Circulating tumor cells (CTCs) are the rare cells found in the peripheral blood, which detached from solid tumors and entered into the vasculature.[1] Although the mechanism of CTC generation is still unclear, CTCs shed from the primary tumor are thought to be the key to understanding the initiation of metastatic progression in cancer.[2] In addition to their potential significance in the cancer biology, their clinical applications in cancer diagnosis, predicting therapeutic responses, and studying the evolution of tumor cell heterogeneity have an even greater importance with respect to clinical applications. It has widely been recognized that the malignant tumors exhibits possess intrinsic heterogeneity, which evolves throughout the time course of disease progression. As such, by providing a continuous and readily accessible source of tumor cells, CTCs are regarded as a “liquid biopsy” of the primary and disseminated tumors, which may be a promising alternative for comprehensive molecular profiling and timely monitoring of cancer progression.. More importantly, recent advances in next generation sequencing (NGS) technologies, which are capable of profiling minute quantities of genetic materials even to the single cell level, provide an opportunity to investigate the genomic alterations in CTCs.
Due to either the absence or limitation of technologies that can characterize and/or isolate rare cell populations, studies relevant to CTC biology have been relatively stagnant until very recently. In 2004, the development and subsequent FDA clearance of the CellSearch™ assay has allowed for the routine measurement of CTCs in the clinic. This assay employs an EpCAM-based immunomagnetic agent capable of recognizing epithelial cells followed by subsequent multiplexed immunocytochemistry studies for CTC identification. This technology, however, is constrained by its low capture efficiency and has limited ability for post-capture molecular analysis.[3] Other approaches fall mostly into one of the three main categories: flow cytometry-based sorting,[4] marker protein-based immunoaffinity capture,[5] and size-based isolation.[6] Many of these approaches reported higher CTC capture efficiencies than the currently employed CellSearch™ assay, but few technologies enable the characterization of CTCs beyond their number. Enumeration of CTCs does not reflect the molecular signatures that can provide insights into tumor biology and heterogeneity and identify driver mutations that can guide therapy in real time. Moreover, the molecular profiling of CTCs may identify novel biomarkers that could guide the choice of therapies for individual patients as well as facilitate the development of new drugs. One of the critical limitations of existing CTC technologies relates to the contamination introduced by non-specifically captured white blood cells (WBCs), which may obfuscate CTC signals, introduce false negatives and generally complicate readout analysis.[7] Detailed analysis of CTC molecular signatures has remained a great challenge requiring a high purity CTC isolation technique, which keeps cells amenable to biological characterization.
Previously, our group demonstrated the utility of “NanoVelcro” Chip,[8] which is capable of efficiently capturing CTCs in patient blood samples using anti-EpCAM coated silicon nanowire (SiNW) substrates. Other types of nanostructured substrates, including electrochemically deposited conjugated polymer nano-features[9] and horizontally packed TiO2 nanofibers,[10] have also demonstrated improved CTC capture efficiency due to enhanced local topographic interactions[11] between the anti-EpCAM coated nanosubstrates and nanoscaled cellular surface components (e.g., microvilli) on a CTC. Moreover, we have added an overlaid polydimethyl-siloxane (PDMS) chaotic mixer,[12] which increases the contact frequency between flow-through CTCs and the substrate, which further improves CTC capture efficiency.[8] Irrespective of these NanoVelcro chip modifications, WBCs are co-captured with CTCs, and in fact, WBCs represent the vast majority of captured nucleated cells. When attempting to apply NGS technology to this system, this contamination requires a significant sequencing depth to detect genetic alterations of importance in these CTC-WBC admixtures. Using only cancer marker-based enrichment methods it is not currently possible to eliminate the WBC signal. As such an additional purification process is required to eliminate WBC noise when sequencing.
