Abstract
Candida albicans and non-C. albicans Candida species are increasingly being isolated from patients in high-risk categories, most notably, those who have undergone stem cell transplantation (SCT). Identification of the presence of non-C. albicans Candida species early in the course of the transplant procedure is important, as these species exhibit different sensitivities to the available antifungal treatments and cause mortality at rates that vary from those for C. albicans. Amplified fragment length polymorphism (AFLP) analysis has been shown to be a reliable method of reproducibly identifying medically important Candida species. We investigated the use of serial AFLP analysis of 54 routine surveillance cultures for the identification and epidemiological examination of Candida sp. colonization in five consecutive children undergoing allogeneic SCT. One child became colonized with a C. albicans strain and remained colonized with this strain during the whole admission period. Another child had persistent colonization with a C. albicans strain with striking variations in its AFLP patterns over time, which was considered indicative of microevolution. Candida dubliniensis, Candida lusitaniae, and Saccharomyces cerevisiae were identified in the three remaining patients, with two children being simultaneously and transiently colonized with different species. These findings show that colonization with yeasts during transplantation is a complex and dynamic interaction between the host and the organism(s). In our study three strains from eight separate time points were incorrectly identified as C. albicans by a rapid enzyme test. AFLP analysis of surveillance cultures allowed more accurate and informative epidemiological evaluations of pathogenic yeasts in children during transplantation.
In patients undergoing allogeneic stem cell transplantation (SCT), fungal infections are an increasing cause of morbidity and are associated with a high mortality rate (16). Although Candida albicans is the most important cause of superficial candidosis and colonization of the gastrointestinal (GI) tract, at least 12 other members of the genus are recognized as human pathogens (13, 23). Infection tends to be derived from an individual's own endogenous flora, with the GI tract acting as a reservoir of infection (8). Patients with impaired humoral and cell-mediated immunity, especially those undergoing SCT, are at risk of dissemination (26). Among pediatric recipients of bone marrow transplants, dissemination is rarely documented in those receiving prophylactic oral antimycotic agents, even though colonization is common (9). The introduction of prophylactic oral fluconazole has led to clear reductions in the rates of both colonization and invasive infections with C. albicans (20, 26). However, in the last decade between 40 and 70% of cases of candidemia in adult patients with hematological malignancies and bone marrow transplant recipients have been reportedly caused by non-C. albicans Candida species (NACs) (13, 38). Similarly, in some pediatric oncology units, NACs are now the most common cause of candidemia, even in azole-naïve patients (19).
The levels of virulence and the consequent rates of mortality, as well as the sensitivities to the available antimycotic agents, vary among the different species. The four most commonly isolated species are Candida parapsilosis (20 to 40% of all reported episodes of candidemia), Candida tropicalis (10 to 30%), Candida krusei (10 to 35%), and Candida glabrata (5 to 40%). However, other NACs have emerged, such as Candida lusitaniae, which has been documented in up to 8% of all cases (13). The occurrence of these species is associated with inherent or acquired resistance to fluconazole in up to 75% of all C. krusei infections (38) and 35% of all C. glabrata infections. Conversely, at present C. lusitaniae remains highly susceptible to azole antifungal agents (13, 24). Candida dubliniensis has emerged as an opportunistic pathogen closely related to C. albicans and shares many diagnostic characteristics with C. albicans but differs from C. albicans with respect to its epidemiology, virulence, and the ability to develop fluconazole resistance (7, 33). The inherent variability in the response to treatment and the associated mortality rates, which can exceed those of C. albicans infections, demand correct and timely identification. This calls for appropriate or species-directed treatment strategies for at-risk patients.
Molecular techniques, based on the demonstration of variations in the conserved DNA sequences in yeast genomes, have led to identification methods based entirely on the detection of naturally occurring DNA polymorphisms (6, 17, 27, 32, 35). These techniques have been applied in a limited manner to studies of Candida colonization in immunocompromised children (5, 34). Amplified fragment length polymorphism (AFLP) analysis is a relatively new technique (1, 12, 36, 39). It is suitable for identification as well as strain typing. This technique has proved to be robust and reproducible and is an almost ideal high-resolution genotyping method suitable for clinical use. As first described by Zabeau and Vos (39), AFLP analysis belongs to the category of selective restriction amplification techniques that are based on the ligation of adapters to genomic restriction factors with PCR amplification of the adapter-specific primers. The restriction fragments analyzed are small enough for mutations of 1 bp to be detected (12). The use of fluorescent instead of radionucleotide labeling of primers means that a computer-based automated sequence analyzer can read the polyacrylamide gel electrophoretic patterns (29).
In the present study, fluorescent AFLP analysis was used to study the variations in medically important Candida species isolated from serial routine surveillance cultures of samples from children undergoing SCT and to assess the potential for a role of AFLP analysis in determining therapeutic interventions in these high-risk patients.
MATERIALS AND METHODS
Patients.
