Abstract
porB PCR-amplicon restriction endonuclease analysis is a rapid, simple method developed to assess porB variation in nonserotypeable meningococci isolated during New Zealand's epidemic of meningococcal disease. Most nonserotypeable meningococci isolated between 1990 and 1999 inclusively either were type 4 (40.5%) or contained the porB variable region 1 (VR1)-19, VR2-D, VR3-7, and VR4-14a sequences (45.1%).
New Zealand has experienced increased levels of meningococcal disease since mid-1991 (3). Most case isolates were phenotype B:4:P1.4 (serogroup B, serotype 4, serosubtype P1.4), although 13.1% (155 of 1,183) of the B:P1.4 meningococci isolated from 1990 through 1999 were not serotypeable by the use of serotype 1, 2a, 2b, 4, 14, and 15 antibodies (2, 3). It was important to determine why these meningococci were nonserotypeable and if the B:nonserotypeable (nt):P1.4 meningococci were nonserotypeable variants of the B:4:P1.4 meningococci or if they encoded different porB sequences.
Restriction fragment length polymorphism (RFLP) analysis of the porA and porB PCR products is a simple, rapid method that enables all meningococci to be typed (1, 5, 8, 9). Most RFLP-based typing methods are an alternative to serotyping and serosubtyping, although porA PCR-amplicon restriction endonuclease analysis (porA PCR-AREA) generated subtype-specific restriction profiles (D. R. Martin and S. J. Walker, Proc., Eleventh Int. Pathogenic Neisseria Conf., p. 234, 1998). This report describes the development and application of porB PCR-AREA to investigate the variability in porB in nonserotypeable meningococci isolated during New Zealand's epidemic.
porB PCR products from 30 New Zealand case isolates with diverse serotypes were sequenced to determine the porB variation (Table 1). Primers PorB F (5′-ATCCGCCCTTCAAAATACACATC-3′) and PorB R (5′-TGGCGCAGACCGACACC-3′) were used for amplification and sequencing. The amplification reaction mixtures were incubated at 94°C for 2 min, followed by 30 cycles of 94°C for 40 s, 55°C for 40 s, and 70°C for 80 s. Extension was completed at 72°C for 3 min.
TABLE 1.
Phenotypes and PorB VR sequences of 30 meningococci isolated from patients with clinical cases of meningococcal disease during 1999
| Phenotype | No. of isolates | PorB VR amino acid sequencea
|
porB VR typeb (VR1,VR2,VR3,VR4) | |||
|---|---|---|---|---|---|---|
| VR1 | VR2 | VR3 | VR4 | |||
| B:4:P1.4 | 8 | EHNGGQVVSVE | QDVDDVK | VEDNY | SFDDADLSND | 4,D,7,14a |
| B:4:nstc | 4 | EHNGGQVVSVE | QDVDDVK | VEDNY | SFDDADLSND | 4,D,7,14a |
| B:4:nstd | 1 | EHNGGQVVSVE | QDVDDVK | VEDNY | SFDDADLSND | 4,D,7,14a |
| B:4:P1.7 | 1 | EHNGGQVVSVE | QDVDDVK | VEDN | SVDDAKRDNT | (4)e,D,7b,21 |
| B:4:P1.14 | 1 | EHNGGQVVSVE | VRVDKNVN | VEDNY | SFDDADLSND | 4,undeff,7,14a |
| B:4:P1.14 | 1 | EHNGGQVVSVE | VRVDENVN | VEDNY | SFDDADLSND | 4,B,7,14a |
| B:14:P1.4 | 3 | AHNGAQAASVE | QNVDNVK | VKDN | SFDDADYTND | 19,Db,7c,14 |
| B:1:P1.4 | 4 | AHNGAQAASVE | HQVQENVN | VEENY | SFDATNYNND | 19,Ac,7a,1 |
| B:15:P1.4 | 1 | AHNGAQAASVE | HRVQEDIN | VEDNY | LVDSADLSND | 19,Ab,7,A |
| B:nt:P1.4 | 1 | EHNGGQVVSVE | QDVDDVK | VEDNY | SFDDADLSND | 4,D,7,14a |
| B:nt:P1.4 | 1 | AHNGAQAASVE | QNVDNVK | VKDN | SFDDADLSND | 19,Db,7c,14a |
| B:nt:P1.4 | 1 | AHNGAQAASVE | QDVDDVK | VEDNY | SFDATNYNND | 19,D,7,1 |
| B:nt:P1.4 | 1 | AHNGAQAASVE | HQVQEDLN | ALPNDN | SFDDADLSND | 19,Aa,10,14a |
| B:nt:P1.4 | 1 | DYQDGQVYSVE | QDVDNVK | VEDNY | SFDDADLSND | Undef,Da,7,14a |
| B:nt:P1.4 | 1 | DYQDGQVYSVE | QDVDNVK | VEDN | SFDDADLSND | Undef,undef,7b,14a |
VR sequences are represented by single-letter amino acid codes.
