Abstract
Background
Accurate identification of hepatocellular cancer (HCC) cases from automated data is needed for efficient and valid quality improvement initiatives and research. We validated HCC ICD-9 codes, and evaluated whether natural language processing (NLP) by the Automated Retrieval Console (ARC) for document classification improves HCC identification.
Methods
We identified a cohort of patients with ICD-9 codes for HCC during 2005–2010 from Veterans Affairs administrative data. Pathology and radiology reports were reviewed to confirm HCC. The positive predictive value (PPV), sensitivity, and specificity of ICD-9 codes were calculated. A split validation study of pathology and radiology reports was performed to develop and validate ARC algorithms. Reports were manually classified as diagnostic of HCC or not. ARC generated document classification algorithms using the Clinical Text Analysis and Knowledge Extraction System. ARC performance was compared to manual classification. PPV, sensitivity, and specificity of ARC were calculated.
Results
1138 patients with HCC were identified by ICD-9 codes. Based on manual review, 773 had HCC. The HCC ICD-9 code algorithm had a PPV of 0.67, sensitivity of 0.95, and specificity of 0.93. For a random subset of 619 patients, we identified 471 pathology reports for 323 patients and 943 radiology reports for 557 patients. The pathology ARC algorithm had PPV of 0.96, sensitivity of 0.96, and specificity of 0.97. The radiology ARC algorithm had PPV of 0.75, sensitivity of 0.94, and specificity of 0.68.
Conclusion
A combined approach of ICD-9 codes and NLP of pathology and radiology reports improves HCC case identification in automated data.
Keywords: Hepatocellular cancer, natural language processing
Introduction
In the United States, approximately 20,000 new cases of hepatocellular cancer (HCC) are diagnosed per year.1 As the incidence of HCC continues to rise and treatment options evolve, large scale studies are necessary to characterize HCC-related epidemiology, health care utilization, and health outcomes.2,3 Furthermore, quality improvement initiatives require efficient methods of extracting data from the medical record and administrative data.
Several studies have utilized administrative data to identify and evaluate outcomes in patients with HCC.4,5 Administrative databases typically include demographic information, procedure codes, and International Classification of Diseases, 9th Revision (ICD-9) diagnosis codes, which can be used to identify patients diagnosed with HCC. However, administrative claims data may be inaccurate due to limited clinical data, diagnostic errors by providers, or miscoded data.6 ICD-9 codes indicative of HCC have been previously validated and found to have a positive predictive value (PPV) equal to 86% although study findings were limited by a small sample size.4
An alternative strategy is to identify HCC patients directly from the electronic health record. In contrast to administrative databases, the electronic health record is the medical record that contains laboratory data and clinical notes (e.g. health care provider progress notes, procedure notes, pathology reports, and radiology reports). However, establishing an HCC diagnosis using unstructured text data from the electronic health record requires manual review which is costly, time consuming, and impractical in large cohorts. Natural language processing (NLP) is a sub-discipline of computer science and linguistics that adds structure to otherwise unstructured free text. NLP can be used for automated document-level classification to identify diseases or treatments in which standard diagnostic or procedural codes are not available or inaccurate. The Automated Retrieval Console (ARC) is NLP-based software that allows investigators without programming expertise to design and perform NLP assisted document-level classification.7 ARC works by combining features derived from NLP pipelines with supervised machine learning classification algorithms.8 ARC creates an algorithm to classify documents based on specific NLP features, such as noun phrases, verb phrases, or negating words.7 ARC has been used successfully in a number of studies to facilitate electronic health record based research including studies to classify colon and prostate cancer based on pathology reports, indications for colonoscopy in inflammatory bowel disease, lung cancer based on radiology reports, and psychotherapy treatment from physician notes.7,9,10
The objectives of this study were to identify the accuracy of ICD-9 codes for HCC using VA administrative databases and to propose a hybrid ICD-9 and NLP based algorithm to identify patients with HCC using pathology and radiology text reports available through the electronic health record.
