Abstract
Virus infection may cause a multiplicity of symptoms in their host including discoloration, distortion and growth retardation. Hibiscus chlorotic ringspot virus (HCRSV) infection was studied using kenaf (Hibiscus cannabinus L.), a non-wood fiber-producing crop in this study. Infection by HCRSV reduced the fiber yield and concomitant economic value of kenaf. We investigated kenaf growth retardation and fluctuations of four selected miRNAs after HCRSV infection. Vegetative growth (including plant height, leaf size and root development) was severely retarded. From the transverse and radial sections of the mock and HCRSV-infected kenaf stem, the vascular bundles of HCRSV-infected plants were severely disrupted. In addition, four conserved plant developmental and defence related microRNAs (miRNAs) (miR165, miR167, miR168 and miR171) and their respective target genes phabulosa (PHB), auxin response factor 8 (ARF8), argonaute 1 (AGO1) and scarecrow-like protein 1 (SCL1) displayed variation in expression levels after HCRSV infection. Compared with the mock inoculated kenaf plants, miR171 and miR168 and their targets SCL1 and AGO1 showed greater fluctuations after HCRSV infection. As HCRSV upregulates plant SO transcript in kenaf and upregulated AGO1 in HCRSV-infected plants, the expression level of AGO1 transcript was further investigated under sulfite oxidase (SO) overexpression or silencing condition. Interestingly, the four selected miRNAs were also up- or down-regulated upon overexpression or silencing of SO. Plant growth retardation and fluctuation of four conserved miRNAs are correlated to HCRSV infection.
Introduction
Hibiscus chlorotic ringspot virus (HCRSV) belongs to the genus Carmovirus, family Tombusviridae [1]. It is an icosahedral, positive sense single-stranded RNA plant virus. It has a (+)-sense ss RNA of 3911 nt, containing seven open reading frames (ORFs). HCRSV has been reported to cause systemic infection in Hibiscus cannabinus (kenaf), H. sabdariffa and H. trionum, and results in chlorotic local lesions on several members of Chenopodium spp. [1-5]. Successful infection with HCRSV results in severe symptoms and stunted growth on kenaf [2]. Symptoms have been characterized to be variable; however, chlorotic ringspots on the leaves of infected plants are considered the most characteristic symptom of HCRSV infection [1]. Other symptoms include mosaic patterns, rings, vein-clearing and vein-banding [6]. Regarding the host-virus interaction, HCRSV coat protein (HCRSV-CP) interacts with sulfite oxidase (SO) [7] which in turn, triggers sulfur enhanced defense (SED) and leads to enhanced plant resistance [8].
MicroRNAs (miRNAs) are small RNAs (~22 nucleotides) which are derived from non-protein-coding RNAs and they negatively regulate gene expression [9,10]. Many miRNAs, with their syntheses and regulation affected by the signals generated from environmental stress, are conserved in plants [11]. MiRNAs are involved in plant development, signal transduction, protein degradation and response to environmental stress and pathogen invasion [12-15]. The targets of most plant miRNAs are transcription factors which play important roles in plant defense responses [16,17]. It is well studied that the targets of miR165, miR167, miR168 and miR171 are phabulosa (PHB), auxin response factor 8 (ARF8), argonaute 1 (AGO1) and scarecrow-like protein 1 (SCL1) respectively, playing essential roles in regulating plant development [18-21]. Furthermore, miRNAs are known to modulate plant viral diseases [22,23]. After virus infection, plant conserved miR165, miR167, miR168 and miR171 showed different expression levels at different stages [20]. Thus, miR165, miR167, miR168 and miR171 were selected for investigation in this study.
Among the targets of four selected miRNAs, AGO1 was known to be involved in the feedback regulation of miRNAs [24,25]. Furthermore, since AGO1 is the key component of RNA induced silencing complex (RISC), it plays essential roles in the small RNA induced silencing pathway [26]. Downregulation of some host genes have been speculated to elicit disease symptoms [27]. Indeed, disease symptoms caused by Cucumber mosaic virus (CMV) satellite RNA are the consequence of siRNA-directed RNA silencing of chlorophyll gene CHLI [28,29]. Furthermore, plant developmental abnormalities were caused through misregulation of the miR167 targeting ARF [21].
As a non-wood fiber producing crop, the reduction in fiber yield of kenaf is considered to be of economic significance. In this study, we compared the gross morphology and cross sections of the stem in mock and HCRSV-infected kenaf plants. Since plant growth retardation was related to certain plant developmental genes expression profiles which are regulated by miRNAs, the four selected plant conserved developmental related miRNAs (miR165, miR167, miR168 and miR171) were investigated after HCRSV infection. This study showed that the plant developmental related miRNAs fluctuated after HCRSV infection, which displayed plant growth retardation. Analyzing the plant developmental related miRNAs will improve the understanding of viral infection and present strategies to prevent infection in future.
Materials and Methods
1: Plant materials and growth condition
Kenaf seeds (cultivar Everglades 41) were obtained from Mississippi State University, USA) and germinated on potting mixture (Tref, Netherlands) for 7 days. Kenaf seedlings were transferred into the same potting mixture after emergence of true leaves. All plants were grown in a plant growth room under 16 h light and 8 h dark at 25 °C with fluorescent tube (Model:CS-22, FoFo Shan Light, China).
2: Plasmid construction
The SO coding region was subcloned from pSAT6-cEYPF-C1-SO vector into binary vector of 35SpGreen [30,31] to form a construct 35SpGreen-SO. For the SO silencing study, artificial-miRNA amiRSO was engineered into the miR319a precursor (plasmid pRS300) by site-directed mutagenesis, following the protocol by Rebecca Scheab Max-Plank Institute for Developmental Biology, Tuebingen, Germany (2005) (http://wmd.weigelworld.org/cgi-bin/mirnatools.pl. Ossowski Stephan, Fitz Joffrey, Schwab Rebecca, Riester Markus and Weigel Detlef, personal communication). The artificial miRNAs amiRSO was also cloned into the pGreen vector. Both of the overexpression and silencing vectors were transferred into Agrobacterium tumefaciens GV3101 using electroporation and the transformed colonies were verified by colony PCR and sequencing.