PorB VR types as defined by Sacchi et al. (7).
The porA P1.7.4 VR sequence was identified by DNA-DNA hybridization. Not serosubtypeable.
The porA P1.7 VR sequence was identified by DNA-DNA hybridization. Not serosubtypeable.
The amino acid sequence is identical to that of VR1-4, but a different nucleotide sequence is present.
Undef, the VR sequence was not described previously.
All serotype 4 isolates with the P1.4 PorA serosubtype (determined by serosubtyping or DNA-DNA hybridization) had identical porB sequences (Table 1). Three type 4 meningococci expressing a PorA serosubtype other than P1.4 had distinct porB sequences, suggesting that they are unrelated to the epidemic strain. All B:P1.4 isolates with the same serotype had identical porB variable-region (VR) sequences, whereas nonserotypeable isolates had a variety of porB sequences (Table 1).
The porB characterization achieved by sequencing cannot be achieved by other methodologies, although time and cost prohibit the use of sequencing for large-scale investigations. In this study the availability of porB sequence data enabled the common porB sequences to be determined and the restriction enzymes that differentiate between these sequences to be selected. As certain combinations of sequences in VR1 and VR2 were associated with particular serotypes, the porB type could be predicted.
Restriction analysis of each of the 30 porB PCR products sequenced (Table 1) showed that PCR-AREA could be used as an alternative to sequencing. Restriction of the porB PCR product (4.0 μl) was performed in 8-μl reaction mixtures containing 4 U of enzyme. The reaction mixtures were incubated in a 37°C water bath for 4 h. The digestion products were electrophoresed on a 2% agarose gel with 0.5× Tris-borate-EDTA buffer at 5 V/cm for 90 min, stained with 1 μg of ethidium bromide per ml, and visualized under UV light.
AluI and SspI were used in a double digest to differentiate between sequences in porB VR1 and VR2 (Fig. 1). AflII was subsequently used to restrict porB PCR products containing porB VR1-19 and VR2-D to discriminate between porB VR4-14 and VR4-14a. Two distinct profiles were found in meningococci with the VR1-19 and VR2-A profile (Fig. 1). The different profiles were due to a silent point mutation at the beginning of porB and not to different VR sequences.
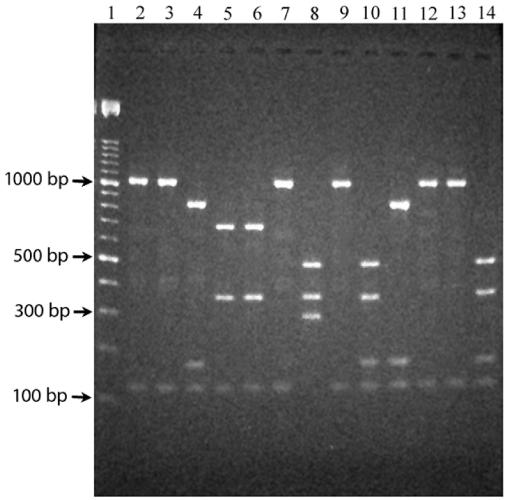
FIG. 1.
porB PCR-AREA patterns observed following gel electrophoresis of AluI and SspI double digests of the porB PCR product. The PCR product was amplified from meningococci with the porB VR1-4 and VR2-D sequences (lanes 2, 3, 7, 9, 12, and 13), the porB VR1-19 and VR2-D sequences (lanes 4 and 11), the porB VR1-19 and VR2-A sequences (lanes 8, 10, and 14), and the porB VR1-4 and VR2-B sequences (lanes 5 and 6). Molecular size markers (100 bp; Roche) are shown in lane 1.
When porB PCR-AREA was applied to 76 meningococci isolated in 1999, the serotype obtained by porB PCR-AREA concurred with the previously established serotype. The restriction profiles for all nonserotypeable meningococci matched the profiles obtained for meningococci whose porB sequences were known. Sequencing of porB from 12 of the 22 meningococci with the B:nt:P1.4 phenotype confirmed the accuracy of porB PCR-AREA, although it was determined that more than two enzymes are required to accurately assess sequences in meningococci containing porB VR2-A due to the variability in these porB sequences.
porB typing.