Herein, we introduced a modified NanoVelcro Chip coupled with ArcturusXT™ laser capture microdissection (LCM) technology that allows for isolation of CTCs from patients with prostate cancer (PC) well suited for NGS.[13] PC is known to have a long natural history on the order of 10-15 years in most cases.[14] As it is known that heterogeneity in the tumor evolves over time, there are concerns that historic tissue from diagnosis may not reflect the biology of a newly-developed metastatic lesion. Since the majority of PC patients have only bone metastasis[15] and the biopsy of bone is both invasive and technically difficult, serial samplings are nearly impossible by conventional biopsy. For proof-of-concept, we performed whole exome sequencing (Exome-Seq) to demonstrate the feasibility of a streamlined process starting from CTC enrichment, isolation, all the way to pure CTCs sequencing, as Exome-Seq is a suitable method in terms of its cost and comprehensive data acquisition for studying the complex tumor heterogeneity of PC.[16]
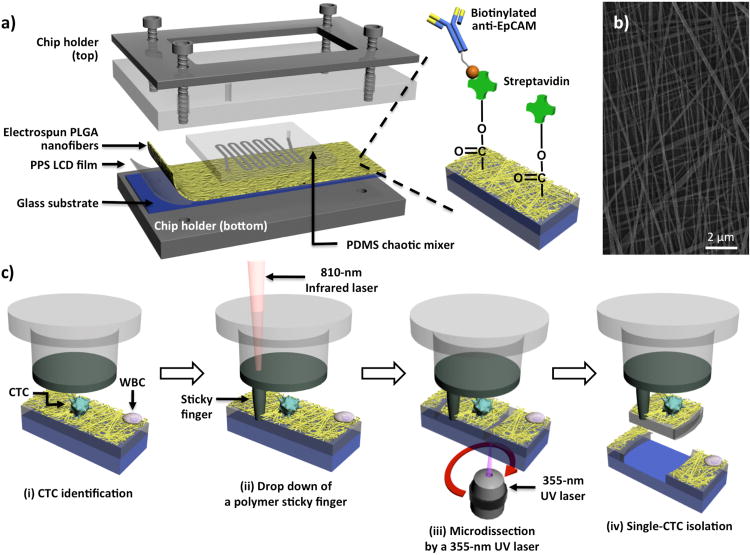
In order to construct the NanoVelcro substrate compatible with the LCM system, poly(lactic-co-glycolic acid) (PLGA), a polymer material with excellent biocompatibility, was electrospun onto the LCM membrane slides. The resultant new PLGA-Nanofiber (PN)-NanoVelcro Chip (Figure 1a) is composed of an overlaid PDMS chaotic mixer chip, and a triple layers' anti-EpCAM coated transparent PN-NanoVelcro substrate. The PN-NanoVelcro substrate consists of three layers: first, the electrospun PLGA nanofiber coated with anti-EpCAM. The anti-EpCAM specifically binds to EpCAM on the CTCs, while the PLGA nanofiber enhances the topographical interaction between the substrate and CTCs (Figure 1b); the second layer is the PPS LCM film, which allows the 355-nm UV laser cutting to isolate the captured cells from the substrate surface; and the third layer is the glass substrate to support the polymer layers. After capturing CTC on the substrate, a 4-step LCM process (Figure 1c) was utilized to isolate single CTCs from the background WBCs.
Figure 1.
a) The configuration of PLGA nanofiber (PN)-NanoVelcro Chip for the high-purity isolation of circulating tumor cells (CTCs) from prostate patients' peripheral blood samples. A custom-designed chip holder is employed to sandwich a PN-NanoVelcro Chip that is composed of (i) an overlaid PDMS chaotic mixer chip and (ii) a transparent PN-NanoVelcro substrate, prepared by depositing electrospun PLGA nanofibers onto a commercial laser microdissection (LMD) glass slide (with a pre-deposited 1.2-μm-thick PPS membrane). Streptavidin is covalently attached onto the PLGA nanofibers for conjugation of the capture agent (i.e., biotinylated anti-EpCAM). b) A SEM image of the electrospun PLGA nanofibers. c) Schematic illustration of the workflow for isolation of high-purity CTCs using a 4-step laser capture microdissection (LCM) technique: (i) CTCs (CK+/CD45-) are first identified from surrounding WBCs (CK-/CD45+), and an LCM cap (with a polymer membrane on bottom) is then aligned over the substrate, (ii) an IR-laser is used to melt the polymer membrane, dropping down a “sticky finger” for adhering onto the PN-NanoVelcro substrate, (iii) a 355-nm UV laser is utilized to dissect the substrates underneath the identified CTC, and (iv) by removing the LCM cap high-purity CTCs can be obtained for subsequent molecular analysis.
