1 is the most prevalent in the countries analyzed [10].
In Argentina, the available information is limited and heterogeneous. Although a Consensus Conference estimated an overall HCV prevalence of around 2% (Consenso Argentino de Hepatitis C, 2007), marked differences have been noted along the country. The highest reported prevalence has been observed in population-based studies performed in small urban and rural towns [11]–[15]. More precisely, a recent study in the small rural town of Wheelwright (Santa Fe, Argentina), which has a 4.9% prevalence of infection, revealed a monophyletic clade and a time for the most recent common ancestor (tMRCA) dated around 1950. The study showed that HCV infection was more frequent in people older than 50 years, with the highest prevalence (22%) in the group aged between 70–79 years [16]. A similar situation has been reported from ÓBrien, another small rural town of approximately 2300 inhabitants located in Buenos Aires Province [14]. The overall prevalence of HCV infection in O’Brien was found to be 5.6%, 0.6% in individuals aged <40 years and 12.6% in those >40 years, with a peak of 23% in the group aged 60–70 years. All patients from O’Brien were infected with HCV genotype 1b. However, the information about the origin of HCV infection was evaluated from the epidemiological forms and phylogenetic analysis was restricted. Therefore, it would be interesting to perform a strong phylogenetic analysis and complement it with a coalescence analysis in order to compare the diversity and origin of samples from small towns and a large cosmopolitan city.
Evolutionary analyses of virus sequences are now an important tool in molecular epidemiology. These methods have been previously used to evaluate the relationships between viral gene sequences and/or to estimate epidemiological history [16]–[26]. Therefore, the aim of this study was to compare the origin and diversification of HCV-1b in two populations of Argentina with different epidemiological characteristics, by means of phylogenetic and Bayesian coalescent analysis.
Materials and Methods
Design and Study Population
From June 1996 to July 2010, the patients infected with HCV genotype 1b who received their first therapy against HCV infection were prospectively followed at four tertiary care centers from Buenos Aires (a large city with 13 million inhabitants). A basal serum sample was collected from each patient and stored at −80°C for subsequent determinations. A total of 46 individuals from this cohort were randomly selected from the four hospitals: CEMIC (n = 10), Italian Hospital (n = 21), Lanari Hospital (n = 8) and Muñiz Hospital (n = 7). On the other hand, 37 samples randomly selected from 93 patients infected with HCV genotype 1b from O’Brien, a small rural town in Buenos Aires province, epidemiologically described by Picchio et al. [14], were also included in this study. Written informed consent to participate in this study was obtained from all patients of the participating hospitals. The protocol of this study was approved by the ethics committee of the Facultad de Farmacia y Bioquímica of the Universidad de Buenos Aires (Exp #701283).
RNA Extraction, cDNA Synthesis and DNA Amplification
RNA was extracted from 150 ml serum samples using a commercial reagent (Trizol, Invitrogen) following the manufacturer’s instructions. Reverse transcription reactions and the E1/E2 (672 nt, positions 1416–2087) and NS5B (486 nt, positions 8139–8624) genes were amplified by a hemi-nested PCR and the amplicons sequenced as described previously [16]. The D90208 (HCV-J) was used as reference sequence.
Phylogenetic Analysis
Phylogenetic analysis was performed using the E1/E2 and NS5B genomic regions by separate and concatenated datasets from Buenos Aires and O’Brien (except from samples from Lanari and Muñiz Hospitals, where only NS5B was analyzed due to lack of serum availability). On the other hand, 352 HCV-1b sequences from GenBank were added as reference [including 55 sequences from the Wheelwright outbreak previously described in [16]]. Additionally, two sequences of HCV-1a and two of HCV-1c were included as out-group. Thus, the total population included in the phylogenetic analysis consisted of 439 sequences. These sequences were aligned with ClustalX v2.0.10 program [27].
Phylogenetic trees were constructed using distance Neighbor-Joining, Maximum Likelihood (ML) and Parsimony methods. Evolutionary models were inferred according to the Akaike Information Criterion (AIC) statistics obtained with the jModelTest v0.1.1 program [28]. The distance analysis was performed with the PAUP* v4.0b10 program [29]. The ML analysis was performed with the RAxML v7.2.8 program [30] and the parsimony analysis was performed with the TNT v1.1 program [31], [32]. The robustness of the reconstructed phylogenies was evaluated by bootstrap analysis (1000 replica), and the phylogenetic trees were analyzed using the Dendroscope v2.7.4 program [33].