3: Staining of transverse and radial sections from mock and HCRSV-infected plants
Young stems were flash frozen with liquid nitrogen. The frozen stems of mock and HCRSV-infected kenaf plants were cut into 50-60 µm thin sections using Leica Cryostat Model 1850. The slices were transferred to glass slides, stained in a drop of 0.05% aqueous toluidine blue O for 3 minutes and washed with 30% glycerol. After drying on the slides, the stained sections were observed using a light microscope (Model DP72, Olympus, USA).
4: Virus inoculation
Before virus inoculation, young seedlings at the cotyledon stage were verified to be healthy and virus-free. Fully developed young leaves from HCRSV-infected kenaf plants exhibiting chlorotic spots (0.1 g) were homogenized in 0.5 ml of virus inoculation buffer (0.01 M potassium phosphate buffer, pH 7.0) (using Protocol online at http://www.protocol-online.org/biology-forums/posts/16381.html) and used for inoculation. Mock-inoculation was carried out by rubbing inoculation buffer alone onto the cotyledons. Fully-developed young leaves were harvested at 3, 6, 9, 12, 15, 20, 25 and 30 days post inoculation (dpi) to quantify the transcript levels of miRNAs and their respective target genes. This experiment was carried out thrice.
5: RNA extraction and cDNA synthesis
Fully-developed young leaves showing severe symptoms were collected for RNA extraction at previously mentioned time points post-inoculation with HCRSV. RNA was extracted using TRIzol® reagent (Invitrogen, USA). The integrity and quality of the extracted RNA were determined by non-denaturing agarose gel electrophoresis. Total RNA (~2 μg) was used to generate cDNAs through reverse transcription, using oligo(dT)15 or specific stem-loop primers (Table 1) [20] and SuperScript® III Reverse Transcriptase kit (Invitrogen, USA).
Table 1. Primers used in this study.
| Primers | Sequences (5’ to 3’) |
|---|---|
| miR165-RT | CTCAGCGGCTGTCGTGGACTGCGCGCTGCCGCTGAGGGGGRATG |
| miR165-F | CTGTGTCGGACCAGGCTTC |
| miR165-R | GGCTGTCGTGGACTGCG |
| miR167-RT | GCGTGGTCCACACCACCTGAGCCGCCACGACCACGCTAGATCAT |
| miR167-F | CGTGCGTGAAGCTGCCA |
| miR167-R | TCCACACCACCTGAGCCG |
| miR168-RT | CTCAGCGGCTGTCGTGGACTGGGTGCTGCCGCTGAGTTCCCGAC |
| miR168-F | CGTGTGTCGCTTGGTGCA |
| miR168-R | GGCTGTCGTGGACTGGGTG |
| miR171-RT | GCGTGGTCCACACCACCTGAGCCGCCACGACCACGCGATATTGG |
| miR171-F | CGTGCGTGATTGAGCCGT |
| miR171-R | TCCACACCACCTGAGCCG |
| I. amiR-So F | GATTTTAATTGACTGCACGTCTTTCTCTCTTTTGTATTCC |
| II. amiR-So R | GAAAGACGTGCAGTCAATTAAAATCAAAGAGAATCAATGA |
| III. amiR-SO* F | GAAAAACGTGCAGTCTATTAAATTCACAGGTCGTGATATG |
| IV. amiR-SO* R | GAATTTAATAGACTGCACGTTTTTCTACATATATATTCCT |
| A | CTGCAAGGCGATTAAGTTGGGTAAC |
| B | GCGGATAACAATTTCACACAGGAAACAG |
| Hc.qARF8F312 | ATCGTTGTGTCCTGAAAGCAG |
| Hc.qARF8R464 | ACTTGTGGAACTGATGTTGAGC |
| Hc.qPHBF14 | CGGATTCTATTGGCATTGTTGC |
| Hc.qPHBR151 | ATCGGCAGTCACGAAACCA |
| Hc.qAGO1F375 | ATGTCACCCATCCTCACCCT |
| Hc.qAGO1R481 | AGCACAAACCAGACCAGCA |
| Hc.qSCL1F403 | TCGTAAAGCAGTTGTCACCCA |
| Hc.qSCL1R537 | TCACAGCATCAAGGGACTCG |
| HcAct-qF603 | ACGAGCAGGAACTGGAGACT |
| HcAct-qR734 | TGAGTGATGGCTGGAAGAGGA |
| ORFHcAGO1F | CTCTTTCTCTGCGTAGTGACTGGTGACT |
| ORFHcAGO1R | TAGCGAGGAACGATAAAGGCTTGTA |
6: Gene expression analysis of four conserved miRNAs and their respective target genes using qRT-PCR
Expression levels of selected transcripts were analyzed via quantitative real time reverse-transcription PCR (qRT-PCR) after cDNA synthesis. The qRT-PCR was set up in a total volume of 5 μl, and was carried out in the CFX384TM real-time PCR detection system (Bio-Rad, USA). Actin gene was used as the internal control, which showed similar expression profiles in the mock and HCRSV-infected plants. In addition, no difference was observed for actin gene expression in healthy and agro-infiltrated kenaf leaves, respectively. The expression levels of the miRNAs were detected following previously described protocols [20,32]. The experiment was carried out thrice. Each test consisted of three biological repeats and each sample contained three technical repeats. The relative gene expression amount was analyzed using the 2-∆∆C T method. The values of miRNAs and their respect target genes in HCRSV-infected plants were calculated by subtracting the values from mock controls which were all set to 1 for standardization. Means of three independent biological repeats were shown with standard deviations.