Methods
Data sources
This was a retrospective study of patients with HCC identified from national VA administrative databases between fiscal years 2005 and 2010. The Outpatient Clinic File contains the ICD-9 codes for principal diagnoses for all outpatient VA visits since 1996. The Patient Treatment File captures the ICD-9 codes for up to ten discharge diagnoses for all inpatient treatment visit at the VA since 1970.11 These two administrative databases were used to identify the study cohort. The national VA electronic health record is contained in the Compensation and Pension Records Interchange. HCC diagnoses were verified by manual review of provider notes, laboratory data, radiology reports, and pathology reports in the electronic health record.
ICD-9 code sampling algorithms
We performed two algorithms of ICD-9 code sampling. For Algorithm 1, we identified a random sample of patients based on the occurrence of HCC code (155.0) in two outpatient or inpatient encounters in the absence of a cholangiocarcinoma code (155.1) during 2005 to 2010. Ancillary claims files were not used, because patients included in these files only typically do not have complete HCC diagnostic information in the medical record. For Algorithm 2, we modified Algorithm 1 by excluding patients who had a non-hepatobiliary cancer code (colorectal cancer (153.1–153.9, 154.0), lung cancer (162.0), or breast cancer (174.0)) prior to their HCC diagnosis code during 1997 to 2010. The purpose of Algorithm 2 was to test if the predictive value of the sampling algorithm improved by excluding patients with liver metastases misclassified as HCC. We also reviewed a random sample of 612 patients with ICD-9 codes for cirrhosis (571.2, 571.5, 571.6) and without HCC codes to calculate the sensitivity of HCC codes. For this cirrhosis comparator group, we performed manual chart review and identified 40 cases of HCC during the study period.
The PPV, sensitivity and specificity of the ICD-9 code sampling algorithms were determined by comparing the diagnosis of HCC by ICD-9 codes to comprehensive medical record review of radiology reports, pathology reports, and clinician notes in the electronic health record. For patients who did not have a diagnosis of HCC based on medical record review, their true diagnosis was recorded if present (metastases from non-HCC primary, cirrhosis, benign liver lesion, no liver-related diagnosis, epithelioid hemangioendothelioma).
Natural language processing algorithms
NLP with ARC
ARC classifies text documents using NLP pipelines to parse documents into structured fragments based on parts of speech, negated terms and a library of medical and non-medical terms.7 The Clinical Text Analysis and Knowledge Extraction System is an Unstructured Information Management Architecture - based analysis pipeline created to parse medical documents.8,12 ARC performs document classification using conditional random fields implementation from the Machine Learning for Language Toolkit, which is a text-based information retrieval tool. ARC does not require custom software development. Multiple combinations of linguistic features for a given classification problem, such as the presence or absence of HCC in a document, are automatically generated by ARC. ARC calculates how each classification model performs against the training set using 10-fold cross-validation. Investigators are shown the recall, precision, and harmonic mean (F-measure) of all models generated by ARC. An individual classification model can then be selected and tested on a separate set of documents.
Manual Classification
Pathology and radiology reports were chosen for this study because imaging or biopsy can be used for HCC diagnosis.13 NLP algorithms were developed using a 70%/30% split validation method for pathology reports and radiology reports separately.10 Documents were randomly divided into a 70% training set for ARC to generate algorithms and a 30% testing set to validate algorithms. For the pathology algorithm, only liver biopsy reports were used. For the radiology algorithm, abdominal CT and abdominal MRI reports were used. All biopsies and images were performed at a VA facility.
Pathology reports from all liver biopsies within 1 year of the first ICD-9 code for HCC were identified within the medical record. All pathology reports were independently classified as HCC or non-HCC by two physicians (JH and YS) who were blinded to the remainder of the medical record and to the classification of the other reader. Disagreement was resolved by a third physician (HES).