Divergence Time and Population Dynamics
Bayesian coalescent-based methods were used to estimate the tMRCA and the population dynamics in: a) Buenos Aires dataset and b) O’Brien dataset (n = 36, without sample O529, which did not belong to O’Brien cluster). This analysis was performed for the E1/E2 and NS5B regions and the concatenated matrix of both regions. The estimates of the tMRCA and the population dynamics were obtained by means of the Bayesian Markov Chain Monte Carlo techniques implemented in the BEAST v1.6.1 program [34]. Both strict and relaxed uncorrelated lognormal molecular clock were enforced. The demographic histories of Buenos Aires and O’Brien datasets were reconstructed using a flexible model known as Bayesian Skyline Plot (BSP) implemented in the BEAST v1.6.1 program [34], [35].
As the O’Brien datasets were isochronous and the Buenos Aires datasets presented a weak temporal structure assessed by Path-O-Gen v1.4 program, we could not estimate the tMRCA parameter reliably, and so used external information to estimate the tMRCA and population dynamics. For this reason, it was necessary to incorporate an external nucleotide substitution rate as previously described by Magiorkinis et al. [21]. Thus, as prior, the regions were calibrated with a normal distribution. The mean rate of substitution obtained and introduced as a prior for E1/E2 was 2.28×10−3 s/s/y (stdev = 4.17×10−4), that for NS5B was 6.10×10−4 s/s/y (stdev = 1.96×10−4) and that for the concatenate analysis was 1.22×10−3 s/s/y (stdev = 2.64×10−3). For the Buenos Aires dataset, the sampling time was incorporated in the analysis since the inclusion do not affect the time of the tMRCA. On the other hand, the tMRCA for the samples from O’Brien was inferred using a normal distribution with a mean value of 50 years and a standard deviation of 20 years as prior distribution, based on our belief about the history of the epidemic.
Finally, the best substitution model inferred according to the AIC statistics obtained with the jModelTest v0.1.1 program [28] was selected for each dataset and implemented in the BEAUti program. The convergence of the parameters to a stationary distribution was assessed with the TRACER v1.5.0 program [36] , and the models compared by a Bayes Factor analysis [37].
Nucleotide Sequence Accession Numbers
GenBank accession numbers for the sequences obtained in this study are KF733833–KF733900 for the E1/E2 sequences and KF733901–KF733983 for the NS5B sequences.
Results
Main Characteristics of the Patients Studied
Forty-six patients from Buenos Aires and 37 from O’Brien were analyzed. Twenty-seven (58.7%) patients from Buenos Aires and 19 (51.3%) from O’Brien were male. The median (Interquartile Range) age for Buenos Aires was 58 (50–66) years and for O’Brien 63 (53–68). Among Buenos Aires patients, 44 (95.7%) were HCV-monoinfected and 2 (4.3%) were HIV/HCV-coinfected, whereas all patients from O’Brien were HCV-monoinfected. Risk factors for HCV infection in patients from Buenos Aires were blood transfusions in 10 of them (21.7%), use of intravenous drugs (IDU) in 7 of them (15.2%), and sexual contact in 2 of them (4.3%). The route of infection in the remaining 25 patients (54.3%) was unknown.
All 37 (100%) HCV-positive patients from O’Brien had received injections with non-disposable and inadequately sterilized syringes and needles administered by the same health care professional. In addition, 29 out of the 37 patients (78.4%) had undergone dental care with the same professional who had used non-disposable and re-sterilized materials. Other risk factors included surgical procedures in 25 (67.6%) patients and blood transfusions in 9 (24.3%).
