Abstract
Leishmania donovani in India causes visceral infection (kala-azar) and dermal infection (post-kala-azar dermal leishmaniasis). We report here the identification of polymorphism in a well-defined genetic locus among the Leishmania parasites causing the visceral and dermal manifestations, in a comparison of 15 post-kala-azar dermal leishmaniasis and 12 kala-azar patient isolates.
Visceral leishmaniasis (VL), or kala-azar (KA), caused by Leishmania donovani, is responsible for severe mortality and morbidity with 500,000 new cases every year (2). In India the disease is endemic in eastern parts of the country, primarily in the state of Bihar. Post-KA dermal leishmaniasis (PKDL) is an unusual dermatosis that develops as a sequel of KA in 10 to 20% of VL cases in India and in 60% of VL cases in Sudan (11, 19). KA transmission in India is thought to be anthroponotic, and PKDL patients are believed to be the sole source of the parasites (17). Studies of parasite biology in PKDL are few, primarily because of the difficulty of culturing the parasite. Intrinsic differences are reported to exist between the KA and PKDL isolates (1).
One of the hallmarks of these pathogenic protozoa is the diversity of tropism and disease resulting from infection. It is hypothesized that reactivation of persistent infections underlies some of the most severe forms of leishmaniasis, including PKDL. Little is known about parasite factors required for persistent infections (16). Moreover, humoral immune responses in PKDL patients are distinct from those in KA patients (12). Therefore, we sought to identify molecular differences between parasites isolated from KA and PKDL patients that may underlie the diversity in clinical manifestations of the disease.
The nuclear gene probe LdP13 allowed discrimination between L. donovani and L. infantum and between geographical isolates of L. donovani (9, 10). We utilized this probe to examine the genetic variation in parasites isolated from Indian KA and PKDL patients.
Parasite isolates.
Parasite isolates from PKDL and KA patients were prepared from clinical samples as described previously (13, 14). The KA and PKDL patients came from different regions within Bihar, categorized as being of high, moderate, and low endemicity. An immunofluorescence assay with monoclonal antibodies D2, T-1, and T-10 (kind gifts from Tropical Disease Research, World Health Organization) specific to L. donovani, L. major, and L. tropica, respectively, and analysis of isoenzyme profiles with a panel of five enzymes (6-phosphogluconic dehydrogenase, nucleoside hydrolase, glucose-6-phosphate dehydrogenase, malate dehydrogenase, and malate esterase) identified all the isolates as L. donovani (5, 6).
Description of the probe and Southern blot analyses.
The DNA fragment LdP13 has been cloned previously and mapped to a chromosome of 0.8 Mb in size (10). The nucleotide sequence of the LdP13 probe was found to be identical to that of the ζ and ɛ subunit regions of the 28S rRNA gene of L. donovani MHOM/SD/00/Khartoum (18). The 774-bp probe used spanned the region from bp 674 to 1448, reported in the GenBank database (accession no. AF115465).
This probe (LdP13) was used to investigate intraspecific DNA polymorphisms among 15 isolates of L. donovani obtained from skin lesions of PKDL patients and 12 isolates from bone marrow of KA patients and identified clear molecular variations between Indian KA and PKDL patient isolates. Southern blots were hybridized with 32P-labeled LdP13 probe under standard conditions (9).