Five consecutive, nonselected children entered into the Leiden University Medical Center transplant program (all boys; age range, 3 to 15 years) were included in the study. The patient characteristics are summarized in Table 1. Children received standard myeloablative conditioning regimens. All but one child (patient 505) received in vitro T-cell modulation with antithymocyte globulin (Imtex; Sangstat) as additional immune suppression. Prophylaxis for graft-versus-host disease (GvHD) (cyclosporine and methotrexate) was administered for the duration of the study to all children except the child undergoing syngeneic transplantation.
TABLE 1.
Patient characteristicsa
| Patient no. | Age | Disease | Donor type | Conditioning |
|---|---|---|---|---|
| 499 | 14 yr 10 mo | ALL, 2CR | MUD T+ | VP16, Cyclo, TBI, ATG |
| 501 | 7 yr 8 mo | X-LPD | MUD T+ | Bu, Cyclo, ATG |
| 502 | 12 yr 5 mo | ALL, 3CR | MUD T+ | VP16, Cyclo, TBI, ATG |
| 503 | 3 yr 0 mo | X-LPD | MUD T+ | Bu, Cyclo, ATG |
| 505 | 8 yr 11 mo | ALL, 2CR | ID twin | VP16, Cyclo, TBI |
Abbreviations: ALL, acute lymphoblastic leukemia; X-LPD, X-linked lymphoproliferative disease; CR, complete remission; MUD, matched unrelated donor; T+, non-T-cell-depleted stem cells; ID, identical; Bu, busulfan (Busulfex); Cyclo, cyclophosphamide; TBI, total-body irradiation; ATG, antithymocyte globulin; 2CR, second complete remission; 3CR, third complete remission; and VP16, etoposide.
All patients were nursed in positive-pressure isolation rooms and received sterile food and beverages. All patients received total gut decontamination with oral nonabsorbable antimicrobials (piperacillin and tazobactam) and antifungals (amphotericin B suspension) in order to reduce the risk of disseminated infections (37). Three children additionally received oral fluconazole (3 mg/kg of body weight/day) at 2 to 3 weeks postadmission due to overgrowth of Candida in the GI tract, as determined by fecal cultures, which contained >104 colonies.
Swab specimens of the nose, throat, rectum (feces), perineum, and preputium for surveillance cultures were collected on admission to the transplant unit and twice weekly thereafter until discharge. All isolates of Candida were analyzed further by standardized AFLP procedures. The clinical isolates were assigned a random number and later batch analyzed with the investigator blinded to the identity of the patient and the source of the isolate, both in location and in time.
Isolation and phenotypic identification of yeasts.
Swabs from the throat, preputium, and perineum were streaked onto Sabouraud dextrose agar medium (containing penicillin at 20 U/ml and streptomycin at 40 mg/liter) and 2% malt extract agar. One gram of fecal sample was dissolved in 10 ml of 0.9% phosphate-buffered saline and further diluted to a 1:10 suspension, from which 10 μl was taken with a calibrated loop to inoculate Sabouraud agar and malt extract agar. Following incubation for 2 days at 35°C, the cultures were macroscopically inspected for the growth of different colony types. When the growth of different colony types was detected, all suspected colonies were investigated separately. A wet preparation and a rapid enzyme test (Murex Diagnostics, Norcross, Ga.) (4) with positive (C. albicans) and negative (C. glabrata) controls were used to identify C. albicans. The API 32C kit was used for further subtyping and confirmation of the identities of the NACs.
Extraction of DNA.
DNA was extracted from approximately 107 CFU by using a DNeasy tissue kit (Qiagen, Crawley, England) according to the manufacturer's instructions (protocol for isolation of genomic DNA from yeasts). DNA was eluted in 100 μl of the elution buffer provided with the kit and stored at −20°C.
AFLP analysis. (i) Restriction and ligation of adapters.
The sequences of the adapters and preselective primers used for AFLP analysis were as described previously (1). After DNA extraction, 5.5 μl of the sample was added to 5.5 μl of restriction ligation mixture (1× T4 DNA ligase buffer, 0.05 M NaCl, 0.5 μg of bovine serum albumin, 10 pmol of the EcoRI adapter, 20 pmol of the MseI adapter, 30 U of T4 DNA ligase, 10 U of EcoRI, 2 U of MseI) and incubated overnight at 37°C. All enzymes were obtained from New England BioLabs (Beverly, Mass.). The mixture was diluted by adding 25 μl of sterile double-distilled water.
(ii) Preselective and selective PCRs.