7: Agrobacterium-mediated transient expression
Agrobacterium tumefaciens liquid cultures containing each of the pGreen-SO, pGreen-amiRSO plasmids and 35SpGreen (negative control) plasmids, were grown to an OD reading (at 600 nm) between 1.0-1.5 and harvested. The cell pellet was resuspended in infiltration buffer (pH 7) containing 10 mM each of MgCl2 and 2-(N-morpholino) ethanesulfonic acid (MES), and 100 µM acetosyringone and infiltrated into kenaf leaves using a syringe without needle [6]. The selected kenaf leaves used for agro-infiltration were taken from the youngest fully expanded leaves at 14 days after seed emergence. The infiltrated regions were cut out and used for RNA extraction. The experiment was carried out thrice. The relative gene expression amount was analyzed using the 2-∆∆C T method. Significant differences among silenced-SO, mock-infiltration and overexpressed-SO were calculated using the one sample Student’s t-test analysis.
8: Cloning of four conserved miRNAs and their respective target genes
Since the conserved miR165, miR167, miR168 and miR171 were not verified in H. cannabinus previously, stem-loop reverse primer (Table 1) was used to carry out the reverse transcription. The PCR products of the four conserved miRNAs were cloned into pGEM TA vector (Promega, USA) for sequencing and verification.
The H. cannabinus PHB, ARF8, AGO1 and SCL1 genes were not available in the GenBank database and were thus amplified and sequenced in this study. Using degenerate primers, the four target genes were amplified from kenaf. Using the DNAMAN v5.0 sequence analysis program, multiple-sequence alignment (MSA) was performed with four different miRNA target genes. A set of forward and reverse degenerate PCR primers were designed based on conserved sequences. The PCR primers were tested using cDNAs obtained from kenaf plants. The PCR products containing the conserved sequences from each of the four genes were cloned into the pGEM TA Vector (Promega, USA) and sequenced. The qRT-PCR primers (Table 1) were designed accordingly using the gene sequences obtained.
9: RACE PCR to amplify the complete sequence of AGO1 from kenaf
Based on the conserved sequence of AGO1 gene, RACE kit GeneRacerTM kit (Invitrogen, USA) was used to amplify the respective 5’- and 3’-sequences. Nested PCR was performed to amplify the specific PCR product. The 5’- and 3’-PCR products obtained were cloned into pGEM TA Vector (Promega, USA) and sequenced. After the sequences were blasted with PUBMED and confirmed, a pair of primers complementary to each of the 5’- and 3’-termini were designed and used to amplify the complete ORF of the AGO1. All primers used were listed in Table 1.
Results
1: Morphology of HCRSV-infected kenaf plants
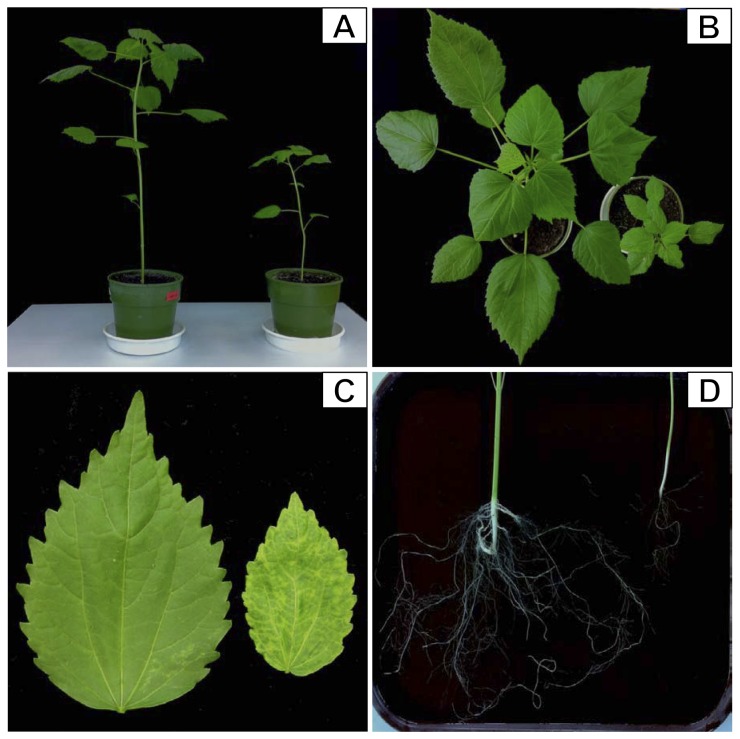
In this study, the morphological changes of HCRSV-infected kenaf plants were compared with mock plants. Our results showed that the plant height, leaf size and root growth were significantly reduced in HCRSV-infected plants (Figure 1A-D). In particular, development of the main and adventitious roots of kenaf was stunted and less dense, as compared to the mock plant (Figure 1D). The vegetative growth of HCRSV-infected plants was severely retarded (Figure 1).
Figure 1. Morphological changes in HCRSV-infected kenaf plants at 15 days post inoculation (dpi).
Growth retardation was observed in the HCRSV-infected plants (plant on the right side of each panel; mock plant was displayed on the left). (A) Side view showing plant height. (B) Top view showing plant canopy. (C) Leaf size and (D) Main and adventitious roots. Note the severe reduction in length and branch of HCRSV-infected kenaf roots.
2: Disruption of vascular bundle formation in HCRSV-infected kenaf stem
Transverse and radial sections were analysed using kenaf stems at 15 dpi with HCRSV. Overall the structure of HCRSV-infected stems and the epidermal cells of HCRSV-infected plants were disorganized (Figure 2). The development of vascular bundles including the fiber and the cortex was severely disrupted (Figure 2A) and the vessels appeared to be collapsed (Figure 2B). The brown colored spaces in the pith region were artifacts that resulted from reflection of illumination from the damaged tissues (Figure 2). Detrimental changes in the water- and food-conducting tissue of HCRSV-infected kenaf were most likely related to its reduced growth. As a non-wood fibrous crop, the reduction in growth (fiber yield) of kenaf is of economic importance.