Similarly, CT and MRI text reports within 6 month before or after the index HCC ICD-9 code were tested using a random sample of the national cohort. All radiology reports were independently classified as definite, probable or no HCC by two physicians (JH and YS) who were blinded to the remainder of the medical record and to the classification of the other reader. Manual classification of radiology reports was based on classic imaging features of arterial enhancement or contrast wash-out on delayed phases, lesion size, clinical history, and final assessment reported by the radiologist. Disagreement was resolved by a third physician (HES).
Statistical analysis
We calculated the PPV, sensitivity, and specificity of the ICD-9 code sampling algorithm alone for identifying HCC cases in automated administrative data. The PPV reflects how well the ICD-9 code sampling algorithm correctly predicts the presence of hepatocellular cancer in the electronic health record. A weighting scheme was applied to adjust for skewed sampling. Subjects were weighted based on the inverse of the probability of being sampled within a stratified sampling scheme. Stratified sampling was necessitated by the low counts of HCC positive compared to those of HCC negative subjects. Because each code pattern stratum had different sampling probabilities, it is necessary for inferential validity to accordingly rescale the sample counts to the full set of patients from which the sample is derived to calculate sensitivity, specificity and positive predictive value.
For manual classification of pathology and radiology reports, the Cohen’s Kappa for agreement in pathology and radiology report classification was calculated. Cross validation was performed on the ARC training cohort. The training set is divided into 10 equal groups, and a classification algorithm is created based on the first 9 of 10 groups. Then, each classification algorithms is validated on the tenth group. The classification algorithm is similarly applied to the other 9 groups and the performance parameters are averaged. The ARC algorithm was validated by calculating the precision, recall, and specificity of the algorithm on the test cohort (Figure 1). The precision of an ARC algorithm estimates PPV, which indicates how well ARC document classification predicts the presence of HCC in the electronic health record for patients with an ICD-9 code for HCC. Recall is an estimate of sensitivity. Statistical analysis was performed using SAS (Version 9.3, Cary, NC).
Figure 1.
Automated Retrieval Console calculations for precision, recall, and specificity
The study was approved by the Baylor College of Medicine Institutional Review Board.
Results
We identified 1138 patients with at least two ICD-9 codes for HCC from VA administrative files. These patients represented 117 VA clinical sites. Table 1 shows the demographic characteristics of the cohort. The median age was 62 years. We identified 773 cases of HCC on manual review. Algorithm 1 had a moderate PPV of 67% and a high of sensitivity of 95%. Table 2 shows the PPV, sensitivity, and specificity of each algorithm. There were 365 patients with two ICD-9 codes for HCC who did not have evidence of HCC on chart review. The most common non-HCC diagnosis was metastasis from non-HCC primary (42%), followed by liver lesion of unknown etiology (27%), benign liver lesion (10%), no liver-related diagnosis (7%), cholangiocarcinoma (6%), cirrhosis (5%), and surveillance for HCC (3%).
Table 1.
Demographic characteristics of patients with two administrative codes for HCC (n=1,138)
| HCC (n=773) | No HCC (n=365) | ||||
|---|---|---|---|---|---|
| n | % | n | % | P-value | |
| Age | |||||
| < 55 | 113 | 15 | 58 | 16 | 0.10 |
| 55–64 | 360 | 47 | 143 | 39 | |
| 65–74 | 144 | 18 | 85 | 23 | |
| ≥ 75 | 156 | 20 | 79 | 22 | |
| Gender | |||||
| Male | 772 | 99 | 355 | 97 | <0.01 |
| Female | 1 | 1 | 10 | 3 | |
| Race | |||||
| Black | 190 | 25 | 73 | 20 | 0.13 |
| White | 556 | 72 | 283 | 78 | |
| Other | 27 | 3 | 9 | 2 | |
| Ethnicity | |||||
| Hispanic | 121 | 16 | 50 | 14 | 0.66 |
| Not Hispanic | 647 | 84 | 312 | 85 | |
| Unknown | 5 | 1 | 3 | 1 | |
| Geographic region | |||||
| Central | 109 | 14 | 55 | 15 | 0.08 |
| East | 174 | 23 | 91 | 25 | |
| South | 314 | 41 | 160 | 44 | |
| West | 176 | 23 | 59 | 16 | |
| Additional insurance | |||||
| Yes | 398 | 51 | 204 | 56 | 0.16 |
| No | 375 | 49 | 161 | 44 | |
| Cirrhosis | |||||
| Yes | 530 | 69 | 68 | 19 | <0.01 |
| Barcelona stage | |||||
| A | 81 | 10 | * | ||
| B | 166 | 22 | |||
| C | 193 | 25 | |||
| D | 133 | 17 | |||
| Unknown | 200 | 26 | |||
Patients without hepatocellular cancer (HCC) cannot be staged
Table 2.