Preselective PCR was undertaken with the primers without extensions (core sequences). The AFLP primers, core mixture, and internal size standard were supplied by Applied Biosystems (Nieuwerkerk aan de IJssel, The Netherlands). Four microliters of the diluted restriction ligation product was added to 15 μl of the AFLP amplification core mixture, 0.5 μl of the EcoRI core sequence, and 0.5 μl of the MseI core sequence. The resulting mixture underwent amplification with the GeneAmp PCR system 9700 with the following standardized conditions: 2 min at 72°C, followed by 20 cycles of 20 s at 94°C, 30 s at 56°C, and 2 min at 72°C. The PCR product was then diluted by the addition of 25 μl of sterile double-distilled water. A second PCR with the more selective primers (6-carboxyfluorescein labeled) EcoRI-AC and MseI-C was subsequently undertaken. The conditions were 2 min at 94°C, followed by 10 cycles consisting of 20 s at 94°C and 30 s at 66°C (with this temperature decreasing 1°C with each successive cycle) and a final extension of 2 min at 72°C. Following this sequence of cycles were 25 cycles consisting of 20 s at 94°C, 30 s at 56°C, and 2 min at 72°C, with a final incubation of 30 min at 60°C.
(iii) Capillary electrophoresis and data analysis.
The samples were prepared for capillary electrophoresis by the addition of 2 μl of the selective PCR product to 24 μl of deionized formamide and 1 μl of GeneScan 500 (labeled with 6-carboxy-X-rhodamine), which functioned as an internal size standard. Following 30-min runs on an ABI 310 genetic analyzer, the data obtained were interpreted by use of the BioNumerics software package (version 2.5; Applied Maths, Kortrijk, Belgium), with the Pearson correlation used as a similarity coefficient in combination with the unweighted pair-group methodology with arithmetic mean cluster analysis. The statistical reliability of the clusters was investigated by the use of cophenetic values, which calculate the correlation between the calculated and the dendrogram-derived similarities.
Sequencing.
For some samples the D1/D2 (large subunit) part of ribosomal DNA as well as the internal transcribed spacers, including the 5.8S rRNA gene, were sequenced. Standard sequencing protocols were followed by using the DYEnamic ET Terminator Cycle Sequencing kit (Amersham Biosciences) in combination with the ABI PRISM 3700 DNA analyzer (Applied Biosystems). The sequences obtained were then analyzed by using the Seqman program (version 3.61) of the Lasergene software package (DNAstar Inc.) and checked for identity by comparing them with the sequences in the National Center for Biotechnology Information database by use of the BLAST program as well as by use of the Yeasts of the World CD-ROM (ETI-Biodiversity Center, Amsterdam, The Netherlands).
RESULTS
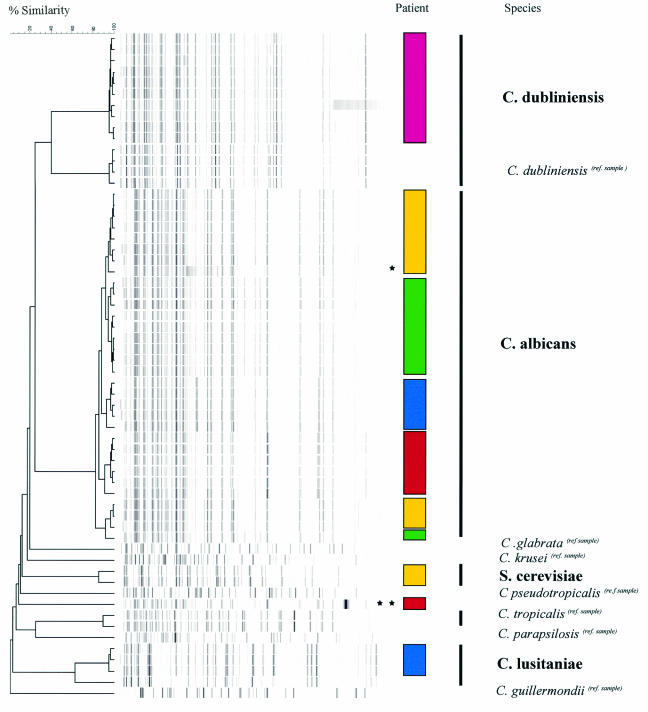
After completion of the study, we found the isolates obtained from the preputial, vulval, and nose swab samples to be of limited additional benefit. Few isolates yielded Candida, and when Candida isolates were found, the results for the rectal (fecal) swab samples were identical to those for the preputial and vulval swab samples. No nasal Candida isolates were obtained. As such, a total of 54 clinical isolates of Candida obtained from the throat, rectal (fecal), and perineal sampling sites form the basis of this study. A dendrogram of all clinical isolates together with the reference strains is depicted in Fig. 1. The AFLP patterns show that each specific species forms a distinct pattern. Known cophenetic values, as described previously (1), allowed genotypic species identification over the time course of the individual patient's transplant.
FIG. 1.
