Abstract
A multilocus restriction typing (MLRT) method was developed to reduce the number of sequencing reactions required to determine the clonal relationships among serogroup B meningococci causing an epidemic in New Zealand. MLRT was a rapid, simple, and inexpensive method, and the results had an excellent correlation with multilocus sequence typing results.
A limited number of clonal complexes of Neisseria meningitidis cause the majority of disease worldwide (2, 9). Methods to identify these hypervirulent complexes are important in surveillance of meningococcal disease.
Multilocus sequence typing (MLST) is the “gold standard” method to determine the genetic relatedness of meningococci (7). The sequence data generated are unambiguous, easily standardized, and can discriminate between millions of sequence types (STs). Despite the valuable information afforded by MLST, the cost, time, and expertise required are prohibitive for its routine application to case isolates. Semiautomation reduces the time required to process each sample (3), although this method is still expensive. It has been reported that MLST could only be justified when high sample numbers were processed, even using semiautomated MLST (3).
The limitations of MLST have led to the development of fluorescent-amplified fragment length polymorphism analysis (4, 6), denaturing gradient gel electrophoresis MLST (5), and MLST-denaturing high-performance liquid chromatography (10). All these methods have significant cost benefits in identifying the clones distinguished by MLST, although all require expensive equipment and a high level of technical expertise to perform.
More recently, Bennett and Cafferkey (1) reported a multilocus restriction typing (MLRT) methodology for N. meningitidis. MLRT typed meningococci based on restriction fragment length polymorphism analysis of the seven MLST PCR products. MLRT was a simple, rapid, inexpensive method, and its results correlated well with serological typing results. This report describes an MLRT method similar to that described by Bennett and Cafferkey (1) and compares MLRT and MLST results.
The MLRT method described in this report was developed using three reference strains from cases of disease in New Zealand (strains NZ91/40, NZ98/53, and NZ98/58). Each had the phenotype of the epidemic strain (B:4:P1.4), and before the commencement of this study, these were the only New Zealand case isolates with a known ST. Strains NZ91/40, NZ98/53, and NZ98/58 were subclones of the ST-44 complex with ST-42, ST-155, and ST-154, respectively.
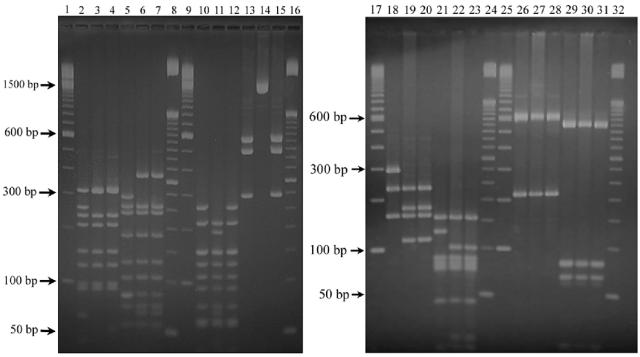
MLST PCR products were amplified as described previously (7), with the addition of primers to amplify fumC (fumC-A1 5′-CACCGAACACGACACGATGG-3′ and fumC-A2 5′-ACGACCAGTTCGTCAAACTC-3′). The PCR products were restricted with enzymes determined to differentiate between the common MLST alleles found in meningococci belonging to the ST-44 complex (Table 1). MLST PCR products (4 μl) were restricted for 4 h in 8-μl single digests containing 4 U of restriction enzyme (New England Biolabs, Beverly, Mass.). MLRT digestion products were electrophoresed on a 2% agarose gel with 0.5× Tris-borate-EDTA buffer at 5 V/cm for 2.5 h before being stained with ethidium bromide (1 μg/ml) and visualized under UV light. The sizes of digestion products were compared to 100- and 50-bp molecular size markers (Invitrogen, Melbourne, Australia). Differences in the abcZ, fumC, and gdh MLRT profiles, which discriminate between ST-42, ST-154, and ST-155, could be visualized on an agarose gel (Fig. 1).
TABLE 1.
Restriction enzymes used in MLRT to differentiate between MLST alleles found in meningococci belonging to the ST-44 clonal complex
| MLST allele | Restriction enzymes used in MLRT |
|---|---|
| abcZ | HhaI and TaqI |
| adk | MspI and TaqI |
| aroE | HhaI and MspI |
| fumC | HhaI and StyI |
| gdh | AluI and HhaI |
| pdhC | AluI and HaeIII |
| pgm | HhaI and MspI |
FIG. 1.