Positive predictive value (PPV), sensitivity and specificity for HCC case identification for each ICD-9 code sampling method
| Algorithm | PPV (%) | Sensitivity (%) |
Specificity (%) |
|---|---|---|---|
| Algorithm 1: Two 155.0 codes (n=1,138) | 67 | 95 | 93 |
| Algorithm 2: Two 155.0 codes and no other malignancy (n=922) | 73 | 84 | 95 |
Pathology document classification by NLP
We randomly selected 619 patients to assess the hybrid ICD-9 code and ARC document classification algorithm. Among these patients, 323 patients had 471 biopsy pathology reports, and 557 patients had 943 radiology reports available. The pathology training set included 359 reports, and 45% of the pathology training set reports were manually classified as definite HCC. The Cohen’s Kappa was 0.98 for agreement between judges on manual classification of pathology reports.
ARC automatically tests 80 algorithms using different combinations of NLP feature types and classifiers. Algorithms are selected based on high precision or high recall depending on the intended use of the classification algorithm. For our study, we selected the algorithm with the highest F-measure, the harmonic mean of precision and recall. In the training cohort, the best performing ARC algorithm for pathology document classification had an F-measure of 0.91, precision of 0.90, and recall of 0.93 on cross-validation. Recall of 0.93 indicated that 93% of pathology reports were correctly identified as consistent with HCC by ARC. Precision of 0.90 indicated that 90% of the pathology reports classified as consistent with HCC by ARC were also manually classified as consistent with HCC.
The pathology test set included 112 biopsy reports from 78 patients, of which 52 were manually classified as definite HCC. When the best performing ARC algorithm was applied to the pathology test set, document classification by ARC had a sensitivity of 0.96, specificity of 0.97, and PPV of 0.96 in comparison to manual classification for the test set (Table 3).
Table 3.
Positive predictive value (PPV), sensitivity and specificity for HCC case identification from pathology and radiology reports using the ARC algorithm
| Method | PPV | Sensitivity | Specificity |
|---|---|---|---|
| Pathology reports (n=471) | 0.96 | 0.96 | 0.97 |
| Radiology reports (n=943) | 0.75 | 0.94 | 0.68 |
Radiology document classification by NLP
The radiology training set included 664 CT and MRI reports, and 429 (65%) of the radiology training set reports were manually classified as suspicious for HCC. The Cohen’s Kappa was 0.81 for agreement between judges on manual classification. The best performing ARC algorithm for radiology document classification had an F-measure of 0.78, precision of 0.76, and recall of 0.80. The radiology test set included 279 reports representing 119 patients. Within the radiology test set, 207 radiology reports were manually classified as consistent with HCC. When the best performing ARC algorithm was applied to the radiology test set, document classification by ARC had a sensitivity of 0.75, specificity of 0.94, and PPV of 0.68 in comparison to manual classification (Table 3).