Dendrogram obtained by AFLP analysis. Patients are coded by color blocks, as follows: patient 449, green; patient 501, yellow; patient 502, blue; patient 503, pink; and patient 505, red. The AFLP dendrogram clearly shows the clustering of bands for each species and strain that can be identified as specific for individual patients (in boldface) and identified by matching to a reference strain (ref. sample). The isolates from no two patients had identical patterns, clearly illustrating that nosocomial spread was not evident in the study population. DNA variations were seen within individual patient isolates, although the sequences of these isolates were mostly 90 to 95% similar to that of the original isolate, which is indicative of homology. *, one of the minimal additional bands associated with a fluconazole-resistant C. albicans isolate (patient 501); **, DNA variation, with the sequence of the isolate being <90% similar to that of the original C. albicans isolate from this patient (patient 505), suggestive of microevolution unrelated to the administration of antimicrobials.
Although systemic dissemination of yeasts did not occur during the administration of total gut decontamination medication, all patients had positive surveillance cultures for Candida at various times during the transplant period. All strains isolated were phenotypically identified as C. albicans by the Murex assay. AFLP analysis recognized C. albicans, C. dubliniensis, and C. lusitaniae as well as Saccharomyces cerevisiae by comparison with known reference strains (Fig. 1). Sequence analysis of the D1/D2 domain of the 26S ribosomal DNAs of a selection of the yeast strains confirmed the data obtained by AFLP analysis. In this study, because of the positive Murex reaction, C. lusitaniae and C. dublienisis isolates were initially not investigated with the API 32C system. However, when the results of AFLP analysis became available, strains of both species were investigated with and correctly identified by the API 32C system.
On admission, two patients (patients 501 and 505) were colonized with C. albicans and one patient (patient 503) was colonized with C. dubliniensis. Throat, fecal, and perineal swab samples from all three patients were positive, and colonization with these strains was maintained in all three patients throughout the study period. The two remaining patients (patients 499 and 502) became colonized with C. albicans shortly after admission, but the colonization was unrelated to the start of gut decontamination or systemic antibiotic administration. In addition, one of these children (patient 502) developed perineal colonization with C. lusitaniae. The transient presence of various additional yeasts species was documented simultaneously with the presence of C. albicans in two of the five children: in patient 502, C. lusitaniae was found at two locations in two episodes, and in patient 501, S. cerevisiae was found at one location in one episode. These episodes were of short duration, however, i.e., 2 and 3 weeks, respectively.
The Candida isolates were genotypically identifiable by AFLP analysis. They were distinct and specific to the individual patient, with the isolates from no two patients having exactly the same DNA fingerprint. For three patients, variations within the DNA fingerprint of the same Candida isolate were observed over the study period. The sequences of these isolates remained from 90 to 95% similar to those of the isolates in the original specimen. This was considered to reflect minor technical variations rather than true clonal evolution. Conversely, the C. albicans isolate from one patient (patient 505) showed a more striking variation over the study period. This was considered indicative of microevolution, which for the purpose of this study was defined as similarity of <90%.
These observations showed no evident relationship to a particular diagnosis or previous treatment. All children received systemic broad-spectrum antibiotics, including vancomycin, which was unrelated to the isolates temporally or to the species identified.
Of the three children who received oral fluconazole (at 2 to 3 weeks postadmission), one (patient 501) developed transient colonization with S. cerevisiae and another (patient 502) was colonized with C. lusitaniae. The third patient (patient 499) had persistent C. albicans colonization, but no additional organisms were identified. All Candida isolates except those from patient 501 remained sensitive to fluconazole (MICs, 0.19 to 0.25 mg/liter); 5 days after the start of therapy patient 501 excreted fluconazole-resistant C. albicans (MIC, 64 mg/liter) in the feces.
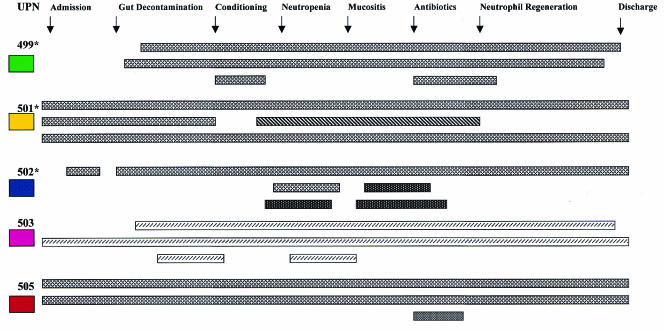
These results are summarized in Fig. 2. As isolates from the preputium or vulva did not differ from those from the rectum (feces) and no Candida strains were isolated from any nasal samples, results for samples from these sites are not included.
FIG. 2.
Diagrammatic representation of the isolates obtained from various sites over time in relation to the occurrence of specific transplant events. The source of each isolate is (from top to bottom for each patient) throat, fecal, and perineal swab specimen cultures (specimens were obtained two times weekly). The color codes for the patients are as described in the legend to Fig. 1. *, patients who received treatment with oral fluconazole. The yeasts are represented as follows: C. albicans, ▩ and ; S. cerevisiae, ▧; C. lusitaniae, ; C. dubliniensis, ▨. UPN, patient number.