MLRT profiles observed following gel electrophoresis of digestion products from NZ91/40, NZ98/53, and NZ98/58. Lanes: 2 to 4, abcZ HhaI digests for NZ91/40, NZ98/53, and NZ98/58, respectively; 5 to 7, abcZ TaqI digests for NZ91/40, NZ98/53, and NZ98/58, respectively; 10 to 12, fumC HhaI digests for NZ91/40, NZ98/53, and NZ98/58, respectively; 13 to 15, fumC StyI digests for NZ91/40, NZ98/53, and NZ98/58, respectively; 18 to 20, gdh AluI digests for NZ91/40, NZ98/53, and NZ98/58, respectively; 21 to 23, gdh HhaI digests for NZ91/40, NZ98/53, and NZ98/58, respectively; 26 to 28, pdhC AluI digests for NZ91/40, NZ98/53, and NZ98/58, respectively; and 29 to 31, pdhC HaeIII digests for NZ91/40, NZ98/53, and NZ98/58, respectively. Digestion products were compared to 100-bp markers (lanes 1, 9, 17, and 25) and 50-bp markers (lanes 8, 16, 24, and 32).
Restriction patterns from all seven loci were used to define the restriction type (RT). Restriction profiles from meningococci with known MLST alleles were assigned numbers according to the known allele. For example, strain NZ91/40 contains the gene coding for abcZ-10 and the abcZ restriction profile generated was defined as abcZ-10. Restriction profiles different from those found in meningococci with a known ST were arbitrarily assigned a letter. RTs from meningococci with a known ST were assigned the same number as the ST. For example, the RT found in strain NZ91/40 (ST-42) was defined as RT-42.
Before MLRT was applied to meningococci of unknown ST, both MLRT and MLST were applied to 18 meningococci with diverse phenotypes (Table 2). A number of different MLRT profiles were found, although the majority of restriction profiles were the same as profiles found for strains NZ91/40, NZ98/53, and NZ98/58 (Table 2), suggesting that the alleles found in the sample were the same.
TABLE 2.
MLRT and MLST results for a sample of 18 meningococci isolated during New Zealand's epidemic
| ID no.a | Strain no. | Phenotype | Method | MLRT/MLST allele no.b
|
RT/STb | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| abcZ | adk | aroE | fumC | gdh | pdhC | pgm | |||||
| 653 | NZ91/40 | B:4:P1.4 | MLRT | 10 | 6 | 9 | 5 | 9 | 6 | 9 | 42 |
| MLST | 10 | 6 | 9 | 5 | 9 | 6 | 9 | 42 | |||
| 1004 | NZ98/53 | B:4:P1.4 | MLRT | 3 | 6 | 9 | 9 | 11 | 6 | 9 | 155 |
| MLST | 3 | 6 | 9 | 9 | 11 | 6 | 9 | 155 | |||
| 1003 | NZ98/58 | B:4:P1.4 | MLRT | 3 | 6 | 9 | 5 | 11 | 6 | 9 | 154 |
| MLST | 3 | 6 | 9 | 5 | 11 | 6 | 9 | 154 | |||
| 3351 | NZ93/74 | B:nt:P1.4 | MLRT | 10 | 6 | 9 | 5 | 9 | 6 | 9 | 42 |
| MLST | 10 | 6 | 9 | 5 | 9 | 6 | 9 | 42 | |||
| 3353 | NZ95/65 | B:4:P1.4 | MLRT | 10 | 6 | 9 | 5 | 9 | 6 | 9 | 42 |
| MLST | 10 | 6 | 9 | 5 | 9 | 6 | 9 | 42 | |||
| 3361 | NZ97/187 | B:nt:P1.4 | MLRT | 10 | 6 | 9 | 5 | 9 | 6 | 9 | 42 |
| MLST | 10 | 6 | 9 | 5 | 9 | 6 | 9 | 42 | |||
| 3709 | NZ98/203 | B:15:P1.4 | MLRT | 10 | 6 | 9 | 5 | 9 | 6 | 9 | 42 |
| MLST | 10 | 6 | 9 | 6 | 9 | 6 | 9 | 42 | |||
| 3364 | NZ99/109 | B:4:P1.4 | MLRT | 10 | 6 | 9 | 5 | 9 | 6 | 9 | 42 |
| MLST | 10 | 6 | 9 | 5 | 9 | 6 | 9 | 42 | |||
| 3705 | NZ91/49 | B:4:P1.4 | MLRT | 10 | 6 | 9 | C | 9 | 6 | 9 | UTDc |