Figure 2. Comparison of transverse and radial sections of mock and HCRSV- infected kenaf stem at 15 dpi.
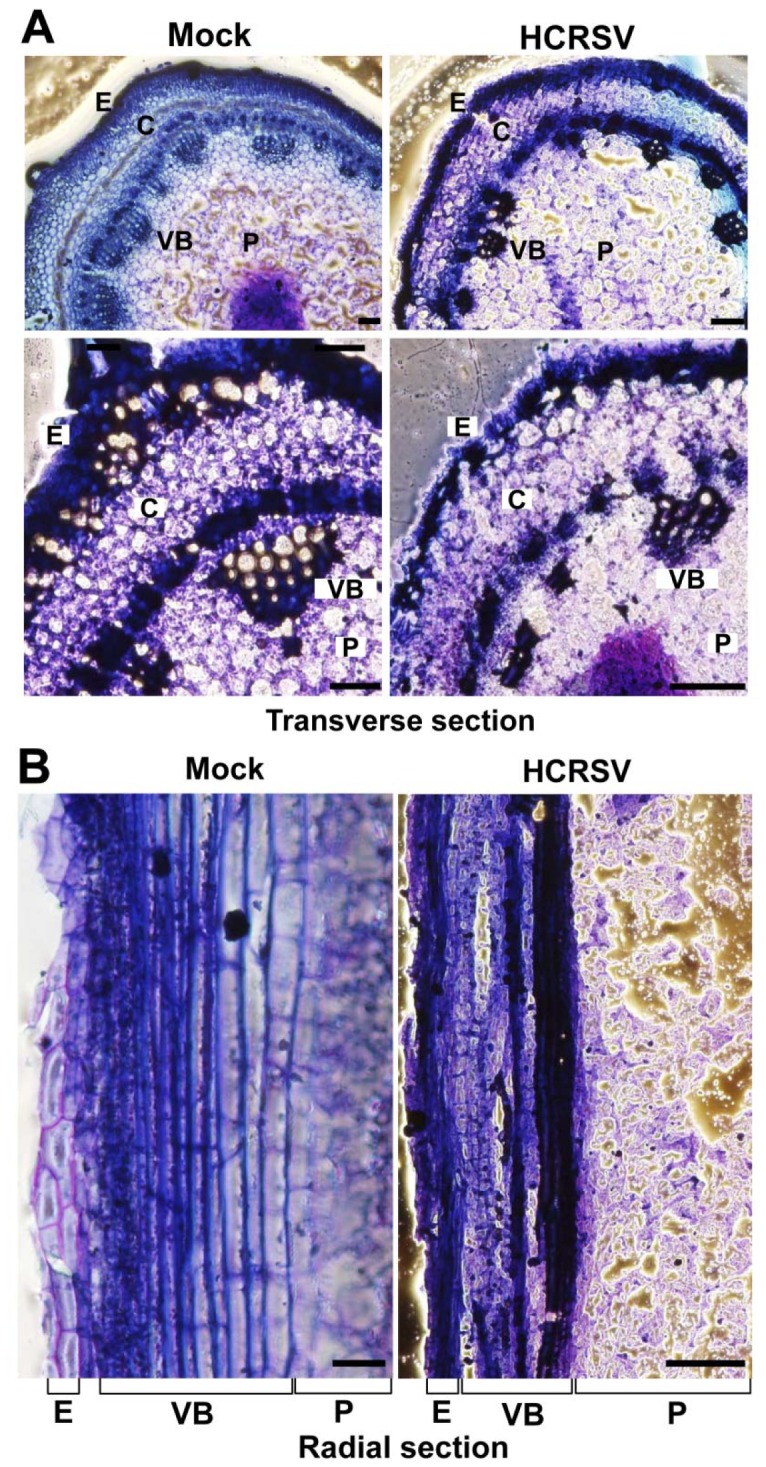
(A) Transverse section of mock and HCRSV-infected kenaf stems. The pith is located in the middle of the transverse section, surrounded by vascular bundles. (B) Partial radial sections of mock and HCRSV-infected kenaf stems. E, epidermis; VB, vascular bundle; C, cortex; P, pith. Bar = 100 µm.