Phylogenetic Analysis
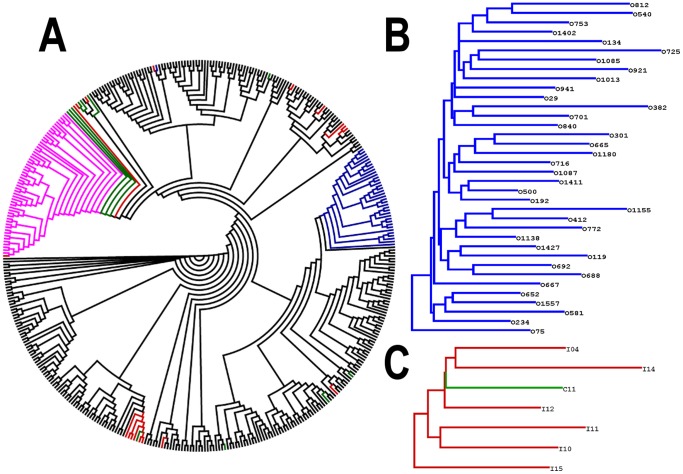
In the separate analyses, principally for the NS5B region, the bootstrap values were lower than those in the concatenated analysis. Thus, the increase in the bootstrap values for the concatenated analysis could be the result of lower phylogenetic signal in the datasets from separate analyses (data not shown). For this reason, the phylogenetic analysis of concatenated regions was the one taken into account. This analysis showed that all sequences from O’Brien, except one (O529), formed a single monophyletic cluster. In addition, this cluster was independent of the Wheelwright cluster previously described by our group [16] (Figure 1). In contrast, most samples from Buenos Aires (CEMIC and Italian Hospital) appear intermingled among the reference sequences, showing a polyphyletic behavior (Figure 1). However, seven of them form a small cluster (BA7) that includes one sequence from CEMIC and six sequences from the Italian Hospital (Figure 1).
Figure 1. Phylogenetic tree from the concatenated analysis for ML.
A) Corresponds to the global analysis of concatenated sequences. B) O’Brien cluster pruned from A (Bootstrap = 100%). C) Cluster BA7 pruned from A (Bootstrap = 98%). Red branches correspond to Italian Hospital, green branches to CEMIC, magenta branches to Wheelwright and black branches to the reference sequences.
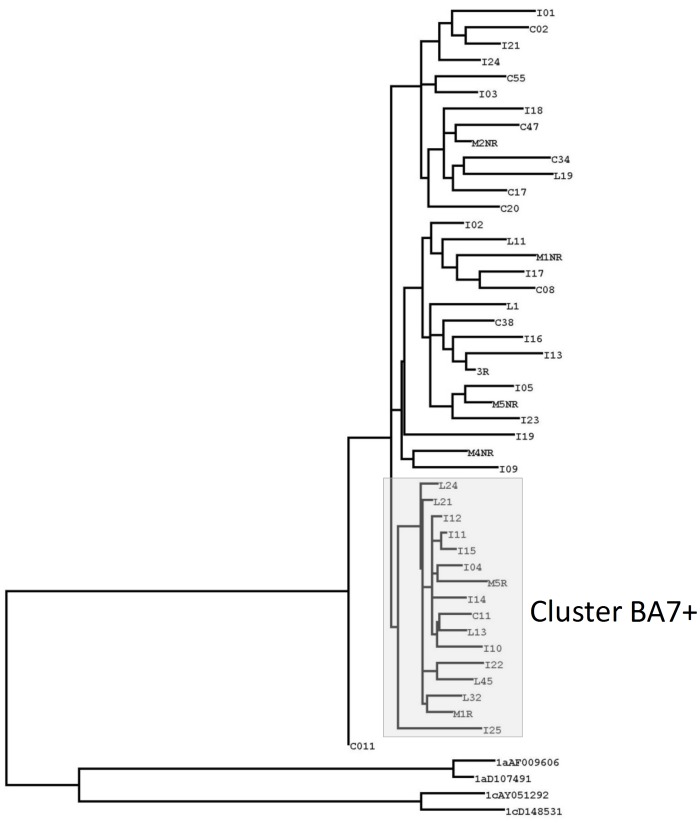
To extend our phylogenetic study, we performed an analysis for NS5B, which included more samples from Buenos Aires. Here, we observed that cluster BA7 incorporates seven sequences from Lanari and Muñiz Hospitals and was denominated cluster BA7+ (Figure 2). However, the I22 sequence of Italian Hospital was incorporated into this cluster. This sequence was not grouped in cluster BA7 in the phylogenetic analysis shown in figure 1.