| MLST | 10 | 6 | 9 | 9 | 9 | 6 | 9 | 2136 | |||
| 3708 | NZ98/75 | B:1:P1.4 | MLRT | 10 | 6 | D | 5 | 9 | 6 | 9 | UTD |
| MLST | 10 | 6 | 15 | 5 | 9 | 6 | 9 | 2673 | |||
| 3350 | NZ93/8 | B:4:P1.4 | MLRT | 3 | 6 | 9 | 5 | 11 | 6 | 9 | 154 |
| MLST | 3 | 6 | 9 | 5 | 11 | 6 | 9 | 154 | |||
| 3352 | NZ95/24 | B:14:P1.4 | MLRT | 3 | 6 | 9 | 5 | 11 | 6 | 9 | 154 |
| MLST | 3 | 6 | 9 | 5 | 11 | 6 | 9 | 154 | |||
| 3359 | NZ97/106 | B:14:P1.4 | MLRT | 3 | 6 | 9 | 5 | 11 | 6 | 9 | 154 |
| MLST | 3 | 6 | 9 | 5 | 11 | 6 | 9 | 154 | |||
| 3363 | NZ99/38 | B:4:P1.4 | MLRT | 3 | 6 | 9 | 5 | 11 | 6 | 9 | 154 |
| MLST | 3 | 6 | 9 | 5 | 11 | 6 | 9 | 154 | |||
| 3706 | NZ98/51 | B:1:P1.4 | MLRT | 3 | 6 | 9 | 5 | 9 | 6 | D | UTD |
| MLST | 3 | 6 | 9 | 5 | 9 | 211 | 18 | 2671 | |||
| 3711 | NZ99/254 | B:4:P1.7 | MLRT | D | 6 | 9 | 9 | 9 | 6 | 9 | UTD |
| MLST | 9 | 6 | 9 | 9 | 9 | 6 | 9 | 44 | |||
| 3349 | NZ92/1 | B:14:P1.14 | MLRT | A | A | A | A | A | A | A | UTD |
| MLST | 4 | 10 | 11 | 17 | 6 | 10 | 12 | 457 | |||
| 3365 | NZ99/144 | B:4:P1.14 | MLRT | A | A | A | A | A | A | A | UTD |
| MLST | 4 | 10 | 11 | 17 | 6 | 10 | 12 | 457 | |||
| 3358 | NZ97/43 | B:nt:nstd | MLRT | E | 6 | E | E | E | E | 9 | UTD |
| MLST | 7 | 6 | 2 | 72 | 3 | 87 | 9 | 2345 | |||
| 3355 | NZ96/59 | C:2a:P1.4 | MLRT | B | A | B | B | B | B | B | UTD |
| MLST | 132 | 10 | 19 | 40 | 62 | 21 | 32 | 2344 | |||
| 3356 | NZ96/211 | C:2b:P1.4 | MLRT | C | B | C | 9 | C | C | C | UTD |
| MLST | 2 | 3 | 7 | 6 | 8 | 5 | 2 | 66 | |||
Identification number in the MLST database.
MLST alleles and STs as defined on the MLST web site (http://neisseria.org/nm/typing/mlst).
UTD, unable to determine using MLRT, as restriction profiles were different from those found in strains NZ91/40, NZ98/53, and NZ98/58. Letters A to E indicate restriction profiles different from those found in meningococci from either ST-42, ST-154, or ST-155.
P1.7 porA sequence was identified by DNA-DNA hybridization.
MLST was carried out as described previously (7), with the addition of primers to sequence the fumC amplicon (fum-S1, 5′-TCGGCACGGGTTTGAACAGC-3′; and fumC-S2, 5′-CAACGGCGGTTTCGCGCAAC-3′). To assign MLST allele numbers, data were submitted to the MLST database (http://neisseria.org/nm/typing/mlst).
Sequence data confirmed MLRT predictions for all 18 meningococci. Meningococci with MLRT profiles that differed at five or more loci (strains NZ92/1, NZ96/59, NZ96/211, NZ97/43, and NZ99/144) when compared to MLRT profiles from the three reference strains were determined by MLST not to belong to the ST-44 complex (Table 2). Strains NZ98/51, NZ98/75, and NZ99/254 had identical restriction profiles, when compared to the three reference strains, at six loci (Table 2), and MLST showed they belonged to the ST-44 complex, yet were ST-2671, ST-2675, and ST-44, respectively (Table 2).
The excellent correlation between MLRT and MLST data means that MLRT could be used to determine the clonal complex causing disease, yet MLST data could still be used to compare results between laboratories. Therefore, MLRT retains the portability of MLST without a high level of dependence on sequence data.