DISCUSSION
In our study, conventional surveillance cultures demonstrate that colonization with Candida spp. is extremely common, despite the administration of oral nonabsorbable antifungal agents. All children were colonized with Candida during the course of transplantation. No child had documented systemic yeast infections, and as such, the effect of the total gut decontamination was clinically acceptable. Molecular techniques can also be useful for the identification of yeasts in the clinical setting, and as expected, AFLP analysis proved to be more discriminative than conventional phenotypic identification of Candida species in all of our patients. As considerable overlaps in admission dates and treatment durations occurred among the study patients, phenotypic identification alone would have raised the concern of nosocomial spread. However, molecular identification by AFLP analysis confirmed that no cross-infection occurred among the members of the study group. Colonization with more than one species of Candida at a specific site can exist in as many as 40% of patients with hematological malignancies (21, 22). However, the literature on this topic dates from the late 1980s, when molecular typing techniques were not available on a large scale. The results for our small study group concur with the published data that colonization with multiple species is not infrequent. Colonization in individual patients has been described as clonal in origin for C. albicans, but small genetic rearrangements can arise by microevolution (14). However, we were not able to demonstrate a frequent occurrence of microevolution in this study. This may reflect the relatively short duration of the study, in which, although microevolution of yeasts might have taken place, microevolution was not a major feature. What was not apparent in our study was any relationship of colonization to any form of treatment administered. Host immunity is an important determinant of infection in transplant patients and thus may have played a more crucial role in species evolution and viability than any of the therapeutic interventions.
Thus, AFLP analysis offers important new epidemiological insights into the process of colonization of the GI tract with Candida in immunocompromised children. We have shown, even in these few patients, that this is a highly dynamic process. Our patients, even those children who were azole naïve, transiently demonstrated colonization with multiple Candida species, reflecting the complexity of host factors in children undergoing SCT. These children are often predisposed to colonization before their transplant episode, as seen for the children with positive cultures on admission to the Leiden University Medical Center prior to any transplant-related intervention. The underlying illness or the treatments received, inclusive of steroids and/or chemotherapy, may influence the risk of colonization but, as was demonstrated in our patients, not the development of colonization with additional and multiple yeasts, often simultaneously.
Our study confirms that the emergence of NAC species may go undetected if one relies on phenotypic identification alone. Although the germ-tube test is a simple and economic method for the rapid identification of C. albicans, false-positive reactions can occur with C. tropicalis and C. parapsilosis (2). Alternative tests based on enzymatic methods, such as the Murex test, have been developed (3). The sensitivities and specificities of these commercial tests for the identification of C. albicans and NAC yeasts were greater than 97% (9). As the germ-tube test is time-consuming and laborious and due to the misidentification of C. tropicalis and C. parapsilosis, we have used the Murex assay for the routine laboratory identification of C. albicans since 1999. We use this assay for the presumptive identification of C. albicans for yeasts isolated from patients with invasive infections. In these circumstances, a definite identification is performed with the API 32C system. However, when yeasts are isolated from surveillance cultures of samples from hematology patients, the Murex assay is not followed by a test with the API 32C system, unless the physician is considering treatment intervention.
The Murex enzyme test misidentified C. lusitaniae and C. dubliniensis. Although C. albicans has been reported to be the only species that expresses both N-acetyl-β-d-galactosaminidase and l-proline aminopeptidase, C. dubliniensis was not included in the published evaluation of the test (4, 10). Only six isolates of C. lusitaniae were tested, and they were reported to be negative. We therefore advise that C. albicans-like strains isolated from blood or other sterile compartment be investigated further by additional phenotypic assays (e.g., assays for β-glucosidase) (30) or DNA-based methods.
C. parapsilosis, C. tropicalis, and C. glabrata are among the most common NACs found in SCT recipients but, perhaps unexpectedly, were not present in our study population. What is striking is the occurrence of the less common variants that colonized almost all of our patients at various times during the transplant episode.
C. dubliniensis has emerged as an opportunistic pathogen commonly associated with recurrent oral candidiasis in human immunodeficiency virus-positive patients (11, 31) and has only recently been reported to occur in association with disseminated infections in patients with hematological malignancies and those undergoing bone marrow transplantation (15). A retrospective analysis of all Candida isolates in immunocompromised children concluded that 0.5% of all Candida fungemias are attributable to C. dubliniensis (3).
C. lusitaniae is an infrequent cause of fungemia, causing approximately 2 to 8% of cases of fungemia in adults with cancer. Its emergence is associated with the use of oral polyenes in susceptible patients (13), which existed in all our patients. These NACs show variable sensitivities and may be resistant to amphotericin B but at present remain susceptible to fluconazole (18, 24, 28). However, in a recent study of 67 C. lusitaniae isolates from blood cultures, only 1 strain was found to have a high level of resistance to polyenes (MIC, ≥8 mg/liter). All 67 strains tested were susceptible to fluconazole and voriconazole (24). C. lusitaniae is a species that is frequently reported in the literature to be capable of developing resistance to amphotericin B during the course of treatment. Consequently, persistent colonization with C. albicans-like strains during selective gut decontamination with oral amphotericin B should alert one to the possibility of C. lusitaniae.