Figure 2. ML Phylogenetic tree from NS5B region.
The box shows cluster BA7+ (Bootstrap value = 31%). C, I, L and M corresponds to the sequences from CEMIC, Italian Hospital, Lanari Hospital and Muñiz Hospital, respectively. Two sequences of HCV-1a and two sequences of HCV-1c were used as an out-group.
Coalescent Bayesian Analysis
The E1/E2 and NS5B partial sequence genes from the samples from Buenos Aires and O’Brien were used to estimate both the tMRCA and population dynamics. From the Bayes Factor analysis, the lognormal relaxed molecular clock model was the one that best fit the data for E1/E2 and concatenated matrices. Nonetheless, there was no significant difference between the strict and lognormal relaxed molecular clock for the NS5B region. Therefore, for the NS5B region, the strict molecular clock was selected to avoid over-parameterization. Table 1 shows the tMRCA values obtained from the samples from Buenos Aires calculated from the BSP model. Similar results were obtained when analyzing the other demographic models (constant population size, exponential growth, expansion growth, logistic growth) that achieved convergence, suggesting that these results are robust to demographic history (data not shown).
Table 1. Estimates of the tMRCA for Buenos Aires and O’Brien samples by Bayesian coalescent analysis under the BSP model.
| Datasets | Concatenated | E1/E2 | NS5B |
| Year (HDP 95%) | Year (HDP 95%) | Year (HPD 95%) | |
| Buenos Aires | NA | NA | 1887 (1805–1935) |
| Buenos Aires (without L and M) | 1905 (1838–1933) | 1939 (1917–1956) | 1900 (1795–1957) |
| Buenos Aires (without BA7+) | NA | NA | 1866 (1784–1933) |
| BA7+ | NA | NA | 1959 (1941–1978) |
| O’Brien | 1950 (1934–1964) | 1961 (1949–1972) | 1961 (1940–1998) |
HPD = highest posterior density (95%HPD).
NA = Not available.
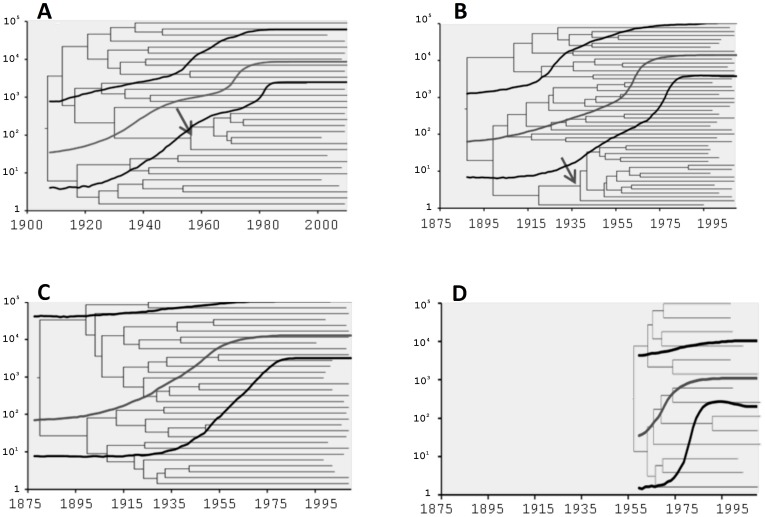
Additionally, the population dynamics for all four Buenos Aires datasets were analyzed through the different demographic profiles obtained with the BSP model (Figure 3). The first dataset showed an exponential growth in the diversity measured as effective number of population multiplied by the generation length (Ne×τ) from around 1920 until about 1980 and then remained constant until the present (Figure 3A).
Figure 3. Demographic profiles and coalescence trees obtained by the BSP model.
A) CEMIC and Italian Hospital, concatenated sequences. B) Buenos Aires, NS5B sequences. C) Buenos Aires without cluster BA7+. D) BA7+ sequences. The lines in the demographic curves correspond to the median value and the estimated HPD95%. The arrow in A and B represents the node of clusters BA7 and BA7+, respectively. The length of the branches of the tree represents the time (year).