Discussion
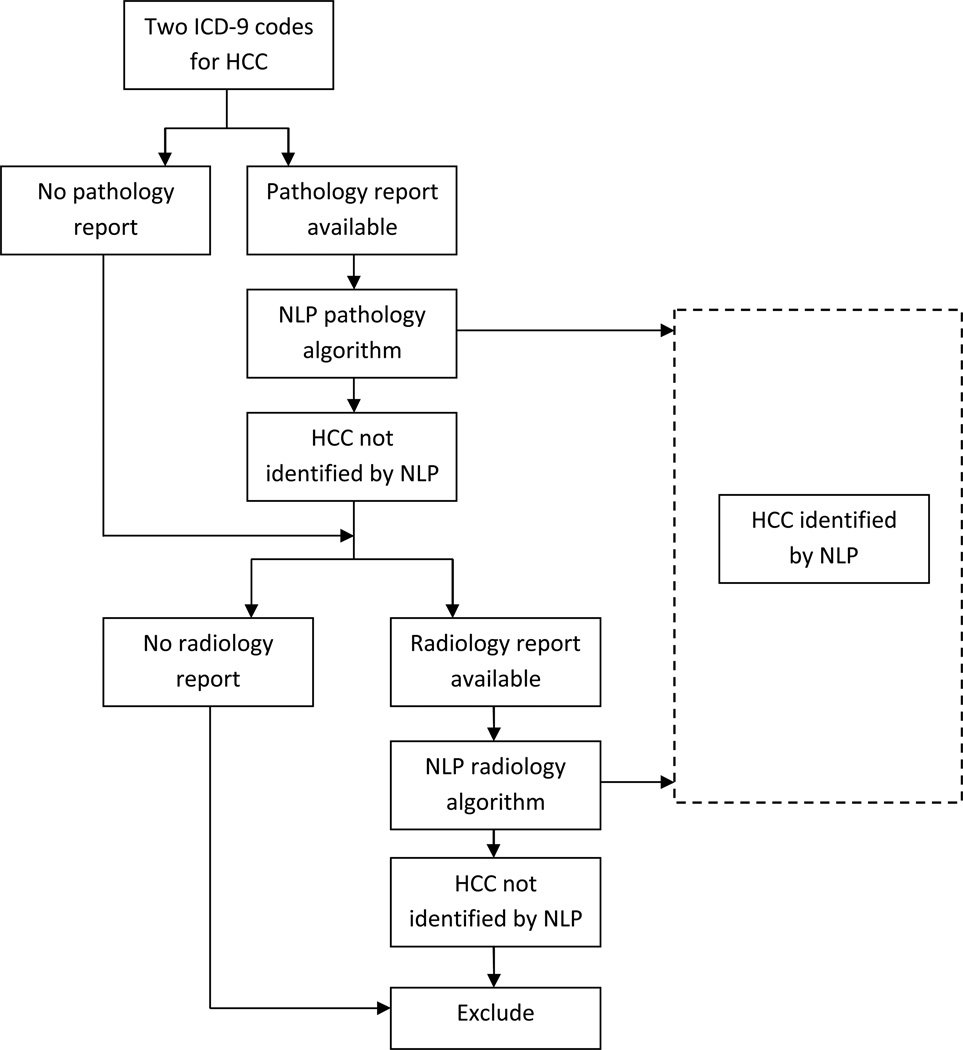
This study demonstrates that a combined approach using administrative data and NLP document classification by ARC is a feasible and accurate method to identify patients with HCC who have pathology or radiology reports in the electronic health record (Figure 2). We found that ICD-9 codes from administrative data alone have a sensitivity of 95% and specificity of 93% for HCC case identification and that NLP can accurately retrieve information from pathology and radiology documents consistent with HCC. The ARC pathology algorithm achieved high sensitivity of 96% and specificity of 97%, but the ARC radiology algorithm did not perform as well with a sensitivity of 94% and specificity of 68%. All patients with HCC should have a pathology or radiology report performed for diagnostic purposes and available for review, unless a patient was diagnosed at an outside facility and the medical records were not transferred.
Figure 2.
Model of sequential process to identify HCC cases by natural language processing in patients with two administrative codes for hepatocellular cancer
Our previous validation study comparing ICD-9 codes for HCC in the VA administrative database to medical chart review showed a PPV of 86% for the HCC ICD-9 code.4 The study included 157 patients diagnosed with HCC at three VA facilities (Houston, Nashville, Kansas City) from 1998 to 2003. However, our current study found a lower PPV of 67% in a larger and more diverse sample. The difference in PPV between the two studies may be due to our larger sample size, more variability in HCC ICD-9 coding among 117 VA facilities, and implementation of new guidelines for HCC diagnosis by imaging in 2005. This demonstrates that using ICD-9 alone codes may lead to significant misclassification.