Candidemia caused by C. dubliniensis and C. lusitaniae has been reported previously. The children colonized with the NACs were highly vulnerable to dissemination during the transplant procedure. It is known that prolonged neutropenia following delayed engraftment or treatment with steroids for GvHD increases the risk of dissemination (26). Our patients all engrafted successfully, with neutrophil regeneration at a median of 21 days post-SCT and with no clinical evidence of GvHD. This may have contributed to the control of infection and the lack of fungemia.
S. cerevisiae is an ascomycetous yeast that is widely distributed in nature and may colonize the GI tracts of healthy humans. Although the pathogenic nature of this organism had been questioned, it is now evident that virulent strains can be isolated from clinical specimens in association with dissemination and clinical disease in immunocompromised patients, including those undergoing SCT (40).
The relevance of this emerging pathogen in pediatric patients undergoing allogeneic SCT is unknown and until now has not been documented. The complexity of the immune manipulation required for successful engraftment is known to increase the risk of fungal infection (25), and further study of the relevance of these pathogens in pediatric SCT patients is required.
Our study highlights the pitfalls of the sole reliance on phenotypic methodologies, such as the Murex assay, for the identification of Candida species. Molecular techniques have the benefit of being able to determine the true extent of NAC yeast colonization and infection in high-risk patient populations. This was especially useful in the light of our findings of simultaneous colonization with multiple Candida species. Although many of the NACs are intrinsically more resistant to fluconazole treatment, our study had too few patients for it to be possible to study conclusively the effects of the prophylactic or preemptive use of oral azole therapy. The two children who received oral fluconazole and who developed NAC yeast colonization did so prior to the use of fluconazole. These strains were not resistant to fluconazole and were successfully eradicated after azole treatment. Resistance did, however, develop in one patient colonized with C. albicans; and although, interestingly, the resistance was associated with the presence of a DNA variation, as determined by AFLP analysis, it is unlikely that it was the cause of resistance. Resistance to azoles is known to require a much more complex genetic shift than is represented by our findings.
The evolving importance of NAC infections in high-risk patients and the need to develop species-directed treatment modalities require the incorporation of a reliable and universally acceptable method of identification. With the limited data set available, the sources or origins of these NAC yeasts could not be determined. However, our data suggest that the use of AFLP analysis within this setting could provide valuable insights into the identities of fungal pathogens. The AFLP technique offers a very high degree of flexibility, and the AFLP procedure is very easy to perform. By AFLP analysis, a fingerprinting result can theoretically be obtained within 8 h. Another advantage of AFLP analysis is the potential to automate virtually the entire procedure. The disadvantages associated with AFLP analysis are that it depends on expensive computer software and that the patterns obtained are not easy to exchange between different laboratories because of the uniformity of the equipment that is required. Standardization of this technique will certainly be required in order to allow interlaboratory comparisons of AFLP data. At present, the expense and expertise required would probably limit the routine application of AFLP analysis in the clinical setting. AFLP analysis of medically important Candida species is, however, a robust epidemiological research tool. It proved to be capable of identifying various Candida species present concurrently in children at high risk of infection and allowed an increased understanding of the dynamics of fungal colonization.
REFERENCES
- 1.Borst, A., B. Theelen, E. Reinders, T. Boekhout, A. C. Fluit, and P. H. M. Savelkout. 2003. Use of amplified fragment length polymorphism analysis to identify medically important Candida species, including C. dubliniensis. J. Clin. Microbiol. 41:1357-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell, C. K., A. D. Holmes, K. G. Davey, A. Szekely, and D. W. Warnock. 1998. Comparison of a new chromogenic agar with the germ tube method for presumptive identification of Candida albicans. Eur. J. Clin. Microbiol. Infect. Dis. 17:367-368. [DOI] [PubMed] [Google Scholar]
- 3.Climolai, N., J. Davis, and C. Trombley. 2002. Candida dubliniensis fungaemia and vascular access infection. J. Pediatr. Hematol. Oncol. 24:237-239. [DOI] [PubMed] [Google Scholar]
- 4.Crist, A. E., Jr., T. J. Dietz, and K. Kampschroer. 1996. Comparison of the MUREX C. albicans, Albicans-Sure, and BactriCard Candida test kits with germ tube test for presumptive identification of Candida albicans. J. Clin. Microbiol. 34:2616-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Damjanovic, V., C. M. Connolly, H. K. van Saene, R. W. Cook, J. A. Corkhill, A. van Belkum, and D. van Velzen. 1993. Selected decontamination with nystatin for control of a candida outbreak in a neonatal intensive care unit. J. Hosp. Infect. 24:245-259. [DOI] [PubMed] [Google Scholar]