Interestingly, the diversification of cluster BA7, whose emergence would be responsible for the second phase in the effective number of population, occurred in this period. Figure 3B shows a similar curve but in this case, the second step could be due to the formation of cluster BA7+, which includes samples from Lanari and Muñiz Hospitals. In figure 3C, without cluster BA7+, a softer curve was observed with only one step of exponential growth. Finally, figure 3D describes the exponential growth of cluster BA7+.
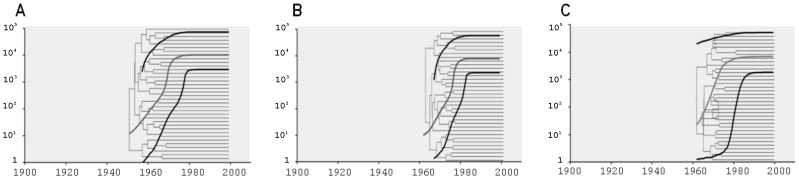
On the other hand, the tMRCA for the samples from O’Brien (HPD: High Posterior Density) was around 1950 (1934–1964) for the concatenated analysis, 1961 (1949–1972) for E1/E2 and 1961 (1940–1998) for NS5B (Table 1). It is important to note that the same results were obtained when the prior for the tMRCA (based on our belief about the history of the epidemic) was not included in the analysis (data not shown). Besides, the demographic profile obtained for O’Brien sequences describes a rapid exponential growth along 15–20 years, which then remains constant until the present (Figure 4).
Figure 4. Demographic profiles and coalescence trees obtained by the BSP model for O’Brien sequences.
A) Concatenated sequences, B) E1/E2 analysis and C) NS5B analysis. The length of the branches of the tree represents the time (year). The lines in the demographic curves correspond to the median value and the estimated HPD95%. The length of the branches of the tree represents the time (year).
Discussion
Demographic differences between cities and small towns are reflected in the molecular epidemiology of viral infections. In this study, analysis of HCV sequences from infected patients from Buenos Aires and O’Brien showed important differences in regard to phylogeny, origin and dispersion. While HCV infection in Buenos Aires had a polyphyletic behavior and an exponential growth in two phases, that in O’Brien showed a monophyletic cluster and an exponential growth in one single step, with a more recent tMRCA.
Kershenobich et al. has recently reported an analysis of previous works on the epidemiology of HCV infection from six countries throughout Latin America (Argentina, Brazil, Mexico, Puerto Rico, Peru and Venezuela). These authors found that the estimated HCV prevalence in the continent ranged from 1.0 to 2.3%, with a predominance of genotype 1 infection. However, the authors highlighted the limitations of the available epidemiological data and the need for further research [10].
In the present study samples from HCV-1b-infected patients from a large and cosmopolitan city such as Buenos Aires were analyzed and compared with samples from HCV-1b-infected patients from O’Brien, a small rural town with a high prevalence of HCV-1b infection. Using the E1/E2 and NS5B regions of HCV genome, we observed different patterns in both populations. Phylogenetic analysis performed on the E1/E2 and/or NS5B regions has been widely used in HCV epidemiological studies and is therefore recommended for this kind of analysis [16], [19], [21], [25], [26], [38], [39].
Most Buenos Aires sequences were intermingled with different reference sequences from other countries (polyphyletic origin), supporting the hypothesis of multiple virus introductions possibly related to the immigration processes that occurred during the 20th century in Argentina. A particular situation was observed for a small monophyletic cluster (BA7+), which was detected in samples from all participating institutions. This monophyletic cluster possibly reflects the spread of a particular lineage in Argentina with a similar source of transmission. In contrast, all samples from O'Brien, with only one exception (O529), were grouped into a single monophyletic clade. Sample O529 belonged to a patient who had received multiple blood transfusions in 1974, before the adoption of anti-HCV control measures in blood banks, and who may have therefore not become infected by the same transmission route. In addition, the O’Brien cluster was reciprocally independent of the Wheelwright cluster previously reported [16], although there was a similar epidemiological situation in both towns. These results suggest that both groups of sequences (ÓBrien and Wheelwright) may possibly reflect a transmission of a particular viral lineage, through a common source of infection, but independent in both towns. In fact, epidemiological data based on patient interviews from ÓBrien strongly suggest that the use of unsafe injection practices was the main risk factor for HCV infection [14].