3: Levels of four conserved miRNAs and their target genes fluctuated in HCRSV-infected kenaf
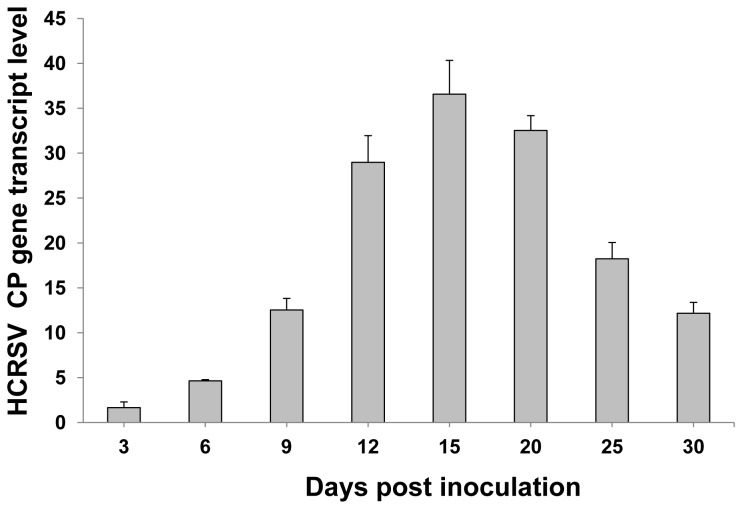
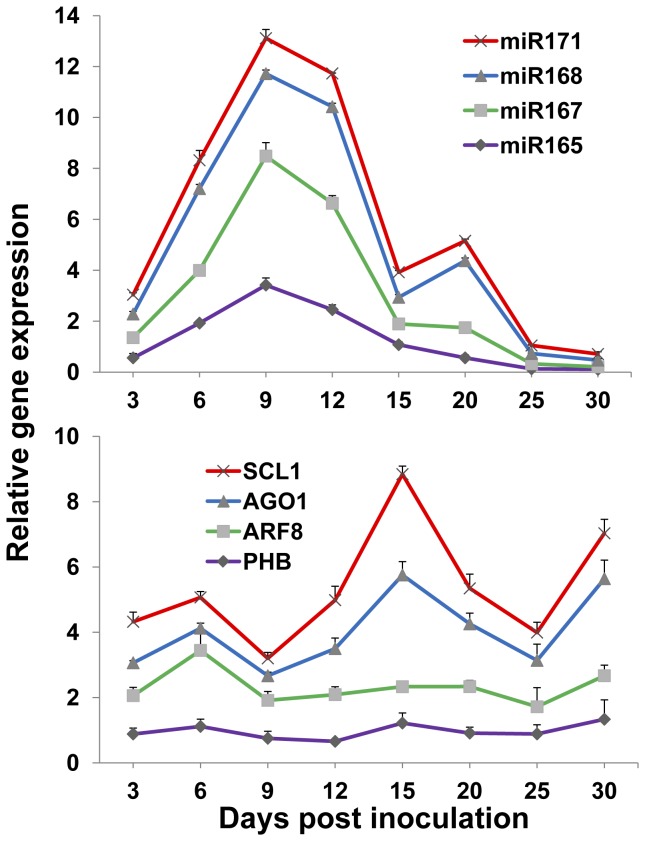
The selected plant miRNAs are also conserved in kenaf plants as compared with Arabidopsis. The sequences are: miR165: UCGGACCAGGCUUCAUCCCCC; miR167: UGAAGCUGCCAGCAUGAUCUA; miR168: UCGCUUGGUGCAGGUCGGGAA; and miR171: UGAUUGAGCCGCGCCAAUAUC. Plant development was regulated by developmental-related genes during plant growth. Target genes PHB, ARF8 and SCL1 of miR165, miR167 and miR171 regulate plant leaf, stem and root development. The expression level of these miRNAs and their respective target genes were further investigated in the HCRSV-infected plants within a period of 30 days. Firstly, the gene transcript of HCRSV-CP was investigated to verify the success of HCRSV infection. The results showed that the gene transcript of HCRSV-CP was increased after HCRSV infection and reached its highest expression level at 15 dpi and decreased from 20 to 30 dpi (Figure 3). Secondly, different from HCRSV-CP, the expression of the four miRNAs and their target genes showed two types of expression. Compared with the mock plants, miR171 and miR168 and their target genes SCL1 and AGO1 showed greater fluctuations after HCRSV infection. The transcript levels of these two miRNAs reached the highest peak at 9 dpi, and a second peak at 20 dpi. (Figure 4, upper graph). Their respective target genes AGO1 and SCL1 showed opposite expression levels, which reached a peak at 6 dpi and a higher peak at 15 dpi. The expression level of SCL1 showed the greatest variations, followed by AGO1 (Figure 4, lower graph). However, even though miR167 and miR165 showed a higher peak at 9 dpi, the expression of their target genes was not significantly affected. These results confirmed that two of the four plant conserved miRNAs targeted the same genes effectively, as reported in other plant species [18-21]. The variations of plant developmental related genes’ expression levels were observed in the HCRSV-infected plants.
Figure 3. Expression of the HCRSV-CP gene transcript after HCRSV infection as determined by qRT-PCR.
The gene transcript level of HCRSV-CP was shown at 3, 6, 9, 12, 15, 20, 25 and 30 days post inoculation (dpi). Relative gene transcript levels (actin as internal control) were analyzed using the 2-∆C T methods.
Figure 4. Expression of miR165, miR167, miR168, miR171 and their respective target genes after HCRSV infection as determined by qRT-PCR.
The expression levels of miRNAs and their respective target genes were shown at 3, 6, 9, 12, 15, 20, 25 and 30 days post inoculation (dpi). Relative gene transcript levels (actin as internal control) were analyzed using the 2-∆∆C T methods. The target genes for miR165, miR167, miR168 and miR171 are phabulosa (PHB), auxin response factor 8 (ARF8), argonaute 1 (AGO1) and scarecrow-like 1 protein (SCL1), respectively. The values of miRNAs and their respect target genes in HCRSV-infected plants were calculated by subtracting the values from mock controls which were all set to 1 for standardization. Means of three independent biological repeats were shown with standard deviations.