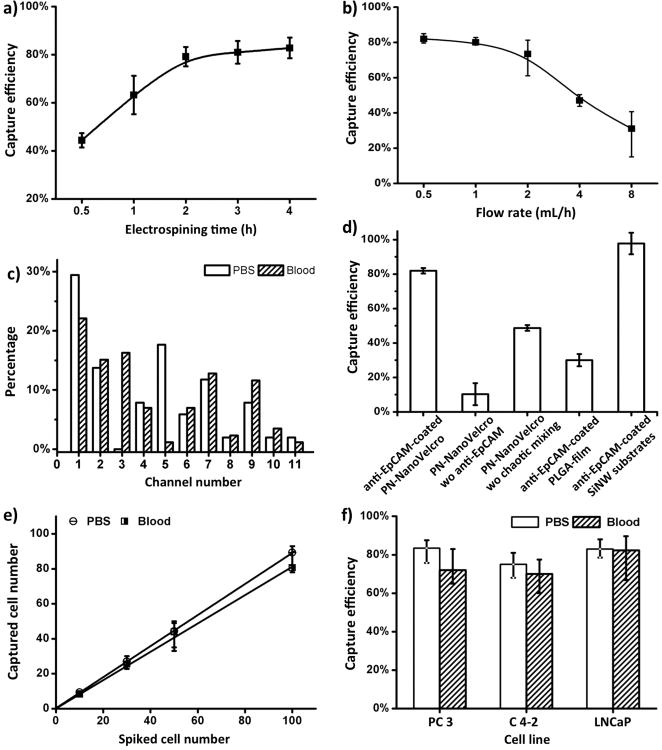
To validate the methodology and optimize experiment conditions, a standard epithelial cell sample, pre-stained SK-BR-3 cells, was spiked into PBS solution for the assessment of CTC capture efficiency. Both the configuration of nano-structured substrate and the vertical flow caused by chaotic mixer may contribute to the enhanced cell capture performance and thus required calibration to optimize the platform. Therefore, we first examined the CTC recovery rate using PN-NanoVelcro substrates with different electrospin deposition times ranging from 0.5 to 4 h. The longer electrospin time led to increased quantity of polymer nanofibers deposited on the Laser MicroDissection (LMD) slide substrates. Consequently, there is a larger surface area for enhanced cell capture efficiency as a result of enhanced topographic interactions between antibody-coated polymer nanofibers and nanoscaled cell surface components. This observation is consistent with our earlier observation, where the longer deposition times of TiO2 nanofibers led to better CTC capture efficiency.[10] During optimization, we found a negligible difference between a 3 h and 4 h deposition time. Moreover, the thinner polymer was more convenient for UV laser dissection. As such, we chose 3 h of deposition as the optimized condition for the following experiments. This deposition condition yielded an average of 3-μm thick PLGA nanostructured membranes (Figure 2a).
Figure 2.
Optimization and validation of the PN-NanoVelcro Chips using CellSearch™ Calibration sample (pre-stained SK-BR-3 breast cancer cells) and cell lines. a) The cell-capture efficiency of PN-NanoVelcro Chip with different electrospining time was assessed. CellSearch™ calibration samples containing 100 pre-stained EpCAM-positive SK-BR-3 cells were spiked into PBS as a model system. b) Cell-capture efficiency of PN-NanoVelcro Chip at flow rates of 0.5, 1, 2, 3, 4 mL h-1. c) The distributions of captured cells on the PN-NanoVelcro Chips were assessed in PBS and normal blood. d) Comparison of the capture performance between PN-NanoVelcro Chip and four different controls: i) PN-NanoVelcro Chip without anti-EpCAM; ii) PN-NanoVelcro Chip without chaotic mixing (flat channel without herringbone structure); iii) Anti-EpCAM coated PLGA thin film with electrospining; iv) SiNW NanoVelcro Chip coated with anti-EpCAM. e) Capture efficiencies at different spiking cell numbers ranging from 10-100 mL-1. f) Capture efficacies of three different PC cell lines, PC3, C4-2 and LNCaP in PBS and normal blood. All error bars show standard deviations (n>=3).
In addition to the preparation technique of PN-NanoVelcro substrate, optimization on flow rate was also conducted. CTC recovery rates at the flow rates of 0.5 to 8 mL h-1 were assessed using optimal PN-NanoVelcro substrates (Figure 2b). As observed, the highest capture performance about 82% was achieved at the flow rate of 0.5 mL h-1, which was selected as the optimized flow rate for subsequent studies. Flow rates lower than 0.5 mL h-1 were not considered to avoid excessive sacrifice as the extended operation time that may have potentially allowed for the apoptosis and adverse effects of subsequent on isolated CTCs. To further validate the system performance, spatial distributions of immobilized CTCs along the PN-NanoVelcro substrate were examined using the optimized conditions (Figure 2c, Supporting Information Figure S1). Side by side comparison between CTCs sample spiked in PBS solution and normal blood showed that in both cases over 50% of the captured CTCs were located in the first 4 channels of the device. We thus concluded that 11 channels were more than adequate for CTC enrichment. Cells spiked into blood were not captured with the same efficiency as those spiked into PBS. This could have been attributed to the existence of peripheral blood cells or plasma proteins producing a negative impact on the interaction between the CTCs and capture agents coated onto the NanoVelcro substrates.