Importantly, in certain analyses, some sequences from Buenos Aires were phylogenetically related to the O’Brien or Wheelwright clusters. For this reason, when assessing a possible outbreak or a single case of transmission, it is important to add the strongest evidence available to the phylogenetic analysis. Therefore, in our study we incorporated the largest number of sequences available from around the world and used several phylogenetic methods.
The tMRCA for Buenos Aires HCV-1b samples was estimated around 1887–1900. As previously mentioned, the most likely explanation for this date is the multiple introductions of HCV-1b in Argentina as a consequence of the immigration from European countries (mainly Spain and Italy) at the beginning of the 20th century. In addition, the tMRCA for Buenos Aires is similar to that calculated for HCV-1b circulating in Brazil, Chile and Spain [19], [26], [40]. On the other hand, the tMRCA for the samples from O’Brien was estimated about 1950–1960 and was similar to that calculated for cluster BA7+ (∼1960) and Wheelwright outbreak (∼1950) [16].
The skyline plot profiles obtained for the samples from Buenos Aires (without considering the samples from Lanari and Muñiz Hospitals) of concatenated sequences showed a two-step curve with an initial period of exponential growth in the Ne×τ from the tMRCA until ∼1960. After this first period, a second and more pronounced period of growth of about 10 years (1970–1980) was observed. Accordingly, the increase in the growth rate of the Ne×τ that occurred between 1970 and 1980 coincides with the diversification of cluster BA7. Furthermore, in the demographic profile obtained from the NS5B sequences from Buenos Aires, we observed an increase in the Ne×τ approximately between 1960 and 1970 that corresponds with the diversification of cluster BA7+. For cluster BA7+, we observed a pronounced and exponential growth in the Ne×τ from ∼1959 (tMRCA) to ∼1975, which then remained constant until the present. These results are consistent with the increase observed in the Ne×τ between 1960 and 1970 at the end of the curve, and allow us to speculate that the two-step curve is the product of the viral diversification of cluster BA7+, while HCV-1b was already expanding in Buenos Aires. Moreover, we observed that the demographic profile and tMRCA obtained for cluster BA7+ were similar to those obtained for the samples from O'Brien. These demographic profiles of viral spread suggest that there was a high rate of initial transmission followed by a process of low or null transmission. In addition, all the datasets analyzed showed that the exponential growth of HCV-1b reached a plateau around 1980, which is consistent with that previously reported by Magiorkinis et al. [21]. This exponential decrease in the growth was observed prior to the implementations of anti-HCV screening in the early 1990s [41].
A limitation of this study is that the E1/E2 region for some samples from Buenos Aires (Lanari and Muñiz Hospitals) was not sequenced. Nevertheless, this fact does not reduce the interest of the findings reported herein, since the use of NS5B has proven to be useful in the phylogenetic analysis of HCV [19]. Another limitation and important point to mention is that most samples from Buenos Aires and O'Brien analyzed were isochronous. However, this inconvenient was solved through the estimation of the tMRCA and population dynamics by the incorporation of an external substitution rate of the virus (a prior with normal distribution and high standard deviation) and the use of epidemiological data for O’Brien dataset. The use of this external substitution rate possibly caused an error in the HPD95% values estimated in the analyses.
In conclusion, in a large cosmopolitan city like Buenos Aires, the polyphyletic origin and the spread of HCV infection are in agreement with a longer period of expansion, while, in small towns like O’Brien, the chances of transmission are strongly restricted. Furthermore, the monophyletic character and the older time of emergence in O’Brien suggest that different HCV-1b ancestors (viral variants) that were in expansion in Buenos Aires had the opportunity to colonize and expand in O’Brien.
Acknowledgments
We thankVictoria Eusevi for enhancing the readability of this paper.