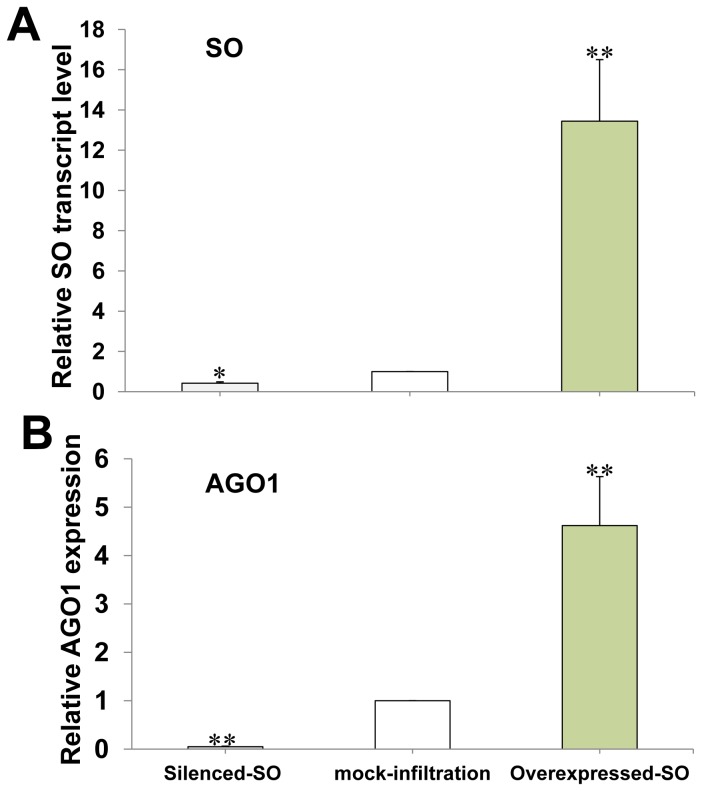
4: Upregulation of the transcript level of AGO1 after SO overexpression
As HCRSV upregulates plant SO transcript and increases sulfate levels in kenaf [7] and upregulated AGO1 in HCRSV-infected plants (Figure 4), the expression level of AGO1 transcript was further investigated after SO overexpression. The efficiency of SO overpression and silencing was first verified in the SO transient expression leaves (Figure 4A). Interestingly, the transcript level of AGO1 gene was also upregulated or downregulated (Figure 4B) after SO was transiently overexpressed or silenced in kenaf plants. In addition, a full-length AGO1 gene was cloned from kenaf using the RACE-PCR. The result validated that the full-length of AGO1 gene from kenaf is 3315 bp and its predicted protein is 121.9 kDa, containing 1104 aa (GenBank accession KF147914).
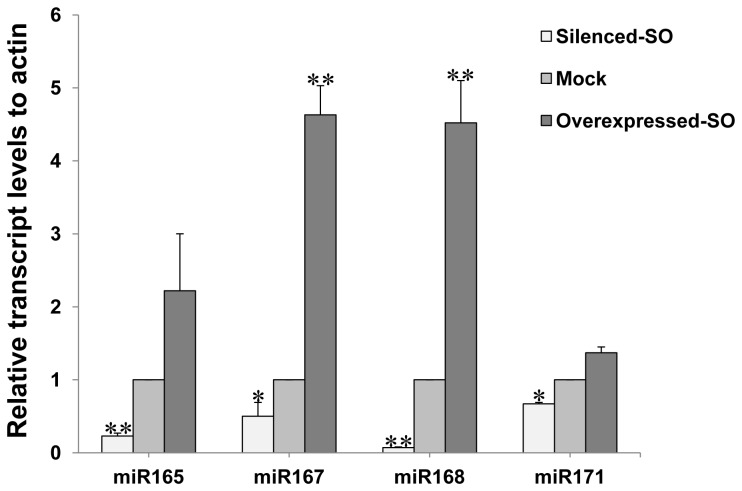
5: Transcript levels of four selected miRNAs after SO overexpression or silencing
The expression levels of four other selected plant conserved miRNAs (miR171, miR168, miR167 and miR165) were also found to be upregulated or downregulated under SO overexpressed or silenced conditions. These results showed that upregulation of SO also increased expression levels of several plant conserved miRNAs, demonstrating a correlation between the fluctuations of the four conserved miRNAs and kenaf plants after infection with HCRSV.
Discussion
Kenaf plant growth retardation and four selected plant conserved developmental related miRNAs were investigated after HCRSV infection. The four conserved miRNAs (miR165, miR167, miR168 and miR171) and their target genes were previously investigated in CMV- and Tomato aspermy virus (TAV)-infected plants. Although their selected miRNAs (including the four miRNAs used in this study) displayed fluctuations after CMV and TAV infections, the changes of their target mRNAs were comparable, suggesting a similar mechanism in perturbing miRNA pathways [20]. Within 30 days of HCRSV infection, two of the four miRNAs and their corresponding target genes indeed fluctuated notably. Two miRNAs (miR171 and miR168) reached the highest expression levels around 10 dpi and dropped until 30 dpi. Their target genes displayed opposite expression patterns corresponding to their regulating miRNAs. Similar expression profiles were also observed in CMV- and TAV-infected plants [20]. On the other hand, the other two miRNAs miR165 and miR167 and their respective target genes displayed less significant changes. Since different miRNAs play different regulation roles during plant development, we did not expect the similar expression profiles for each miRNA. In addition, the regulation between miRNAs and their target genes are a multi-regulation process, meaning one miRNA can target many genes and one gene can be targeted by many miRNAs. It is reasonable that some miRNAs will display different expression profiles after HCRSV infection.
AGO1 is involved in the miRNA pathway which plays essential roles in regulating plant development. The gene transcript of AGO1 was upregulated after SO was transiently overexpressed or in HCRSV-infected kenaf. In this study, for the first time, we have obtained the full-length sequence of AGO1 from kenaf. The fluctuations of the two selected miRNAs miR168 and miR171, together with elevated level of AGO1, regulate the respective target genes. Co-regulation of miR168 and AGO1 gene and stabilization of miR168 by AGO1 are two regulatory processes required to maintain AGO1 at a certain level, and disrupting any of these regulatory processes disturbs a proper functioning of the miRNA pathway [25]. Among the four conserved plant development-related miRNAs, miR168 displayed more significant fluctuation. AGO1, the target gene of miR168, is the main component in the RISC and displayed corresponding fluctuations. The feedback regulation between miR168 and AGO1 causes other related miRNAs and their target genes fluctuations. Because the development-related genes were affected, the plant growth was retarded.