- 6.Forbes, K. J., K. D. Bruce, J. Z. Jordens, A. Ball, and T. H. Pennington. 1991. Rapid methods in bacterial DNA fingerprinting. J. Gen. Microbiol. 137:2051-2058. [DOI] [PubMed] [Google Scholar]
- 7.Gutierrez, J., P. Morales, M. A. Gonzalez, and G. Quindos. 2002. Candida dubliniensis, a new fungal pathogen. J. Basic Microbiol. 42:207-227. [DOI] [PubMed] [Google Scholar]
- 8.Hoppe, J. E., T. Klingebiel, and D. Niethammer. 1995. Orointestinal yeast colonization of pediatric bone marrow transplant recipients: surveillance of quantitative culture and serology. Mycoses 38:51-57. [DOI] [PubMed] [Google Scholar]
- 9.Hoppe, J. E., M. Klausner, T. Klingebiel, and D. Niethammer. 1997. Retrospective analysis of yeast colonization and infections in pediatric bone marrow transplant recipients. Mycoses 40:47-54. [DOI] [PubMed] [Google Scholar]
- 10.Hoppe, J. E., and P. Frey. 1999. Evaluation of six commercial tests and the germ-tube test for presumptive identification of Candida albicans. Eur. J. Clin. Microbiol. Infect. Dis. 18:188-191. [DOI] [PubMed] [Google Scholar]
- 11.Jabra-Rizk, M. A., W. A. Falker, W. G. Merz, A. A. Baqui, J. I. Kelly, and T. F. Meiller. 2000. Retrospective identification and characterization of Candida dubliniensis isolates among Candida albicans clinical laboratory isolates from human immunodeficiency virus (HIV)-infected and non-HIV-infected individuals. J. Clin. Microbiol. 38:2423-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janssen, P., R. Coopman, G. Huys, J. Swings, M. Bleeker, P. Vos, M. Zabeau, and K. Kersters. 1996. Evaluation of DNA fingerprinting method AFLP as a new tool in bacterial taxonomy. Microbiology 142:1881-1893. [DOI] [PubMed] [Google Scholar]
- 13.Krcmery, V., and A. J. Barnes. 2002. Non-albicans Candida spp. causing fungaemia: pathogenicity and antifungal resistance. J. Hosp. Infect. 50:243-260. [DOI] [PubMed] [Google Scholar]
- 14.Lockhart, S. R., J. J. Fritch, A. S. Meier, K. Schroppel, T. Skrikantha, R. Galask, and D. R. Soll. 1995. Colonizing populations of Candida albicans are clonal in origin but undergo microevolution through C1 fragment reorganization as demonstrated by DNA fingerprinting and C1 sequencing. J. Clin. Microbiol. 33:1501-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meis, J. F., M. Ruhnke, B. E. de Pauw, F. C. Odds, W. Siegert, and P. E. Verweij. 1999. Candida dubliniensis candidemia in patients with chemotherapy induced neutropenia and bone marrow transplantation. Emerg. Infect. Dis. 5:150-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyers, J. D. 1990. Fungal infections in bone marrow transplant patients. Semin. Oncol. 17:10-13. [PubMed] [Google Scholar]
- 17.Micheli, M. R., R. Bova, E. Pascale, and E. Dámbrosio. 1994. Reproducible DNA fingerprinting with the random amplified polymorphic DNA (RAPD) method. Nucleic Acids Res. 22:1921-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minari, A., R. Hachem, and I. Raad. 2001. Candida lusitaniae: a cause of breakthrough fungemia in cancer patients. Clin. Infect. Dis. 32:186-190. [DOI] [PubMed] [Google Scholar]
- 19.Mullen, C. A., H. Abd El-Baki, H. Samir, J. J. Tarrand, and K. V. Rolston. 2003. Non-albicans Candida is the most common cause of candidemia in pediatric cancer patients. Supportive Care Cancer 11:321-325. [DOI] [PubMed] [Google Scholar]
- 20.Ninane, J. 1994. A multicentre study of fluconazole versus oral polyenes in the prevention of fungal infection in children with haematological or oncological malignancies. Eur. J. Clin. Microbiol. Infect. Dis. 13:330-337. [DOI] [PubMed] [Google Scholar]
- 21.Odds, F. C. 1987. Candida infections: an overview. Crit. Rev. Microbiol. 15:1-5. [DOI] [PubMed] [Google Scholar]
- 22.Odds, F. C., P. Auger, P. Krogh, A. N. Neely, and E. Segal. 1989. Biotyping of Candida albicans: results of an international collaborative survey. J. Clin. Microbiol. 27:1506-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfaller, M. A., R. N. Jones, S. A. Messer, M. B. Edmond, R. P. Wenzel, et al. 1998. National surveillance of nosocomial blood stream infections due to species of Candida other than Candida albicans: frequency of occurrence and antifungal susceptibility in the SCOPE program. Diagn. Microbiol. Infect. Dis. 30:121-129.9554180 [Google Scholar]
- 24.Pfaller, M. A., D. J. Diekema, S. A. Messer, L. Boyken, R. J. Hollis, and R. N. Jones. 2003. In vitro activities of voriconazole, posaonazole, and four licensed systemic antifungal agents against Candida species infrequently isolated from blood. J. Clin. Microbiol. 41:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pirsch, J. D., and D. G. Maki. 1986. Infectious complications in adults with bone marrow transplantation and T-cell depletion of donor marrow. Increased susceptibility to fungal infections. Ann. Intern. Med. 104:619-631. [DOI] [PubMed] [Google Scholar]
- 26.Prentice, H. G., C. C. Kibbler, and A. G. Prentice. 2000. Towards a targeted, risk-based, anti-fungal strategy in neutropenic patients. Br. J. Haematol. 1110:273-284. [DOI] [PubMed] [Google Scholar]
- 27.Rademaker, J. L. W., and F. J. de Bruijn. 1997. Characterisation and classification of microbes by rep-PCR genomic fingerprinting and computer-assisted pattern analysis, p. 151-171. In G. Caetoan-Anollés and P. M. Gresshoff (ed.), DNA markers: protocols, applications and overviews. John Wiley & Sons, Inc., New York, N.Y.