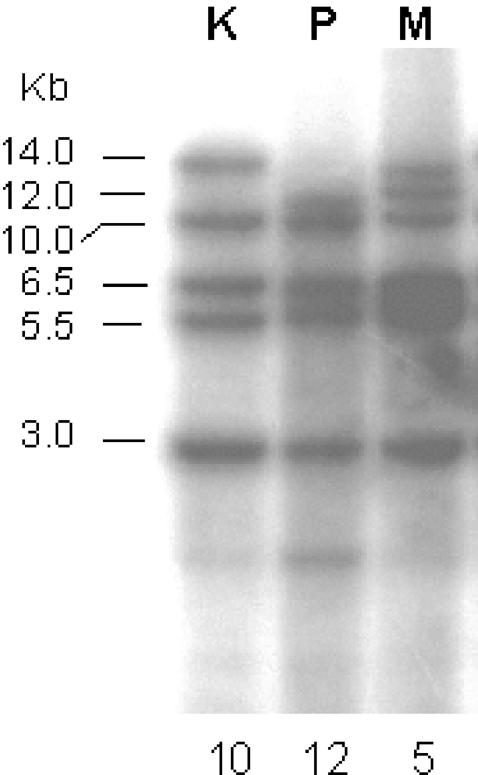
Based on the pattern obtained with MvaI, NaeI, NcoI, and PstI initially on a subset of isolates, subsequent analysis was done with MvaI. MvaI restriction followed by hybridization with LdP13 revealed three discrete restriction patterns in the 27 isolates. Figure 1 shows the three different restriction patterns. A majority of the KA patient isolates and the reference isolate of Indian origin (Ld AG83) gave an identical pattern (termed K), yielding fragments of 14.0, 10.0, 6.5, 5.5, and 3.0 kb (Fig. 1). In contrast, a majority of the PKDL patient isolates gave an identical pattern (termed P) that showed fragments of 12.0, 10.0, 6.5, 5.5, and 3.0 kb. The remaining PKDL and KA patient isolates could be clustered into a third group (termed M for mixed) that showed both the 14.0- and the 12.0-kb fragments, in addition to the smaller common fragments. Taken together, these data suggested that the PKDL patient isolates in the first group have gained an MvaI site that the KA patient isolates lack. The third group, giving both the 14.0- and 12.0-kb fragments, could represent a transition stage between the two groups. The intensity of the 14.0-kb fragment in the mixed group was approximately half of that in the first group, suggesting that only one of the alleles might have gained the MvaI site. Interestingly, the percentage of KA patient isolates falling in the mixed group (16.7%) was quite comparable to the percentage of KA patients who go on to develop PKDL. Further studies are necessary to examine if the observed differences are responsible for the phenotypic differences.
FIG. 1.
Southern blot analysis of representative isolates from the three groups. Genomic DNA (5 μg) of L. donovani from groups K, P, and M was digested for 16 h at 37°C with MvaI (4 U/μg of DNA; MBI Fermentas) and resolved on a 1% agarose gel, and blots were probed with 32P-labeled LdP13 and visualized by autoradiography. The numbers below the lanes indicate the numbers of isolates falling in the respective categories.
No correlation was found among the groups K, P, and M with the geographic origin with respect to KA endemicity, as all groups contained samples from low-, medium-, and high-endemicity areas within the state of Bihar (Table 1). Within PKDL patient isolates, a majority of the P group sample patients had a history of KA in excess of 4 years while all members in patient group M had a comparatively short history (Table 2).
TABLE 1.
Geographic origins of the KA and PKDL isolates in relation to KA endemicity
| Isolatea | Region of origin | Level of KA endemicity in regionb |
|---|---|---|
| K group | ||
| K1 | Vaishali | M |
| K2 | Bhagalpur | M |
| K3 | Munger | L |
| K4 | Siwan | L |
| K5 | Saaran | H |
| K6 | Madhubani | M |
| K7 | Darbhanga | H |
| K8 | Saaran | H |
| K9 | Katihar | H |
| K10 | Saaran | H |
| P group | ||
| P1 | Siwan | L |
| P2 | Siwan | L |
| P3 | Gopalganj | L |
| P4 | Darbhanga | H |
| P5 | Vaishali | M |
| P6 | Saharsa | H |
| P7 | Begusarai | M |
| P8 | Muzaffarpur | H |
| P9 | Araria | H |
| P10 | Saaran | H |
| P11 | Khagaria | H |
| P12 | Saharsa | H |
| M group | ||
| K11 | Saharsa | H |
| K12 | Gopalganj | L |
| P13 | Madhubani | M |
| P14 | Gopalganj | L |
| P15 | Madhubani | M |
Groups K, P, and M refer to KA, PKDL, and mixed groups, respectively, as defined in the text. K1 to K12 and P1 to P15 are isolates from 12 KA and 15 PKDL patients, respectively.