PCR-AREA was used to determine the porB types of all meningococci with the B:nt:P1.4 phenotype that were isolated in New Zealand from 1990 through 1999 (Table 2) except two nonviable case isolates recovered in 1991. It was determined that most meningococci contained the porB VR1-4 and VR2-D (40.5%) or the VR1-19 and VR2-D (47.7%) sequences. Restriction with AflII determined that 69 (94.5%) of the meningococci determined to contain porB VR1-19 and VR2-D contained the 19, D, 7, and 14a (type 19,D,7,14a) sequences (VR1, VR2, VR3, and VR4, respectively).
TABLE 2.
porB PCR-AREA VR sequence predictions for isolates of N. meningitidis with phenotype B:nt:P1.4 isolated in New Zealand from 1990 through 1999
| Yr | No. (%) of type B:nt:P1.4 isolates of the following typesa predicted by porB PCR-AREA:
|
|||||
|---|---|---|---|---|---|---|
| 4,B | 4,D | 19,A | 19,D,14a | 19,D,14 | Total | |
| 1990 | 1 (100) | 1 | ||||
| 1991 | 2 (100) | 2 | ||||
| 1992 | 1 (100) | 1 | ||||
| 1993 | 1 (25) | 3 (75) | 4 | |||
| 1994 | 3 (42.9) | 4 (57.1) | 7 | |||
| 1995 | 12 (57.1) | 9 (42.9) | 21 | |||
| 1996 | 4 (14.8) | 5 (18.5) | 18 (66.7) | 27 | ||
| 1997 | 13 (32.5) | 5 (12.5) | 19 (47.5) | 3 (7.5) | 40 | |
| 1998 | 2 (7.4) | 13 (48.1) | 1 (3.7) | 10 (37.0) | 1 (3.7) | 27 |
| 1999 | 1 (4.3) | 12 (52.2) | 4 (17.4) | 6 (26.1) | 23 | |
| Total | 7 (4.6) | 62 (40.5) | 11 (7.2) | 69 (45.1) | 4 (2.6) | 153 (100) |
VR1,VR2 or VR1,VR2,VR4 types.
Together, serotyping and porB PCR-AREA showed that type 4, type 19,D,7,14a, and type 14 are the most common porB types found among meningococci with the B:P1.4 phenotype (Table 3). A porB PCR product was amplified from all nonserotypeable meningococci, which indicated that they encoded the gene required to express PorB. The inability to type these meningococci is probably not due to the lack of PorB expression, as all 22 B:nt:P1.4 meningococci examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis expressed a band that was consistent with the size of PorB (32 to 35 kDa). Immunoblotting with the type 4 antibody (5DC4C8G8; National Institute for Biological Standards and Control, Potters Bar, England) confirmed that the 32- to 35-kDa bands were PorB. It is most likely that the inability to type 62 of 1,037 (6.0%) meningococci with the porB VR1-4 sequence was due to the inability of the type 4 antibody to recognize the epitope expressed on the surface of the bacterium. This result may be caused by the masking of the type 4 PorB epitope by meningococcal surface structures or the low sensitivity of the serotype 4 antibody (6). By contrast, 45.1% (69 of 153) of the nonserotypeable meningococci contained the type 19,D,7,14a sequences (Table 2). These meningococci were not serotypeable because the panel of monoclonal antibodies did not include serotype 19 and 7 antibodies.
TABLE 3.
porB types of serogroup B meningococci expressing P1.4 PorA isolated in New Zealand from 1990 through 1999 determined by serological typing and porB PCR-AREA
| Yr | No. (%) of type B:P1.4 types of the following porB types:
|
||||
|---|---|---|---|---|---|
| 4 | 14 | 19,D,14a | Other | Total | |
| 1990 | 3 (60.0) | 2 (40.0) | 0 (0.0) | 0 (0.0) | 5 |
| 1991 | 16 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 16 |
| 1992 | 40 (95.2) | 1 (2.4) | 0 (0.0) | 1 (2.4) | 42 |
| 1993 | 48 (90.6) | 2 (3.8) | 3 (5.7) | 0 (0.0) | 53 |
| 1994 | 87 (91.6) | 4 (4.2) | 4 (4.2) | 0 (0.0) | 95 |
| 1995 | 139 (88.5) | 9 (5.7) | 9 (5.7) | 0 (0.0) | 157 |
| 1996 | 162 (85.3) | 6 (3.2) | 18 (9.5) | 4 (2.1) | 190 |
| 1997 | 201 (85.2) | 11 (4.7) | 19 (8.1) | 5 (2.1) | 236 |
| 1998 | 152 (86.9) | 7 (4.0) | 10 (5.7) | 6 (3.4) | 175 |
| 1999 | 182 (85.9) | 12 (5.7) | 6 (2.8) | 13 (6.1) | 212 |
| Total | 1,030 (87.1) | 54 (4.6) | 69 (5.8) | 28 (2.4) | 1,181 (100.00) |
Our use of three restriction enzymes in porB PCR-AREA was justified in the context of New Zealand's epidemic, as it was expected that a limited number of porB types would be found. porB PCR-AREA differentiated between the type 4,D,7,14a (VR1, VR2, VR3, and VR4, respectively) and the type 4,B,7,14a sequences, although it did not differentiate between the type 4,D,7,14a and type (4),D,7b,21 sequences or the type 4,B,7,14a and type 4,undefined,7,14a sequences. Similarly, the type 19,D,7,14a, type 19,Db,7c,14a, and type 19,D,7,1 sequences were not differentiated. However, porB PCR-AREA was able to differentiate between the most common porB types identified (Table 1).