Among patients with a misclassified ICD-9 code for HCC, nearly 40% were due to liver metastases from a non-hepatocellular cancer. When patients with a non-hepatobiliary cancer code were excluded from the sampling algorithm, the PPV of 67% improved to 73%. Conversely, among patients with ICD-9 codes for HCC and a non-HCC cancer, 40% of patients had both cancers. Although excluding subjects with ICD-9 codes for non-hepatobiliary cancers improves the PPV, the criteria will be too restrictive (less sensitive) and inappropriately exclude patients who have multiple malignancies that include HCC. Given the high sensitivity of the ICD-9 code sampling algorithm, NLP for pathology and radiology report classification may improve specificity.
NLP is a potential tool to improve HCC case identification in automated data including electronic health records. Our study found that the ARC algorithm accurately identifies pathology reports (F-measure 0.91), but does not perform as well with radiology reports (F-measure 0.78). This is consistent with D’Avolio’s findings that ARC can identify colon cancer and prostate cancer from pathology reports (F-measure 0.88 and 0.93, respectively), but ARC does not perform as well using radiology reports to identify lung cancer (F-measure 0.75).7 The low F-measure for radiology reports is not unexpected, given that physician agreement on diagnosis of HCC using imaging was 81% when radiology reports were manually judged. The lower agreement among physicians for radiology reports compared to pathology reports is likely due to the linguistic variability of radiology reporting in different sites. ARC will only perform as well as manual review. Pathology reports are easier for physicians and ARC to interpret given standardized language format. Traditional natural language processing development requires document parsing and programming, which can be labor intensive and expensive. This study demonstrates that ARC is a feasible alternative for pathology and radiology, document classification for HCC.
The combined approach of using administrative data in conjunction with NLP has been explored for lung nodules. Danforth et al. proposed an algorithm to identify lung nodules by combining diagnostic codes, procedural codes, and NLP free text searching for key words in radiology reports. The sensitivity was 96% with a specificity of 86% compared to medical record review.14 Our study also supports using a combined method of administrative data and NLP to identify specific diagnoses.
The generalizability of our findings to non-VA datasets is unknown, and the algorithm would need to be validated in non-VA sites. However, since the algorithms were created using reports from multiple VA sites across the United States, we hypothesize that the algorithms are more generalizable than if the algorithms were developed from reports in a single center. Another limitation to generalizability of our findings is the limited availability and diversity of electronic health record systems in non-VA sites. The transition of all health care systems to electronic health records will eventually allow the wide usage of the described NLP tools. The current availability of comprehensive electronic health records in the VA make it an excellent source for the development of novel bioinformatics tools now. Also, the specific components of pathology and radiology reports that contributed to each algorithm could not be specified using ARC software. However, to our knowledge, this is the first study to test pathology and radiology reports for hepatocellular cancer in a national cohort using NLP. An additional strength is that this national sample of pathology and radiology reports likely has greater linguistic diversity than reports obtained from a single site or region.
In summary, ICD-9 codes alone have limited accuracy to identify patients with HCC from administrative data. We recommend a combined approach of administrative data and NLP to improve HCC case identification. This approach uses readily available and highly sensitive ICD-9 codes for HCC as a first level of identification followed by NLP to improve specificity. Manually reviewing the electronic health record for HCC diagnostic confirmation is labor intensive and costly; this hybrid approach would decrease the time used to identify appropriate cases for health outcomes and epidemiology database research. With an increasing focus on improving the quality of healthcare, this algorithm could also be used in real time to harness information in the electronic health record from billing codes and reports to identify diagnostic errors or delays. Further studies are needed to investigate applying NLP to progress notes for HCC, as well as using the combined ICD-9 code sampling and NLP algorithm approach in other malignancies.