- 28.Sanchez, V., J. A. Vazquez, D. Barth-Jones, L. Dembry, J. D. Sobel, and M. J. Zervos. 1992. Epidemiology of nosocomial acquisition of Candida lusitaniae. J. Clin. Microbiol. 30:3005-3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Savelkoul, P. H. M., H. J. M. Aarts, J. de Haas, L. Dijkshoorn, B. Duim, M. Otsen, J. L. Rademaker, L. Schouls, and J. A. Lenstra. 1999. Amplified-fragment length polymorphism: the state of the art. J. Clin. Microbiol. 37:3083-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schoofs, A., F. C. Odds, R. Colebunders, M. Ieven, and H. Gossens. 1997. Use of specialised isolation media for recognition and identification of Candida dubliniensis isolates from HIV infected patients. Eur. J. Clin. Microbiol. Infect. Dis. 16:296-300. [DOI] [PubMed] [Google Scholar]
- 31.Schorling, S. R., H. C. Kortinga, M. Froschb, and F. A. Muhlschlegel. 2000. The role of Candida dubliniensis in oral candidiasis in human immunodeficiency virus-infected individuals. Crit. Rev. Microbiol. 26:59-68. [DOI] [PubMed] [Google Scholar]
- 32.Strand, S., T. A. Prolla, R. M. Liskay, and T. D. Peters. 1993. Destabilisation of tracts of simple repetitive DNA in yeast by mutations affecting DNA mismatch repair. Nature 365:274-276. [DOI] [PubMed] [Google Scholar]
- 33.Sullivan, D., and D. Coleman. 1997. Candida dubliniensis: an emerging opportunistic pathogen. Curr. Top. Med. Mycol. 8:15-25. [PubMed] [Google Scholar]
- 34.Van Belkum, A., W. Mol, H. K. van Saene, L. M. Ball, D. van Velzen, and W. Quint. 1994. PCR-mediated genotyping of Candida albicans strains from bone marrow transplant patients. Bone Marrow Transplant. 13:811-815. [PubMed] [Google Scholar]
- 35.Van Belkum, A., W. van Leeuwen, S. Scherer, and H. Verbrugh. 1999. Occurrence and structure-function relationship of pentameric short sequence repeats in microbial genomes. Res. Microbiol. 150:617-626. [DOI] [PubMed] [Google Scholar]
- 36.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Freijters, J. Pot, J. Peleman, M. Kuiper, and M. Zabeau. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vossen, J. M., P. J. Heidt, H. van den Berg, E. J. Gerritsen, J. Hermans, and L. J. Dooren. 1990. Prevention of infection and graft-versus-host disease by suppression of intestinal microflora in children treated with allogeneic bone marrow transplantation. Eur. J. Clin. Microbiol. Infect. Dis. 9:14-23. [DOI] [PubMed] [Google Scholar]
- 38.Wingard, J. R., W. G. Mertz, M. G. Rinaldi, T. R. Johnson, J. E. Karp, and R. Saral. 1991. Increase in Candida krusei infection among patients with bone marrow transplant and neutropenia treated prophylactically with fluconazole. N. Engl. J. Med. 325:1274-1277. [DOI] [PubMed] [Google Scholar]
- 39.Zabeau, M., and P. Vos. 1993. Selective restriction fragment amplification: a general method for DNA fingerprinting. European Patent Office. Publication 0 534 858 A1 bulletin 93/13.
- 40.Zerva, L., R. J. Hollis, and M. A. Pfaller. 1996. In vitro susceptibility testing and DNA typing of Saccharomyces cerevisiae clinical isolates. J. Clin. Microbiol. 34:3031-3034. [DOI] [PMC free article] [PubMed] [Google Scholar]