To demonstrate the superiority of our PN-NanoVelcro Chip design, the CTC capture efficiency was compared with four control groups, including (i) PN-NanoVelcro Chip without anti-EpCAM coating, (ii) PN-NanoVelcro device without chaotic mixer (flat channel without herringbone structure), (iii) Anti-EpCAM coated PLGA thin film, and (iv) SiNW NanoVelcro chip coated with anti-EpCAM. The experiment results were summarized in Figure 2d. We found the PN-NanoVelcro outperformed three control groups (i), (ii) and (iii) suggesting that the anti-EpCAM capture agent played a crucial role for the immobilization of CTCs, while PLGA nanofibers and the vertical flow introduced by chaotic mixer both contributed to the improved cell capture efficiency. The performance of PN-NanoVelcro Chip was similar to our previously described SiNW NanoVelcro chip, further proving that the enhanced local topographic interaction was the key to NanoVelcro chip capture.
To evaluate the capture efficiency for different CTC numbers, we spiked different number of SK-BR-3, cells into both PBS and blood (Figure 2e). Consistent recovery rates were observed at various numbers of spiked cells as low as 10 cells per mL. Finally, the capability of capturing PC cells was confirmed using three different PC cell lines: PC3, C4-2 and LNCaP (Figure 2f). The cells were pre-stained with DiO and enumerated before being spiked into the blood or PBS (See supporting information for cell enumeration method). The capture efficiencies were determined under a fluorescent microscope by the enumeration of DAPI+/DiO+ cells captured on the substrate divided by the number spiked. The average capture efficiencies for all three cell lines were respectively 80.5% in PBS and 74.7% in blood. This indicated our PN-NanoVelcro Chips were capable of performing CTCs isolation from PC patient samples. The viability of the captured cells was also assessed by Calcein AM/Ethidium bromide fluorescent assay 2 h after captured onto the PN-NanoVelcro substrate. We found >80% of captured cells viable. This suggested that our device is truly biocompatible while minimizing the apoptosis and preserving high quality genomic materials. (Supporting Information Figure S2)
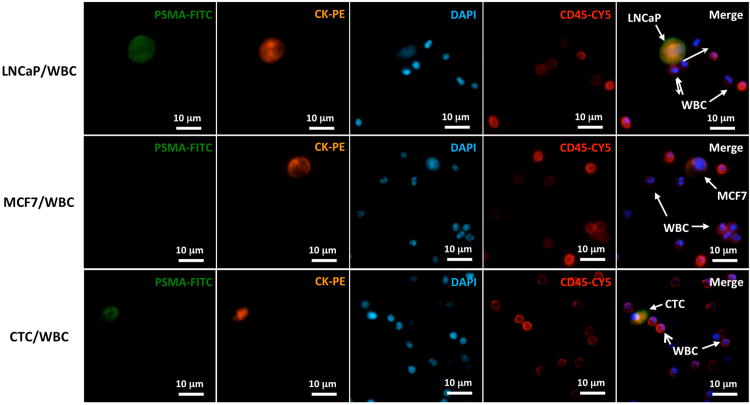
After capturing the CTCs onto the substrate surface, we performed immediate methanol-fixation to preserve the most genomic information. The PN-NanoVelcro Chip was dissembled and the CTC-bearing substrate was stained with a four-color immunocytochemistry (ICC) method. DAPI, anti-prostate specific membrane antigen (PSMA), anti-CK and anti-CD45 were utilized to identify the CTCs from background WBCs. Given the concern of EpCAM and CK positive circulating epithelial cells influencing our purity of capture, PSMA, a prostatic epithelia cell specific marker for cells of prostate origin, was introduced to our used to facilitate our CTC selection. The PN-NanoVelcro substrate was first scanned under a 4× objective of a fluorescent microscope (Nikon 90i) in conjunction with an auto-scan imaging software (Nikon, Element). All DAPI+ cellular events were plotted onto an XY scatter plot by their CK and CD45 intensities and the CK and CD45 threshold was determined by the distribution of cells. The CK+/DAPI+/CD45- cells were defined as CTC candidates and their locations were recorded and double-checked under a 10× objective for their PSMA intensity and cellular morphology. To demonstrate the feasibility of this four-color channel immunocytochemistry method, fluorescent images of a positive control (LNCaP/WBC), a negative control (MCF7/WBC) and a patient sample from Cedars-Sinai Medical Center (CSMC), after PN-NanoVelcro Chip capture, were shown in Figure 3.
