Abstract
A multiplex PCR for detection of three categories of diarrheagenic Escherichia coli was developed. With this method, enterohemorrhagic E. coli, enteropathogenic E. coli, and enterotoxigenic E. coli were identified in fecal samples from patients with hemorrhagic colitis, watery diarrhea, or hemolytic-uremic syndrome and from food-borne outbreaks.
Enteric infections remain an important cause of morbidity in Chile. Incidence rates of diarrhea in Chilean children range from 1.3 to 4.5 per 100,000 inhabitants (8), and in Santiago, the incidence rates of food-borne outbreaks were reported as 3, 4.1, and 5.1 from 1999 to 2001, respectively (7). In an etiologic study in Chilean children with diarrhea, Levine et al. (5) showed that different categories of diarrheagenic Escherichia coli organisms have an important role as the causes of enteric infections. However, these pathogens are probably underestimated due to inappropriate diagnostic methods in clinical practice (4, 5).
Six categories of diarrheagenic E. coli organisms that differ in their virulence factors have been described (6). The most commonly reported diarrheagenic E. coli organisms in Chile are enteropathogenic E. coli strains that have a pathogenicity island (locus of enterocyte effacement), encoding proteins involved in the formation of attaching and effacing lesions on host intestinal cells (3); enterotoxigenic E. coli strains, which produce heat-labile and/or heat-stable enterotoxins (4); and enterohemorrhagic E. coli strains, important pathogens around the world (9), characterized by the presence of the locus of enterocyte effacement (2) and the production of Shiga toxins (Stx1 and Stx2) (10). Our study was undertaken to develop a multiplex PCR to simultaneously detect enterohemorrhagic E. coli, enterotoxigenic E. coli, and enteropathogenic E. coli in different fecal samples.
Diarrheagenic E. coli reference strains 933J (stx1 stx2 eae), C600J (stx1), C600W (stx2), 2348/69 (eae), and H10407 (st and lt) were used as positive controls. We also used Shigella and Salmonella strains to define the accuracy of the method (Table 1). Clinical samples included samples from 20 children with hemolytic uremic syndrome, 27 patients involved in six different food-borne outbreaks in Santiago during 2000, and 1,468 stool samples from sporadic diarrheal episodes in children less than 5 years old. We also incorporated one stool sample from an adult patient diagnosed with hemorrhagic colitis by colonoscopy and histopathological procedures. We used PCR primers specific for stx1 and stx2, as previously described (1), and the primers for eae, bfp, stII, and lt were obtained from sequences available in the GenBank database with the OMIGA software for alignment and Primer3 program for primer design (Table 2).
TABLE 1.
Reference E. coli strains tested by multiplex PCR
| Bacterial straina | No. of strains showing positive PCR result/no. of strains tested
|
|||||
|---|---|---|---|---|---|---|
| stx1 | stx2 | eae | bfp | lt | st | |
| Enterohemorrhagic E. coli | ||||||
| 933J O157:H7 CVD | 10/10 | 10/10 | 10/10 | 0/10 | 0/10 | 0/10 |
| O157, ISP | 0/5 | 5/5 | 5/5 | 0/5 | 0/5 | 0/5 |
| C600J O157, CVD | 10/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| C600W O157 CVD | 0/10 | 10/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| Enteropathogenic E. coli | ||||||
| 2348/69, CVD | 0/10 | 0/10 | 10/10 | 10/10 | 0/10 | 0/10 |
| HS negative control, CVD | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| O142, SP | 0/5 | 0/5 | 5/5 | 5/5 | 0/5 | 0/5 |
| O25, ISP | 0/5 | 0/5 | 5/5 | 5/5 | 0/5 | 0/5 |
| Enterotoxigenic E. coli | ||||||
| O159, MP | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 5/5 |
| O8, MP | 0/5 | 0/5 | 0/5 | 0/5 | 5/5 | 0/5 |
| O6, MP | 0/5 | 0/5 | 0/5 | 0/5 | 5/5 | 0/5 |
| Nt, MP | 0/10 | 0/10 | 0/10 | 0/10 | 3/10 | 10/10 |
| H10407, CVD | 0/10 | 0/10 | 0/10 | 0/10 | 10/10 | 10/10 |
| Salmonella enteriditis, MP | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| Salmonella group D, MP | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| Salmonella group B, MP | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| Salmonella spp., MP | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| Shigella flexneri, MP | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| Shigella sonnei, MP | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| Klebsiella oxytoca, MP | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 | 0/5 |
| Proteus spp., MP | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 |
CVD, Center for Vaccine Development, University of Maryland; ISP, Chilean Institute of Public Health; MP, Microbiology Program, University of Chile; Nt, not typeable.