Acknowledgments
Funding: This project was supported in part by the National Cancer Institute (R01 CA160738, PI: J. Davila) the facilities and resources of the Houston Veterans Affairs Health Services Research and Development Center of Excellence (HFP90-020), Michael E. DeBakey Veterans Affairs Medical Center, and the Dan Duncan Cancer Center, Houston, Texas, United States of America.
The views expressed in this article are those of the authors and do not necessarily represent the views of the Department of Veteran Affairs.
Acronyms
- HCC
Hepatocellular cancer
- NLP
natural language processing
- ARC
Automated Retrieval Console
- VA
Veterans Affairs
- ICD-9
International Classification of Diseases, 9th Revision
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial disclosures: None
References
- 1.El-Serag HB. Hepatocellular carcinoma. N. Engl. J. Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB, Mason AC. Rising incidence of hepatocellular carcinoma in the United States. N. Engl. J. Med. 1999;340:745–750. doi: 10.1056/NEJM199903113401001. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB, Davila JA, Petersen NJ, McGlynn KA. The continuing increase in the incidence of hepatocellular carcinoma in the United States: an update. Ann. Intern. Med. 2003;139:817–823. doi: 10.7326/0003-4819-139-10-200311180-00009. [DOI] [PubMed] [Google Scholar]
- 4.Davila JA, Weston A, Smalley W, El-Serag HB. Utilization of screening for hepatocellular carcinoma in the United States. J. Clin. Gastroenterol. 2007;41:777–782. doi: 10.1097/MCG.0b013e3180381560. [DOI] [PubMed] [Google Scholar]
- 5.Davila JA, et al. Utilization of surveillance for hepatocellular carcinoma among hepatitis C virus-infected veterans in the United States. Ann. Intern. Med. 2011;154:85–93. doi: 10.7326/0003-4819-154-2-201101180-00006. [DOI] [PubMed] [Google Scholar]
- 6.Peabody JW, Luck J, Jain S, Bertenthal D, Glassman P. Assessing the accuracy of administrative data in health information systems. Med Care. 2004;42:1066–1072. doi: 10.1097/00005650-200411000-00005. [DOI] [PubMed] [Google Scholar]
- 7.D’Avolio LW, et al. Evaluation of a generalizable approach to clinical information retrieval using the automated retrieval console (ARC) J Am Med Inform Assoc. 2010;17:375–382. doi: 10.1136/jamia.2009.001412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Savova GK, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc. 2010;17:507–513. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shiner B, et al. Automated classification of psychotherapy note text: implications for quality assessment in PTSD care. J Eval Clin Pract. 2012;18:698–701. doi: 10.1111/j.1365-2753.2011.01634.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hou JK, et al. Automated Identification of Surveillance Colonoscopy in Inflammatory Bowel Disease Using Natural Language Processing. Dig. Dis. Sci. 2012 doi: 10.1007/s10620-012-2433-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyko EJ, Koepsell TD, Gaziano JM, Horner RD, Feussner JR. US Department of Veterans Affairs medical care system as a resource to epidemiologists. Am. J. Epidemiol. 2000;151:307–314. doi: 10.1093/oxfordjournals.aje.a010207. [DOI] [PubMed] [Google Scholar]
- 12.Farwell WR, D’Avolio LW, Scranton RE, Lawler EV, Gaziano JM. Statins and prostate cancer diagnosis and grade in a veterans population. J. Natl. Cancer Inst. 2011;103:885–892. doi: 10.1093/jnci/djr108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruix J, Sherman M. Management of hepatocellular carcinoma: an update. Hepatology. 2011;53:1020–1022. doi: 10.1002/hep.24199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Danforth KN, et al. Automated identification of patients with pulmonary nodules in an integrated health system using administrative health plan data, radiology reports, and natural language processing. J Thorac Oncol. 2012;7:1257–1262. doi: 10.1097/JTO.0b013e31825bd9f5. [DOI] [PMC free article] [PubMed] [Google Scholar]