Figure 3.
Identification of CTCs based on a 4-color immunocytochemistry staining using FITC-conjugated anti-PSMA, PE-conjugated anti-CK, Cy5-conjugated anti-CD45 (a marker for WBC) and DAPI (nuclear specific) was validated on a CSMC patient. LNCaP cells spiked in normal blood were used as a positive control, and MCF7, a breast cancer cell line, was chosen as a negative control.
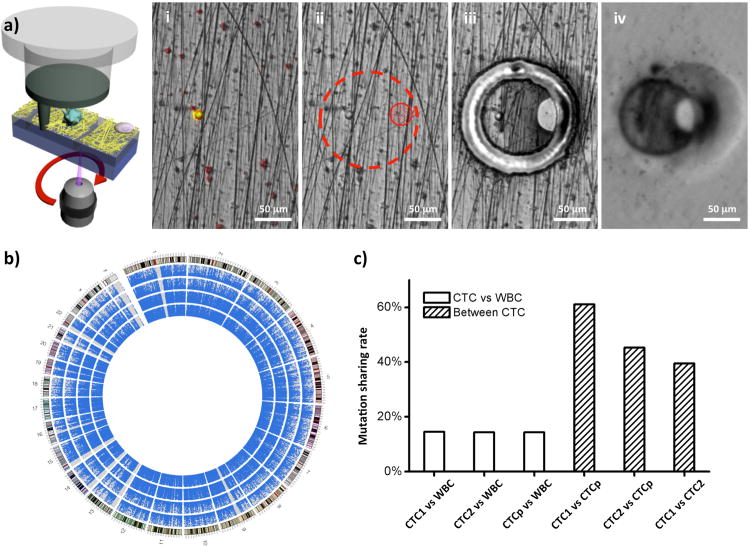
The immunostaining protocols for the samples used for CTC Exome-Seq were slightly modified by removing DAPI staining to avoid error due to DAPI binding to DNA. After confirming the CTCs under 10× objectives by their morphology, the locations of all CTCs were recorded and the PN-NanoVelcro substrate was placed on the LCM microscope (ArcturusXT™). The ArcturusXT™ LCM system is designed for single cell and/or tissue isolation, and was demonstrated in previous publications with the feasibility of subsequent molecular analysis.[17] Using an aforementioned 4-step LCM process (Figures 1c and 4a), first, an LCM cap (with a polymer membrane on bottom) was aligned on a pre-determined CTC location. Second, an 810-nm IR-laser was used to melt the polymer membrane, dropping down a “sticky finger” for adhering onto the PN-NanoVelcro substrate. Third, a 355-nm UV laser was utilized to dissect the substrates underneath the identified CTC. To avoid possible laser-associated damage to the cellular genomic contents, we carefully calibrate the laser beam width (around 10 to 20 μm) and activate laser at least 50 μm away from the target CTC. Finally, by removing the LCM cap, high-purity CTCs were obtained for subsequent whole genome amplification (WGA) and Exome-Seq. This method was a modification of our previously reported laser microdissection (LMD) approach.[18] While our previous method could also isolate CTCs for genetic analysis, the LMD isolation suffered from low cell retrieval rates due to an inability to secure the transfer of CTCs from the LMD slides to the collection PCR. By introducing the IR-laser sticky finger, the isolated polymer films were physically attached to the LCM cap and ensure the presence of CTC genomic materials for WGA.