Although 73% of the B:nt:P1.4 isolates from England and Wales were categorized as having type 4 porB, sequence differences were identified between the isolates (10). By contrast, we found no differences in the sequences of porB from serotype 4 and nonserotypeable meningococci with type 4 porB. Meningococci causing disease in England and Wales belong to a number of clonal complexes and have a number of different phenotypes (4). In New Zealand the strain causing the majority of cases of disease appears to be highly clonal, which may account for the differences observed when the porB sequences from B:nt:P1.4 meningococci in New Zealand and the United Kingdom are compared.
We found that the porB PCR-AREA profiles were reproducible and easy to interpret and that they enabled the porB types of all meningococci to be determined.
Nucleotide sequence accession numbers.
The sequences of type 4,undef,7,14a, type undef,Da,7,14a, and type undef,undef,7b,14a have been submitted to the GenBank database and have been given accession numbers AY333946, AY333947, and AY333948, respectively.
Acknowledgments
We acknowledge financial support for this study from ESR Ltd., the Otago Medical Research Foundation, and the Maurice and Phyllis Paykel Trust.
REFERENCES
- 1.Kertesz, D. A., S. K. Byrne, and A. W. Chow. 1993. Characterization of Neisseria meningitidis by polymerase chain reaction and restriction endonuclease digestion of the porA gene. J. Clin. Microbiol. 31:2594-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin, D., M. Baker, and C. Kieft. 2000. The epidemiology of meningococcal disease in New Zealand in 1999. A report to the New Zealand Ministry of Health. [Online.] www.moh.govt.nz.
- 3.Martin, D. R., S. J. Walker, M. G. Baker, and D. R. Lennon. 1998. New Zealand epidemic of meningococcal disease identified by a strain with phenotype B:4:P1.4. J. Infect. Dis. 177:497-500. [DOI] [PubMed] [Google Scholar]
- 4.Noah, N., and B. Henderson. 2002. Surveillance of bacterial meningitis in Europe 1999/2000. European bacterial meningitis surveillance project. Public Health Laboratory Service, Colindale, London, United Kingdom.
- 5.Peixuan, Z., H. Xujing, and X. Li. 1995. Typing Neisseria meningitidis by analysis of restriction fragment length polymorphisms in the gene encoding the class 1 outer membrane protein: application to assessment of epidemics throughout the last four decades in China. J. Clin. Microbiol. 33:458-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poolman, J. T., P. Kris-Kuzemenska, F. Ashton, W. Bibb, J. Dankert, A. Demina, L. O. Frøholm, M. Hassan-King, D. M. Jones, I. Lind, K. Prakashi, and H. Xujing. 1995. Serotypes and subtypes of Neisseria meningitidis: results of an international study comparing sensitivities and specificities of monoclonal antibodies. Clin. Diagn. Lab. Immunol. 2:69-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sacchi, C. T., A. P. Lemos, A. M. Whitney, C. A. Solari, M. E. Brandt, C. E. Melles, C. E. Frasch, and L. W. Mayer. 1998. Correlation between serological and sequencing analyses of the PorB outer membrane protein in the Neisseria meningitidis serotyping system. Clin. Diagn. Lab. Immunol. 5:348-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Speers, D. J., and J. Jelfs. 1997. Typing of Neisseria meningitidis by restriction analysis of the amplified porA gene. Pathology 29:201-205. [DOI] [PubMed] [Google Scholar]
- 9.Stefanelli, P., C. Fazio, and P. Mastrantonio. 2001. Typing of Neisseria meningitidis isolates from patients with invasive disease by molecular analysis of porin genes. New Microbiol. 24:149-155. [PubMed] [Google Scholar]
- 10.Urwin, R., I. M. Feavers, D. M. Jones, M. C. Maiden, and A. J. Fox. 1998. Molecular variation of meningococcal serotype 4 antigen genes. Epidemiol. Infect. 121:95-101. [DOI] [PMC free article] [PubMed] [Google Scholar]