The differences of the miRNA fluctuation in HCRSV-infected kenaf leaves and Agro-infiltrated kenaf leaves may be due to the differences of in vitro and in vivo systems. In both SO overexpressed/silenced kenaf leaves and HCRSV-infected kenaf, miR168 and its target gene AGO1 showed significant change profiles (Figures 4 to 6). This suggests that miR168 plays a crucial role during HCRSV infection. After HCRSV infection, miR171 and its target gene displayed the greatest changes compared with the mock plants. The variations between the mock and HCRSV-infected plants indicate the severe growth retardation of the HCRSV-infected plants (Figure 1D). The root is the crucial organ for plants to absorb and transport water and nutrients which contribute to the growth and development of the upper portion of the plants. Disrupting the normal root development of HCRSV-infected plants, the development of the whole kenaf including stem and leaf was severely inhibited.
Figure 5. Upregulation of AGO1 after SO overexpression.
(A) Relative SO gene transcript level was used to test the SO gene overexpression and silencing efficiency. (B) Upregulation or downregulation of AGO1 gene transcript after SO overexpression or silencing. Actin gene was used as internal control. The relative gene expression amount was analyzed using the 2-∆∆C T method. Significant differences among silenced-SO, mock-infiltration and overexpressed-SO were calculated using the one sample Student’s t-test analysis. Asterisks (* and **) indicate significance at 0.05 and 0.01 levels of confidence, respectively.
Figure 6. Upregulation or downregulation of plant conserved miRNAs (miR165, miR167, miR168 and miR171) expression levels at 3 dpi of SO overexpressed or silenced leaves as determined by qRT-PCR.
Relative miRNAs gene transcription levels (actin as internal control) were analyzed using 2-∆∆C T method. The value of mock control was subtracted from the overexpressed or silenced HcSO samples. Means of three independent biological repeats were shown with standard deviations. Significant differences between mock and Agroinfiltrated leaves were calculated using the one sample Student’s t-test. Asterisks (* and **) indicate significance at 0.05 and 0.01 levels of confidence, respectively.
This study showed that the plant development-related miRNAs fluctuated after HCRSV infection, which displayed plant growth retardation. This analysis of plant development-related miRNAs fluctuations and plant growth retardation may help to improve the understanding of plant viral infection and present strategies to prevent infection in future. For example, the fluctuation of miR168 can influence the fluctuation of AGO1 which is the main component in RNA-induced silencing complex that functions to prevent infection. However, we have not yet established the strong link to correlate miRNAs fluctuations and plant growth retardation after virus infection. A further study needs to be performed in subsequent research.
In conclusion, kenaf plant growth was severely retarded and four selected plant conserved miRNAs were found to fluctuate after HCRSV infection over time. The fluctuations of certain miRNAs, which target conserved plant development related genes, imply their indirect linkages to the plant growth retardation observed in HCRSV-infected kenaf. Since kenaf is a non-wood fiber-producing crop, the reduction in fiber yield resulting from infection by HCRSV is detrimental to its production and reduces its economic values.
Funding Statement
This work was supported by the National University of Singapore research grant R-154-000-552-112. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Huang M, Koh DCY, Weng LJ, Chang ML, Yap YK et al. (2000) Complete nucleotide sequence and genome organization of Hibiscus chlorotic ringspot virus, a new member of the genus Carmovirus: Evidence for the presence and expression of two novel open reading frames. J Virol 74: 3149-3155. doi: 10.1128/JVI.74.7.3149-3155.2000. PubMed: 10708431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Waterworth HE, Lawson RH, Monroe RL (1976) Purification and properties of Hibiscus chlorotic ringspot virus. Phytopathology 66: 570-575. doi: 10.1094/Phyto-66-570. [DOI] [Google Scholar]
- 3. Brunt AA, Spence NJ (2000) The natural occurrence of Hibiscus chlorotic ringspot virus (Carmovirus; Tombusviridae) in aibika or bele (Abelmoschus manihot) in some South Pacific Island countries. Plant Pathology 49: 798-798. doi: 10.1046/j.1365-3059.2000.00501.x. [DOI] [Google Scholar]
- 4. Li SC, Chang YC (2002) First report of Hibiscus chlorotic ringspot virus in Taiwan. Plant Pathology 51: 803-803. doi: 10.1046/j.1365-3059.2002.00775.x. [DOI] [Google Scholar]
- 5. Tang J, Elliott DR, Quinn BD, Clover GRG, Alexander BJR (2008) Occurrence of Hibiscus chlorotic ringspot virus in Hibiscus spp. in new Zealand. Plant Disease 92: 1367-1367. doi: 10.1094/PDIS-92-9-1367C. [DOI] [PubMed] [Google Scholar]
- 6. Zhou T, Fan ZF, Li HF, Wong SM (2006) Hibiscus chlorotic ringspot virus p27 and its isoforms affect symptom expression and potentiate virus movement in kenaf (Hibiscus cannabinus L.). Mol Plant Microbe Interact 19: 948-957 [DOI] [PubMed]
- 7. Zhang X, Wong SM (2009) Hibiscus chlorotic ringspot virus upregulates plant sulfite oxidase transcripts and increases sulfate levels in kenaf (Hibiscus cannabinus L.). J Gen Virol 90: 3042-3050 [DOI] [PubMed]
- 8. Gao R, Ng FK, Liu P, Wong SM (2012) Hibiscus chlorotic ringspot virus coat protein upregulates sulfur metabolism genes for enhanced pathogen defense. Mol Plant Microbe Interact 25: 1574-1583. doi: 10.1094/MPMI-08-12-0203-R. PubMed: 23134059. [DOI] [PubMed] [Google Scholar]