Funding Statement
This work was supported by grants from Universidad de Buenos Aires (SECyT-UBA 2008, B037, www.ffyb.uba.ar), Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET; PIP2009, 01169, www.conicet.gov.ar) and Agencia Nacional de Promoción Científica y Tecnológica (ANPCyT; PICT2004, 25355, www.agencia.mincyt.gov.ar), Fundación para la Docencia e Investigación de las Enfermedades del Hígado (FUNDIEH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mizokami M, Tanaka Y (2004) Molecular evolutionary analysis predicts the incidence of hepatocellular carcinoma in the United States and Japan. Cancer Chemother Pharmacol 54 Suppl 1S83–86. [DOI] [PubMed] [Google Scholar]
- 2. Pawlotsky JM (2004) Pathophysiology of hepatitis C virus infection and related liver disease. Trends Microbiol 12: 96–102. [DOI] [PubMed] [Google Scholar]
- 3. Robertson B, Myers G, Howard C, Brettin T, Bukh J, et al. (1998) Classification, nomenclature, and database development for hepatitis C virus (HCV) and related viruses: proposals for standardization. International Committee on Virus Taxonomy. Arch Virol 143: 2493–2503. [DOI] [PubMed] [Google Scholar]
- 4. Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, et al. (2005) Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology 42: 962–973. [DOI] [PubMed] [Google Scholar]
- 5. Simmonds P (2004) Genetic diversity and evolution of hepatitis C virus–15 years on. J Gen Virol 85: 3173–3188. [DOI] [PubMed] [Google Scholar]
- 6. Cochrane A, Searle B, Hardie A, Robertson R, Delahooke T, et al. (2002) A genetic analysis of hepatitis C virus transmission between injection drug users. J Infect Dis 186: 1212–1221. [DOI] [PubMed] [Google Scholar]
- 7. Pybus OG, Cochrane A, Holmes EC, Simmonds P (2005) The hepatitis C virus epidemic among injecting drug users. Infect Genet Evol 5: 131–139. [DOI] [PubMed] [Google Scholar]
- 8. Quarleri JF, Bolcic FM, Bouzas MB, Laufer N, Gomez Carrillo M, et al. (2007) HCV genotype distribution among HIV co-infected individuals in Argentina: relationship with host and viral factors. Acta Gastroenterol Latinoam 37: 76–83. [PubMed] [Google Scholar]
- 9. Tan Y, Wei QH, Chen LJ, Chan PC, Lai WS, et al. (2008) Molecular epidemiology of HCV monoinfection and HIV/HCV coinfection in injection drug users in Liuzhou, Southern China. PLoS One 3: e3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kershenobich D, Razavi HA, Sanchez-Avila JF, Bessone F, Coelho HS, et al. (2011) Trends and projections of hepatitis C virus epidemiology in Latin America. Liver Int 31 Suppl 218–29. [DOI] [PubMed] [Google Scholar]
- 11.Bessone F, Campodonico M, Fay F (2005) Elevada prevalencia de infección por HCV en personas mayores de 60 años en una localidad de 5800 habitantes.,. Acta Gastroenterológica Latinoamericana 35.
- 12.Mengarelli S, Correa G, Farias A (2006) Por qué el virus de la Hepatitis C en Cruz del Eje? Acta Gastroenterológica Latinoamericana 36.
- 13.Mengarelli S, Kohn I, Correa G (2006) Circulación del Virus C en la Provincia De Córdoba. Acta Gastroenterológica Latinoamericana 3.
- 14. Picchio GR, Bare PC, Descalzi VI, Bussy MV, Soria SM, et al. (2006) High prevalence of infection with a single hepatitis C virus genotype in a small rural community of Argentina. Liver Int 26: 660–665. [DOI] [PubMed] [Google Scholar]
- 15.Ramadan A, Rossi L, Lura G (2006) Prevalencia de infección por HCV en Rufino, Santa Fe. Acta Gastroenterológica Latinoamericana 36.