Figure 4.
a)Micrograph images recording the process of CTC isolation using a ArcturusXT™ Laser Capture Microdissection (LCM) system; i) CTC identification; ii) Determination of IR sticky finger positions and UV dissection route; iii) UV laser dissection; iv) Isolated CTC immobilized on the LCM cap. b) Circos plots representing the coverage areas of Exome-Seq. With each dot indicating a sequenced area among the captured areas by the exome-enrichment kit, the rings from the inside out represent WBC, pooled CTCs, CTC-1 and CTC-2. The outermost ring represents the karyotype of the human reference genome (hg19), with red areas being centromeres. c) The shared mutations between CTCs and WBC are compared with the shared mutations among CTCs. (CTCp = pooled CTCs)
Using the same patient from CSMC as a pilot, we isolated 25 single CTCs, 9 pooled CTCs of 2-10 cells along with 6 single WBCs and 6 pooled WBCs from 6 mL of blood (Supporting Information Table S1). After WGA and subsequent quality check, the amplified genomic DNA from, four batches including 2 single CTCs, 1 pool of 6 CTCs and 1 pool of 10 WBCs as control, were sequenced by a standard exon-capture targeted sequencing separately. The results showed 25 to 80% of the targeted exome regions were sequenced with a mean coverage of 29 to 48× (Supporting Information Table S2). We also observed that pooled cells had larger areas covered, indicating some genomic materials were missing during the WGA process and this defect could have been partially be compensated by increasing the starting number of CTCs. As indicated in the Circos plot (Figure 4b),[19] the covered areas were scattered homogenously in exome areas in all 23 chromosomes, indicating there was no chromosomal loss during the isolation and sequencing process. Single nucleotide mutations were called from the commonly covered areas in four exomes. As shown in Figure 4c, the shared mutations between CTCs and WBC were much fewer in number compared with the shared mutations among CTCs. As sequenced CTCs were more similar to each other and were different from the WBC control, we confirmed the feasibility of using PN-NanoVelcro Chip as a means to capture pure CTCs and eliminate the contamination of WBCs by LCM. Taken together, these data demonstrated that our PN-NanoVelcro-LCM platform for Exome-Seq is working, but at the same time, there's not enough statistical robustness in performing further mutational analysis until more CTCs are sequenced.
In conclusion, we reported a novel device facilitating the pure CTC isolation and their Exome-Seq by PN-NanoVelcro chip capture and LCM. This PN-NanoVelcro Chip exerted similar capture efficiency and scalable throughput to the original NanoVelcro Chip while enabling LCM for pure CTCs Exome-Seq. We anticipate that using our PN-NanoVelcro-LCM platform, CTC characterization will be feasible on a larger scale allowing for greater numbers of specimens to be characterized, including patients at different time points, under different therapeutic intervention, in patients known to exhibit different clinical phenotype. Additionally, CTCs could be compared to primary and metastatic tissue. Finally, CTCs sampled before and after the resistance developed to a given therapeutic approach could be characterized. At the same time, we would also adapt this platform to characterize changes in DNA methylation and to perform RNA sequencing. Overall speaking, our reported PN-NanoVelcro-LCM platform may enable a means to the potential of resolving questions of clonal evolution and drug resistant mechanisms upon disease progression at a single cell level with the prospect of improving personalized medicine.
Supplementary Material
Acknowledgments
The research endeavors at UCLA were supported by a Creativity Award from Prostate Cancer Foundation (PCF), and research grants (R21 CA151159 and R33 CA157396) from NIH/NCI Innovative Molecular Analysis Technologies (IMAT) Program. The research endeavors at Cedars Sinai Medical Center were supported by DoD Idea Award (W81XWH-11-1-0422) and PCF Young Investigator Award.
Footnotes
Experimental: The experimental section is available in the Supporting information.
Contributor Information
Dr. Libo Zhao, Key Laboratory of Molecular Nanostructure and Nanotechnology, Institute of Chemistry, Chinese Academy of Science, Beiyi Street 2#, Zhongguancun, Beijing, 100190 (P.R. China); Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), University of California at Los Angeles 570 Westwood Plaza, Build 114, Los Angeles, California 90095-1770, USA.
Dr. Yi-Tsung Lu, Urologic Oncology Program & Uro-Oncology Research Program; Department of Medicine, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center 8700 Beverly Blvd. Los Angeles, California 90048, USA.
Fuqiang Li, BGI-ShenZhen, 2nd F, Building No. 11, Beishan Industrial Zone, Yantian District Shenzhen 518083, China.
Dr. Kui Wu, BGI-ShenZhen, 2nd F, Building No. 11, Beishan Industrial Zone, Yantian District Shenzhen 518083, China
Dr. Shuang Hou, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), University of California at Los Angeles 570 Westwood Plaza, Build 114, Los Angeles, California 90095-1770, USA
Dr. Juehua Yu, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), University of California at Los Angeles 570 Westwood Plaza, Build 114, Los Angeles, California 90095-1770, USA
Qinglin Shen, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), University of California at Los Angeles 570 Westwood Plaza, Build 114, Los Angeles, California 90095-1770, USA.