L, M, and H refer to low, medium, and high endemicity, respectively. Information on endemicity is taken from reference 8a.
TABLE 2.
Profile of PKDL patients belonging to the PKDL (P) and mixed (M) groups
| Group | No. of patients with:
|
|||||
|---|---|---|---|---|---|---|
| History of disease (yr)
|
Clinical presentation
|
|||||
| KA
|
PKDL
|
|||||
| <4 | 4-12 | <1 | 1-6 | Polymorphic | Macular | |
| P | 4 | 8 | 3 | 9 | 8 | 4 |
| M | 3 | 0 | 3 | 0 | 3 | 0 |
Molecular techniques such as PCR-restriction fragment length polymorphism, random amplified polymorphic DNA, and single-strand conformation polymorphism analyses have been used to demonstrate the genetic variability within and between different Leishmania species including L. donovani complex (4, 8, 15). Earlier studies to differentiate between VL and PKDL patient isolates in Sudan did not reveal any correlation between sequence polymorphisms and clinical manifestations of human disease (3, 7). No such studies have been reported with isolates of L. donovani from Indian populations. Despite overall similarities there are many important differences between Indian and African forms of KA and PKDL, warranting investigations in this direction (19). Our approach using a probe from an expressed region reported to show intraspecific differences has led to the identification of a correlation between genetic variability among and clinical manifestations of Indian isolates. Whether similar differences occur in Sudanese VL and PKDL patient isolates in the LdP13 locus remains to be investigated.
REFERENCES
- 1.Das Gupta, S., D. K. Ghosh, and H. K. Majumder. 1991. A cloned kinetoplast DNA mini-circle fragment from a Leishmania spp. specific for post-kala-azar dermal leishmaniasis strains. Parasitology 102:187-191. [DOI] [PubMed] [Google Scholar]
- 2.Desjeux, P. 2001. The increase in risk factors for leishmaniasis worldwide. Trans. R. Soc. Trop. Med. Hyg. 95:239-243. [DOI] [PubMed] [Google Scholar]
- 3.El Tai, N. O., M. E. Fari, I. Mauricio, M. A. Miles, L. Oskam, S. H. El Safi, W. H. Presber, and G. Schonian. 2001. Leishmania donovani: intraspecific polymorphisms of Sudanese isolates revealed by PCR-based analyses and DNA sequencing. Exp. Parasitol. 97:35-44. [DOI] [PubMed] [Google Scholar]
- 4.Guerbouj, S., K. Victoir, I. Guizani, N. Seridi, S. N. Nuwayri, M. Belkaid, R. B. Ismail, D. Le Ray, and J. C. Dujardin. 2001. Gp63 gene polymorphism and population structure of Leishmania donovani complex: influence of the host selection? Parasitology 122:25-35. [DOI] [PubMed] [Google Scholar]
- 5.Jaffe, C. L., E. Bennett, G. Grimaldi, Jr., and D. McMahon Pratt. 1984. Production and characterization of species specific monoclonal antibodies against Leishmania donovani for immunodiagnosis. J. Immunol. 133:440-447. [PubMed] [Google Scholar]
- 6.Kreutzer, R. D., and H. A. Christensen. 1980. Characterization of Leishmania spp by isoenzyme electrophoresis. Am. J. Trop. Med. Hyg. 29:199-208. [DOI] [PubMed] [Google Scholar]
- 7.Lewin, S., G. Schonian, N. ElTai, L. Oskam, P. Bastien, and W. Presber. 2002. Strain typing in Leishmania donovani by using sequence-confirmed amplified region analysis. Int. J. Parasitol. 32:1267-1276. [DOI] [PubMed] [Google Scholar]
- 8.Mauricio, I. L., M. W. Gaunt, J. R. Stothard, and M. A. Miles. 2001. Genetic typing and phylogeny of the Leishmania donovani complex by restriction analysis of PCR amplified gp63 intergenic regions. Parasitology 122:393-403. [DOI] [PubMed] [Google Scholar]
- 8a.Ministry of Health and Family Welfare. 2000. Kala-azar facts worthy to know. Ministry of Health and Family Welfare, Government of India, New Delhi, India.