Electrophoresis of MLRT digestion products is relatively time-consuming, although large numbers of meningococci can be assessed simultaneously. Additionally, interpretation of MLRT profiles is rapid, unlike analysis of sequence data, which can be very time-consuming.
Restriction enzymes do not have the ability to differentiate between all alleles in the MLST database. The use of MLRT to assess meningococci causing New Zealand's meningococcal disease epidemic was justified, as serological typing indicated the epidemic was caused by closely related meningococci (8). MLRT did not differentiate between pdhC-6 and pdhC-211 (strain NZ98/51, Table 2). By contrast, strains NZ91/49 and NZ98/53 both contain the fumC-9 allele, yet they yielded different MLRT profiles. This was due to an additional StyI restriction site in the fumC allele in strain NZ91/49 that was outside the 450-bp region assessed by using MLST (result not shown).
The successful application of MLRT in this study and by Bennett and Cafferkey (1) demonstrates the wide range of potential applications for MLRT. The enzymes used by Bennett and Cafferkey (1) had the potential to discriminate between meningococci with a large number of different serological typing results, which were likely to belong to a different clonal complexes. By contrast, the meningococci assessed in our study predominantly belonged to the ST-44 clonal complex and the enzymes used were able to differentiate within the ST-44 clonal complex.
As was found by Bennett and Cafferkey (1), MLRT profiles were reproducible and easy to interpret. Our results determined that there was an excellent correlation between MLRT and MLST. Importantly, MLRT distinguishes between the clones differentiated by use of MLST and would have minimal start-up costs and a reduced cost per sample.
Acknowledgments
We acknowledge financial support for this study from ESR Ltd., the Otago Medical Research Foundation, and the Maurice and Phyllis Paykel Trust.
This research made use of the Multilocus Sequence Typing web site (http://neisseria.org/nm/typing/mlst) located at the University of Oxford and developed by M.-S. Chan and K. Jolley.
REFERENCES
- 1.Bennett, D. E., and M. T. Cafferkey. 2003. Multilocus restriction typing: a tool for Neisseria meningitidis strain discrimination. J. Med. Microbiol. 52:781-787. [DOI] [PubMed] [Google Scholar]
- 2.Caugant, D. A., P. Bol, E. A. Høiby, H. C. Zanen, and L. O. Frøholm. 1990. Clones of serogroup B Neisseria meningitidis causing systemic disease in The Netherlands, 1958-1986. J. Infect. Dis. 162:867-874. [DOI] [PubMed] [Google Scholar]
- 3.Clarke, S. C., M. A. Diggle, and G. F. S. Edwards. 2001. Semiautomation of multilocus sequence typing for the characterization of clinical isolates of Neisseria meningitidis. J. Clin. Microbiol. 39:3066-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goulding, J. N., J. V. Hookey, J. Stanley, W. Olver, K. R. Neal, D. A. A. Ala'Aldeen, and C. Arnold. 2000. Fluorescent amplified-fragment length polymorphism genotyping of Neisseria meningitidis identifies clones associated with invasive disease. J. Clin. Microbiol. 38:4580-4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gurtler, V., H. D. Barrie, and B. C. Mayall. 2002. Denaturing gradient gel electrophoretic multilocus sequence typing of Staphylococcus aureus isolates. Electrophoresis 23:3310-3320. [DOI] [PubMed] [Google Scholar]
- 6.Hookey, J. V., and C. Arnold. 2001. A comparison of multilocus sequence typing and fluorescent fragment-length polymorphism analysis genotyping of clone complex and other strains of Neisseria meningitidis. J. Med. Microbiol. 50:991-995. [DOI] [PubMed] [Google Scholar]
- 7.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin, D. R., S. J. Walker, M. G. Baker, and D. R. Lennon. 1998. New Zealand epidemic of meningococcal disease identified by a strain with phenotype B:4:P1.4. J. Infect. Dis. 177:497-500. [DOI] [PubMed] [Google Scholar]
- 9.Olyhoek, Y., B. Crowe, and M. Achtman. 1987. Clonal population structure of Neisseria meningitidis serogroup A isolated from epidemics and pandemics between 1915 and 1983. Rev. Infect. Dis. 9:665-682. [DOI] [PubMed] [Google Scholar]
- 10.Shlush, L. I., D. M. Behar, A. Zelazny, N. Keller, J. R. Lupski, A. L. Beaudet, and D. Bercovich. 2002. Molecular epidemiology analysis of the changing nature of a meningococcal outbreak following a vaccination campaign. J. Clin. Microbiol. 40:3565-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]