TABLE 2.
Primers used in the multiplex PCR for amplification of diarrheagenic E. coli genesa
| Gene | Primer sequence, 5′-3′ | Size of product (bp) | Reference |
|---|---|---|---|
| eae | TCA ATG CAG TTC CGT TAT CAG TT | 482 | This study |
| GTA AAG TCC GTT ACC CCA ACC TG | |||
| bfp | GGA AGT CAA ATT CAT GGG GGT AT | 254 | This study |
| GGA ATC AGA CGC AGA CTG GTA GT | |||
| stx1 | CAG TTA ATG TGG TGG CGA AGG | 348 | 1 |
| CAC CAG ACA ATG TAA CCG CTG | |||
| stx2 | ATC CTA TTC CCG GGA GTT TAC G | 584 | 1 |
| GCG TCA TCG TAT ACA CAG GAG C | |||
| lt | GCA CAC GGA GCT CCT CAG TC | 218 | This study |
| TCC TTC ATC CTT TCA ATG GCT TT | |||
| stII | AAA GGA GAG CTT CGT CAC ATT TT | 129 | This study |
| AAT GTC CGT CTT GCG TTA GGA C |
Specific primers used for amplification of diarrheagenic E. coli virulence genes were designed from sequences obtained from the indicated GenBank accession numbers: eae, AB040740, AF025311, AF043226, AF065628, AF116899, AF200363, AF449418, AF530555, AF530556, AF530557, Z11541, AJ271407, AJ298279, AJ308550, AJ308551, AJ308552, M58154, U38618, and U66102; bfp, AF119170 and U27184; lt, gi1648865; and stII, AY028790 and M35586.
The DNA sequences of the primers and the sizes of PCR products are shown in Table 2. Fecal E. coli isolates were analyzed by multiplex PCR to detect virulence genes (stx1, stx2, eae, bfp, stII, and lt). Each multiplex PCR assay was performed in 50 μl of reaction mixture containing 1 mM deoxynucleoside triphosphate mix, 10 pmol of each primer, 1.5 mM MgCl2, 1× reaction buffer (10 mM Tris-HCl, 50 mM KCl), 0.25 U of Taq DNA polymerase, and 3 μl of DNA as the template. The crude cell lysate used to obtain the DNA was obtained by boiling five colonies of the E. coli isolate for 20 min in 0.5% Triton X-100. The hot-start technique was used to prevent nonspecific amplification (40 μl of the reaction mixture was heated to 94°C for 5 min before Taq DNA polymerase was added [2 U in 10 μl of reaction mixture]). The samples were amplified for 35 cycles, and each cycle consisted of 1.5 min at 94°C, 1.5 min at 64°C, and 1.5 min at 72°C. The PCR products were separated by electrophoresis in 1.5% agarose gels and stained with ethidium bromide.
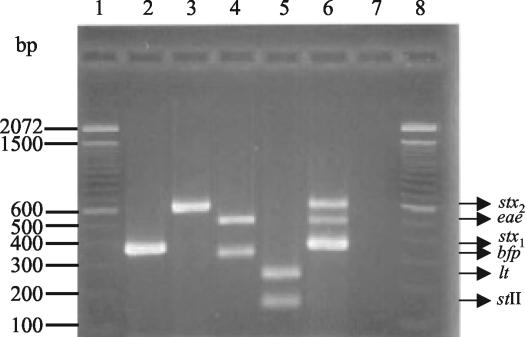
To develop the multiplex PCR, we tested the progressive incorporation of primers corresponding to the different genes and several combinations of melting temperatures and primer concentrations. The sensitivity and specificity of the reaction were assayed with the reference strains (Fig. 1). The PCR products for the stx1, stx2, eae, bfp, stII, and lt genes were 348, 584, 482, 254, 129, and 218 bp, respectively. Multiplex PCR-positive colonies were serotyped by agglutination with commercial antisera (Probac R, Sao Paulo, Brazil), and the results were confirmed at the Laboratory for Foodborne Zoonoses, Canada, or at the Chilean Institute of Public Health.