Dr. Dongxia Wu, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), University of California at Los Angeles 570 Westwood Plaza, Build 114, Los Angeles, California 90095-1770, USA
Dr. Min Song, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), University of California at Los Angeles 570 Westwood Plaza, Build 114, Los Angeles, California 90095-1770, USA
Wei-Han OuYang, CytoLumina Technologies Corp. 21038 Commerce Point Dr. Walnut, California 91789, USA.
Zheng Luo, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), University of California at Los Angeles 570 Westwood Plaza, Build 114, Los Angeles, California 90095-1770, USA.
Tom Lee, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), University of California at Los Angeles 570 Westwood Plaza, Build 114, Los Angeles, California 90095-1770, USA.
Prof. Xiaohong Fang, Key Laboratory of Molecular Nanostructure and Nanotechnology, Institute of Chemistry, Chinese Academy of Science, Beiyi Street 2#, Zhongguancun, Beijing, 100190 (P.R. China)
Prof. Chen Shao, Department of Urology, Xijing Hospital, Fourth Military Medical University, Xi'an, China
Dr. Xun Xu, Email: xuxun@genomics.cn, BGI-ShenZhen, 2nd F, Building No. 11, Beishan Industrial Zone, Yantian District Shenzhen 518083, China.
Dr. Mitch A. Garcia, Email: mgarcia@cytolumina.com, CytoLumina Technologies Corp. 21038 Commerce Point Dr. Walnut, California 91789, USA.
Prof. Leland W. K. Chung, Email: Leland.Chung@cshs.org, Urologic Oncology Program & Uro-Oncology Research Program; Department of Medicine, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center 8700 Beverly Blvd. Los Angeles, California 90048, USA.
Prof. Matthew Rettig, Email: MRettig@mednet.ucla.edu, Departments of Medicine and Urology, Jonsson Comprehensive Cancer Center, University of California at Los Angeles, Chief, Division of Hematology-Oncology, VA Greater Los Angeles Healthcare System.
Prof. Hsian-Rong Tseng, Email: HRTseng@mednet.ucla.edu, Department of Molecular and Medical Pharmacology, Crump Institute for Molecular Imaging (CIMI), California NanoSystems Institute (CNSI), University of California at Los Angeles 570 Westwood Plaza, Build 114, Los Angeles, California 90095-1770, USA.
Prof. Edwin M. Posadas, Email: Edwin.Posadas@csmc.edu, Urologic Oncology Program & Uro-Oncology Research Program; Department of Medicine, Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center 8700 Beverly Blvd. Los Angeles, California 90048, USA.
References
- 1.Kling J. Nat Biotechnol. 2012;30:578. doi: 10.1038/nbt.2295. [DOI] [PubMed] [Google Scholar]
- 2.Kaiser J. Science. 2010;327:1072. doi: 10.1126/science.327.5969.1072. [DOI] [PubMed] [Google Scholar]; Bernards R, Weinberg RA. Nature. 2002;418:823. doi: 10.1038/418823a. [DOI] [PubMed] [Google Scholar]; Criscitiello C, Sotiriou C, Ignatiadis M. Curr Opin Oncol. 2010;22:552. doi: 10.1097/CCO.0b013e32833de186. [DOI] [PubMed] [Google Scholar]
- 3.Attard G, Swennenhuis JF, Olmos D, Reid AH, Vickers E, A'Hern R, Levink R, Coumans F, Moreira J, Riisnaes R, Oommen NB, Hawche G, Jameson C, Thompson E, Sipkema R, Carden CP, Parker C, Dearnaley D, Kaye SB, Cooper CS, Molina A, Cox ME, Terstappen LW, de Bono JS. Cancer Res. 2009;69:2912. doi: 10.1158/0008-5472.CAN-08-3667. [DOI] [PubMed] [Google Scholar]
- 4.Allan AL, Vantyghem SA, Tuck AB, Chambers AF, Chin-Yee IH, Keeney M. Cytom Part A. 2005;65:4. doi: 10.1002/cyto.a.20132. [DOI] [PubMed] [Google Scholar]