- 16. Golemba MD, Di Lello FA, Bessone F, Fay F, Benetti S, et al. (2010) High prevalence of hepatitis C virus genotype 1b infection in a small town of Argentina. Phylogenetic and Bayesian coalescent analysis. PLoS One 5: e8751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Demetriou VL, van de Vijver DA, Kostrikis LG (2009) Molecular epidemiology of hepatitis C infection in Cyprus: evidence of polyphyletic infection. J Med Virol 81: 238–248. [DOI] [PubMed] [Google Scholar]
- 18. Di Lello F, Garcia G, Kott V, Sookoian S, Campos R (2008) Diversity of hepatitis C virus genotype 1b in Buenos Aires, Argentina: description of a new cluster associated with response to treatment. J Med Virol 80: 619–627. [DOI] [PubMed] [Google Scholar]
- 19. Di Lello FA, Pineiro YLFG, Munoz G, Campos RH (2009) Diversity of hepatitis B and C viruses in Chile. J Med Virol 81: 1887–1894. [DOI] [PubMed] [Google Scholar]
- 20. Ferraro D, Genovese D, Argentini C, Giordano V, Pizzillo P, et al. (2008) Phylogenetic reconstruction of HCV genotype 1b dissemination in a small city centre: the Camporeale model. J Med Virol 80: 1723–1731. [DOI] [PubMed] [Google Scholar]
- 21. Magiorkinis G, Magiorkinis E, Paraskevis D, Ho SY, Shapiro B, et al. (2009) The global spread of hepatitis C virus 1a and 1b: a phylodynamic and phylogeographic analysis. PLoS Med 6: e1000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakano T, Lu L, He Y, Fu Y, Robertson BH, et al. (2006) Population genetic history of hepatitis C virus 1b infection in China. J Gen Virol 87: 73–82. [DOI] [PubMed] [Google Scholar]
- 23. Njouom R, Frost E, Deslandes S, Mamadou-Yaya F, Labbe AC, et al. (2009) Predominance of hepatitis C virus genotype 4 infection and rapid transmission between 1935 and 1965 in the Central African Republic. J Gen Virol 90: 2452–2456. [DOI] [PubMed] [Google Scholar]
- 24. Pybus OG, Barnes E, Taggart R, Lemey P, Markov PV, et al. (2009) Genetic history of hepatitis C virus in East Asia. J Virol 83: 1071–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Re VE, Culasso AC, Mengarelli S, Farias AA, Fay F, et al. (2011) Phylodynamics of hepatitis C virus subtype 2c in the province of Cordoba, Argentina. PLoS One 6: e19471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Romano CM, de Carvalho-Mello IM, Jamal LF, de Melo FL, Iamarino A, et al. (2010) Social networks shape the transmission dynamics of hepatitis C virus. PLoS One 5: e11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25: 1253–1256. [DOI] [PubMed] [Google Scholar]
- 29.Swofford DL (2003) PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4: Sinauer Associates, Sunderland, Massachusetts.
- 30. Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- 31.Goloboff P, Farris J, Nixon K (2003) T.N.T Tree Analysis Using New Technology. Version 1.1 ed.
- 32. Goloboff PA, Farris JS, Nixon KC (2008) TNT, a free program for phylogenetic analysis. Cladistics 24: 774–786. [Google Scholar]
- 33. Huson DH, Richter DC, Rausch C, Dezulian T, Franz M, et al. (2007) Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics 8: 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Drummond AJ, Rambaut A, Shapiro B, Pybus OG (2005) Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol 22: 1185–1192. [DOI] [PubMed] [Google Scholar]
- 36.Rambaut A, Drummond AJ (2007) Tracer v1.4. Available: http://beast.bio.ed.ac.uk/Tracer.
- 37. Suchard MA, Weiss RE, Sinsheimer JS (2001) Bayesian selection of continuous-time Markov chain evolutionary models. Mol Biol Evol 18: 1001–1013. [DOI] [PubMed] [Google Scholar]
- 38. Halfon P, Roubicek C, Gerolami V, Quentin Y, Khiri H, et al. (2002) Use of phylogenetic analysis of hepatitis C virus (HCV) hypervariable region 1 sequences to trace an outbreak of HCV in an autodialysis unit. J Clin Microbiol 40: 1541–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Salemi M, Vandamme AM (2002) Hepatitis C virus evolutionary patterns studied through analysis of full-genome sequences. J Mol Evol 54: 62–70. [DOI] [PubMed] [Google Scholar]
- 40. Tanaka Y, Kurbanov F, Mano S, Orito E, Vargas V, et al. (2006) Molecular tracing of the global hepatitis C virus epidemic predicts regional patterns of hepatocellular carcinoma mortality. Gastroenterology 130: 703–714. [DOI] [PubMed] [Google Scholar]
- 41. Armstrong GL, Alter MJ, McQuillan GM, Margolis HS (2000) The past incidence of hepatitis C virus infection: implications for the future burden of chronic liver disease in the United States. Hepatology 31: 777–782. [DOI] [PubMed] [Google Scholar]