FIG. 1.
Multiplex PCR of selected reference strains. Bands corresponding to eae, bfp, stx2, stx1, lt, and stII are indicated. Lane 1, 100-bp size ladder; lane 2, E. coli C600J (stx1); lane 3, E. coli C600W (stx2); lane 4, enteropathogenic E. coli 2348/69 (eae bfp); lane 5, enterotoxigenic E. coli H10407 (lt st); lane 6, enterohemorrhagic E. coli 933J (stx1 stx2 eae); lane 7, reagent control; lane 8, 100-bp size ladder.
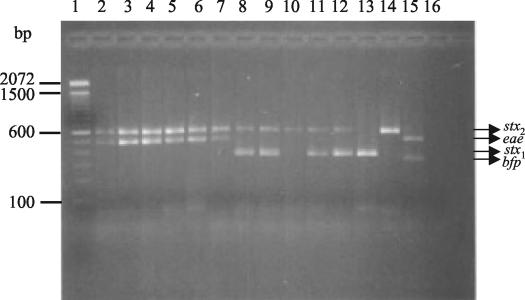
E. coli isolates from all hemolytic-uremic syndrome patients and from one adult patient with hemorrhagic colitis were positive for the stx2 and eae genes (Fig. 2) and corresponded to enterohemorrhagic E. coli serotype O157:H7 (Table 3). Among the stool samples from patients involved in food-borne outbreaks, 122 E. coli colonies were isolated from 27 patients. For samples from two of the six outbreaks, we obtained positive multiplex PCR results. For four of the patients from the first outbreak, an enteropathogenic E. coli strain of serogroup O26 was detected, which was positive for the eae and bfp genes (Table 3). In the other outbreak, enterohemorrhagic E. coli-positive colonies were detected in samples from three of four patients affected. One patient was positive for enterohemorrhagic E. coli O157:H7 (with the genotype stx2 eae), and in samples from another patient, we detected colonies of three different genotypes and serotypes (stx2 eae/O157:H7, stx2/O91:H21, and stx1 stx2/O174:H28). A third patient was positive for stx1 stx2/O174:H28, suggesting that this particular food-borne outbreak was a mixed infection (Fig. 2, Table 3). A third outbreak was caused by Shigella spp., and for the three remaining outbreaks, we did not isolate any enteric pathogen. Analysis of 1,468 E. coli isolates from children with sporadic diarrheal episodes in the metropolitan region of Santiago from November 2002 to April 2003 revealed enterohemorrhagic E. coli strains in nine patients (0.6%), enteropathogenic E. coli strains in 78 children (5.3%), and enterotoxigenic E. coli strains in 45 patients (3.1%) (Table 3).
FIG. 2.
Multiplex PCR analysis of clinical samples. Lane 1, 100-bp size ladder; lanes 2 to 6, samples from hemolytic uremic syndrome patients; lanes 7 to 12, samples from patients involved in food-borne outbreaks; lane 13, E. coli C600J; lane 14, E. coli C600W; lane 15, enteropathogenic E. coli 2348/69; lane 16, reagent control.
TABLE 3.
Results of the multiplex PCR assay with E. coli strains obtained from clinical samples
| Disease | No. of patients | No. of colonies tested | Multiplex PCR results (no. of positive patients/ no. studied) | Genotype detected by multiplex PCR | E. coli serotypea |
|---|---|---|---|---|---|
| Hemolytic-uremic syndrome | 20 | 85 | 5/20 | stx2eae | O157:H7 |
| Hemorrhagic colitis | 1 | 2 | 1/1 | stx2eae | O157:H7 |
| Sporadic diarrhea episodes | 1,468 | 2,936 | 4/1,468 | stx1 | ND |
| 1/1,468 | stx1 | O26 | |||
| 1/1,468 | stx1 | O111 | |||
| 2/1,468 | stx1eae | ND | |||
| 1/1,468 | stx2 | O125 | |||
| 78/1,468 | eae bfp | ND | |||
| 12/1,468 | stII | ND | |||
| 25/1,468 | lt | ND | |||
| 8/1,468 | stII lt | ND | |||
| Food-borne outbreaks | |||||
| 1 | 4 | 17 | 4/4 | eae-bfp | O26 |
| 2 | 4 | 19 | 3/4 | stx1-eae | O157:H7 |
| stx1-stx2 | O174:H28 | ||||
| stx2 | O9:H21 | ||||
| 6b | 6 | 27 | 0/6 | ||
| 9 | 3 | 12 | 0/3 | ||
| 10 | 3 | 11 | 0/3 | ||
| 12 | 7 | 35 | 0/7 |
ND, not done.