- 5.Jacob K, Sollier C, Jabado N. Expert Rev Proteomics. 2007;4:741. doi: 10.1586/14789450.4.6.741. [DOI] [PubMed] [Google Scholar]; Yang L, Lang JC, Balasubramanian P, Jatana KR, Schuller D, Agrawal A, Zborowski M, Chalmers JJ. Biotechnology and bioengineering. 2009;102:521. doi: 10.1002/bit.22066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zheng S, Lin H, Liu JQ, Balic M, Datar R, Cote RJ, Tai YC. J Chromatogr A. 2007;1162:154. doi: 10.1016/j.chroma.2007.05.064. [DOI] [PubMed] [Google Scholar]
- 7.Navin N, Hicks J. Genome medicine. 2011;3:31. doi: 10.1186/gm247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Wang S, Wang H, Jiao J, Chen KJ, Owens GE, Kamei K, Sun J, Sherman DJ, Behrenbruch CP, Wu H, Tseng HR. Angew Chem Int Ed. 2009;48:8970. doi: 10.1002/anie.200901668. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wang S, Liu K, Liu J, Yu ZT, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee YK, Chung LW, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CK, Tseng HR. Angew Chem Int Ed Engl. 2011;50:3084. doi: 10.1002/anie.201005853. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Hou S, Zhao H, Zhao L, Shen Q, Wei KS, Suh DY, Nakao A, Garcia MA, Song M, Lee T, Xiong B, Luo SC, Tseng HR, Yu HH. Adv Mater. 2013;25 doi: 10.1002/adma.201203185. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Shen Q, Xu L, Zhao L, Wu D, Fan Y, Zhou Y, OuYang W, Xu X, Zhang Z, Lee T, Xiong B, Hou S, Tseng HR, Fang X. Adv Mater. 2013;25 doi: 10.1002/adma.201300082. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sekine J, Luo SC, Wang S, Zhu B, Tseng HR, Yu HH. Adv Mater. 2011;23:4788. doi: 10.1002/adma.201102151. [DOI] [PubMed] [Google Scholar]
- 10.Zhang N, Deng Y, Tai Q, Cheng B, Zhao L, Shen Q, He R, Hong L, Liu W, Guo S, Liu K, Tseng HR, Xiong B, Zhao XZ. Adv Mater. 2012;24:2756. doi: 10.1002/adma.201200155. [DOI] [PubMed] [Google Scholar]
- 11.Fischer KE, Aleman BJ, Tao SL, Hugh Daniels R, Li EM, Bunger MD, Nagaraj G, Singh P, Zettl A, Desai TA. Nano letters. 2009;9:716. doi: 10.1021/nl803219f. [DOI] [PMC free article] [PubMed] [Google Scholar]; Curtis ASG, Varde M. J Natl Cancer. 1964;33:15. [PubMed] [Google Scholar]
- 12.Stroock AD, Dertinger SK, Ajdari A, Mezic I, Stone HA, Whitesides GM. Science. 2002;295:647. doi: 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- 13.Gallagher RI, Blakely SR, Liotta LA, Espina V. Methods Mol Biol. 2012;823:157. doi: 10.1007/978-1-60327-216-2_11. [DOI] [PubMed] [Google Scholar]
- 14.Pound CR, Partin AW, Eisenberger MA, Chan DW, Pearson JD, Walsh PC. JAMA-J Am Med Assoc. 1999;281:1591. doi: 10.1001/jama.281.17.1591. [DOI] [PubMed] [Google Scholar]
- 15.Bubendorf L, Schopfer A, Wagner U, Sauter G, Moch H, Willi N, Gasser TC, Mihatsch MJ. Hum Pathol. 2000;31:578. doi: 10.1053/hp.2000.6698. [DOI] [PubMed] [Google Scholar]
- 16.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. N Engl J Med. 2012;366:883. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alatrash G, Mittendorf EA, Sergeeva A, Sukhumalchandra P, Qiao N, Zhang M, St John LS, Ruisaard K, Haugen CE, Al-Atrache Z, Jakher H, Philips AV, Ding X, Chen JQ, Wu Y, Patenia RS, Bernatchez C, Vence LM, Radvanyi LG, Hwu P, Clise-Dwyer K, Ma Q, Lu S, Molldrem JJ. J Immunol. 2012;189:5476. doi: 10.4049/jimmunol.1201221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hou S, Zhao L, Shen Q, Yu J, Ng C, Kong X, Wu D, Song M, Shi X, Xu X, OuYang WH, He R, Zhao XZ, Xiong B, Lee T, Brunicardi C, Garcia MA, Ribas A, Lo RS, R TH. Angew Chem Int Ed Engl. 2013;52 doi: 10.1002/anie.201208452. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Genome Res. 2009;19:1639. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.