- 9. Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215-233. doi: 10.1016/j.cell.2009.01.002. PubMed: 19167326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281-297. doi: 10.1016/S0092-8674(04)00045-5. PubMed: 14744438. [DOI] [PubMed] [Google Scholar]
- 11. Zhang B, Pan X, Cannon CH, Cobb GP, Anderson TA (2006) Conservation and divergence of plant microRNA genes. Plant J 46: 243-259. doi: 10.1111/j.1365-313X.2006.02697.x. PubMed: 16623887. [DOI] [PubMed] [Google Scholar]
- 12. Lu YD, Gan QH, Chi XY, Qin S (2008) Roles of microRNA in plant defense and virus offense interaction. Plant Cell Rep 27: 1571-1579. doi: 10.1007/s00299-008-0584-z. PubMed: 18626646. [DOI] [PubMed] [Google Scholar]
- 13. Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57: 19-53. doi: 10.1146/annurev.arplant.57.032905.105218. PubMed: 16669754. [DOI] [PubMed] [Google Scholar]
- 14. Chen X (2012) Small RNAs in development - insights from plants. Curr Opin Genet Dev 22: 361-367. doi: 10.1016/j.gde.2012.04.004. PubMed: 22578318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kidner CA, Martienssen RA (2005) The developmental role of microRNA in plants. Curr Opin Plant Biol 8: 38-44. doi: 10.1016/j.pbi.2004.11.008. PubMed: 15653398. [DOI] [PubMed] [Google Scholar]
- 16. Mlotshwa S, Pruss GJ, Vance V (2008) Small RNAs in viral infection and host defense. Trends Plant Sci 13: 375-382. doi: 10.1016/j.tplants.2008.04.009. PubMed: 18550416. [DOI] [PubMed] [Google Scholar]
- 17. Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N et al. (2006) A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312: 436-439. doi: 10.1126/science.1126088. PubMed: 16627744. [DOI] [PubMed] [Google Scholar]
- 18. Llave C, Xie Z, Kasschau KD, Carrington JC (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297: 2053-2056. doi: 10.1126/science.1076311. PubMed: 12242443. [DOI] [PubMed] [Google Scholar]
- 19. Zhang J, Zeng R, Chen J, Liu X, Liao Q (2008) Identification of conserved microRNAs and their targets from Solanum lycopersicum Mill. Gene 423: 1-7. doi: 10.1016/j.gene.2008.05.023. PubMed: 18602455. [DOI] [PubMed] [Google Scholar]
- 20. Feng J, Wang K, Liu X, Chen S, Chen J (2009) The quantification of tomato microRNAs response to viral infection by stem-loop real-time RT-PCR. Gene 437: 14-21. doi: 10.1016/j.gene.2009.01.017. PubMed: 19374024. [DOI] [PubMed] [Google Scholar]
- 21. Jay F, Wang Y, Yu A, Taconnat L, Pelletier S et al. (2011) Misregulation of AUXIN RESPONSE FACTOR 8 underlies the developmental abnormalities caused by three distinct viral silencing suppressors in Arabidopsis . PLoS Pathog 7: e1002035 PubMed: 21589905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dunoyer P, Lecellier CH, Parizotto EA, Himber C, Voinnet O (2004) Probing the microRNA and small interfering RNA pathways with virus-encoded suppressors of RNA silencing. Plant Cell 16: 1235-1250. doi: 10.1105/tpc.020719. PubMed: 15084715. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23. Simón-Mateo C, García JA (2006) MicroRNA-guided processing impairs Plum pox virus replication, but the virus readily evolves to escape this silencing mechanism. J Virol 80: 2429-2436. doi: 10.1128/JVI.80.5.2429-2436.2006. PubMed: 16474149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Khraiwesh B, Zhu JK, Zhu J (2012) Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim Biophys Acta 1819: 137-148. doi: 10.1016/j.bbagrm.2011.05.001. PubMed: 21605713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vaucheret H, Mallory AC, Bartel DP (2006) AGO1 homeostasis entails coexpression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Mol Cell 22: 129-136. doi: 10.1016/j.molcel.2006.03.011. PubMed: 16600876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Duan CG, Fang YY, Zhou BJ, Zhao JH, Hou WN et al. (2012) Suppression of Arabidopsis ARGONAUTE1-mediated slicing, transgene-induced RNA silencing, and DNA methylation by distinct domains of the Cucumber mosaic virus 2b protein. Plant Cell 24: 259-274. doi: 10.1105/tpc.111.092718. PubMed: 22247253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moissiard G, Voinnet O (2006) RNA silencing of host transcripts by Cauliflower mosaic virus requires coordinated action of the four Arabidopsis Dicer-like proteins. Proc Natl Acad Sci U S A 103: 19593-19598. doi: 10.1073/pnas.0604627103. PubMed: 17164336. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28. Shimura H, Pantaleo V, Ishihara T, Myojo N, Inaba J et al. (2011) A viral satellite RNA induces yellow symptoms on tobacco by targeting a gene involved in chlorophyll biosynthesis using the RNA silencing machinery. PLOS Pathog 7: e1002021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smith NA, Eamens AL, Wang MB (2011) Viral small interfering RNAs target host genes to mediate disease symptoms in plants. PLoS Pathog 7: e1002022 PubMed: 21573142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu H, Ito T, Wellmer F, Meyerowitz EM (2004) Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat Genet 36: 157-161. doi: 10.1038/ng1286. PubMed: 14716314. [DOI] [PubMed] [Google Scholar]
- 31. Hellens R, Mullineaux P, Klee H (2000) A guide to Agrobacterium binary Ti vectors. Trends Plant Sci 5: 446-451. doi: 10.1016/S1360-1385(00)01740-4. PubMed: 11044722. [DOI] [PubMed] [Google Scholar]
- 32. Varkonyi-Gasic E, Wu R, Wood M, Walton EF, Hellens RP (2007) Protocol: a highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 3: 12. doi: 10.1186/1746-4811-3-12. PubMed: 17931426. [DOI] [PMC free article] [PubMed] [Google Scholar]