- 9.Pogue, G. P., S. Koul, N. S. Lee, D. M. Dwyer, and H. L. Nakhasi. 1995. Identification of intra- and interspecific Leishmania genetic polymorphisms by arbitrary primed polymerase chain reactions and use of polymorphic DNA to identify differentially regulated genes. Parasitol. Res. 81:282-290. [DOI] [PubMed] [Google Scholar]
- 10.Pogue, G. P., M. Joshi, N. S. Lee, D. M. Dwyer, R. T. Kenney, A. A. Gam, and H. L. Nakhasi. 1996. Conservation of low-copy gene loci in Old World leishmanias identifies mechanisms of parasite evolution and diagnostic markers. Mol. Biochem. Parasitol. 81:27-40. [DOI] [PubMed] [Google Scholar]
- 11.Ramesh, V., and A. Mukherjee. 1995. Post kala-azar dermal leishmaniasis. Int. J. Dermatol. 34:85-91. [DOI] [PubMed] [Google Scholar]
- 12.Salotra, P., A. Raina, and V. Ramesh. 1999. Western blot analysis of humoral immune response to Leishmania donovani antigens in patients with post kala-azar dermal leishmaniasis. Trans. R. Soc. Trop. Med. Hyg. 93:98-101. [DOI] [PubMed] [Google Scholar]
- 13.Salotra, P., G. Sreenivas, G. P. Pogue, N. Lee, H. L. Nakhasi, V. Ramesh, and N. S. Negi. 2001. Development of a species-specific PCR assay for detection of Leishmania donovani in clinical samples from patients with kala-azar and post-kala-azar dermal leishmaniasis. J. Clin. Microbiol. 39:849-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salotra, P., G. Sreenivas, K. R. Beena, A. Mukherjee, and V. Ramesh. 2003. Parasite identification by molecular and immunological methods in post kala-azar dermal leishmaniasis patients in India. J. Clin. Pathol. 56:840-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schonian, G., H. Akuffo, S. Lewin, K. Maasho, S. Nylen, F. Pratlong, C. L. Eisenberger, L. F. Schnur, and W. Presber. 2000. Genetic variability within the species Leishmania aethiopica does not correlate with clinical variations of cutaneous leishmaniasis. Mol. Biochem. Parasitol. 106:239-248. [DOI] [PubMed] [Google Scholar]
- 16.Spath, G. F., L. F. Lye, H. Segawa, D. L. Sacks, S. J. Turco, and S. M. Beverley. 2003. Persistence without pathology in phosphoglycan-deficient Leishmania major. Science 301:1241-1243. [DOI] [PubMed] [Google Scholar]
- 17.Thakur, C. P., and K. Kumar. 1992. Post kala-azar dermal leishmaniasis: a neglected aspect of kala-azar control programmes. Ann. Trop. Med. Parasitol. 86:355-359. [DOI] [PubMed] [Google Scholar]
- 18.Yan, S., M. J. Lodes, M. Fox, P. J. Myler, and M. Stuart. 1999. Characterization of the Leishmania donovani ribosomal RNA promoter. Mol. Biochem. Parasitol. 103:197-210. [DOI] [PubMed] [Google Scholar]
- 19.Zijlstra, E. E., A. M. Musa, E. A. G. Khalil, I. M. El Hassan, and A. M. El Hassan. 2003. Post kala-azar dermal leishmaniasis. Lancet Infect. Dis. 3:87-98. [DOI] [PubMed] [Google Scholar]