Shigella spp. found in six of six patients.
In the present study, we designed specific primers for the eae, bfp, stII, and lt genes. For the eae gene, 14 different subtypes have been identified so far, and their differences were incorporated into the primer design so that they can recognize most of the variants that have been described. For the bfp, stII, and lt genes, there are few sequences in the databases, but they were considered in the primer design.
The multiplex PCR showed sensitivity and specificity for the most common categories of diarrheagenic E. coli strains of different clinical origins, genotypes, and serotypes. The results of this study indicate that it is possible to perform simultaneous amplification of virulence genes from differentiate enterohemorrhagic, enterotoxigenic, and enteropathogenic E. coli strains and apply this technique to the diagnosis of patients with hemolytic-uremic syndrome, those involved in food-borne outbreaks, and patients with sporadic enterocolitis.
Acknowledgments
We thank Mohamed Karmali, Laboratory for Foodborne Zoonoses, Guelph, Ontario, Canada, for assistance with serotyping and James P. Nataro, Center for Vaccine Development, University of Maryland at Baltimore, and Alfredo Torres, Medical Branch Department of Microbiology and Immunology, University of Texas, for careful review of the manuscript and helpful discussions.
This work was supported by FONDECYT grant 1000636.
REFERENCES
- 1.Cebula, T. A., W. L. Payne, and P. Feng. 1995. Simultaneous identification of strains of Escherichia coli serotype O157:H7 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J. Clin. Microbiol. 33:248-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jarvis, K. G., and J. B. Kaper. 1996. Secretion of extracellular proteins by enterohemorrhagic Escherichia coli via a putative type III secretion system. Infect. Immun. 64:4826-4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jerse, A. E., J. Yu, B. D. Tall, and J. B. Kaper. 1990. A genetic locus of enteropathogenic Escherichia coli necessary for the production of attaching and effacing lesions on tissue culture cells. Proc. Natl. Acad. Sci. USA 87:7839-7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Levine, M. M. 1987. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J. Infect. Dis. 155:377-389. [DOI] [PubMed] [Google Scholar]
- 5.Levine, M. M., C. Ferreccio, V. Prado, M. Cayazzo, P. Abrego, J. Martínez, L. Maggi, M. M. Baldini, W. Martin, D. Maneval, B. Kay, L. Guers, H. Lior, S. S. Wasserman, and J. P. Nataro. 1993. Epidemiologic studies of Escherichia coli diarrheal infections in a low socioeconomic level peri-urban community in Santiago, Chile. Am. J. Epidemiol. 138:849-869. [DOI] [PubMed] [Google Scholar]
- 6.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prado, V., V. Solari, I. Alvarez, C. Arellano, R. Vidal, M. Carreño, N. Mamani, D. Fuentes, M. O'Ryan, and V. Muñoz. 2002. Situación epidemiológica de las enfermedades transmitidas por alimentos en Santiago de Chile. Período 1999-2000. Rev. Med. Chile 130:495-501. [PubMed] [Google Scholar]
- 8.Sotomayor, V. 2000. Situación de las enfermedades de notificacion obligatorias, p. 16-28. In X. Aguilera, El vigia, vol. 3. Gobierno de Chile, Santiago, Chile.
- 9.Tesh, V. 1998. Virulence of enterohemorrhagic Escherichia coli: role of molecular crosstalk. Trends Microbiol. 6:228-233. [DOI] [PubMed] [Google Scholar]
- 10.Tesh, V. L., and A. D. O'Brien. 1991. The pathogenic mechanisms of Shiga toxin and the Shiga-like toxins. Mol. Microbiol. 5:1817-1822. [DOI] [PubMed] [Google Scholar]