Abstract
Amino acids, especially glutamine (GLN) have been known for many years to stimulate the growth of small intestinal mucosa. Polyamines are also required for optimal mucosal growth, and the inhibition of ornithine decarboxylase (ODC), the first rate-limiting enzyme in polyamine synthesis, blocks growth. Certain amino acids, primarily asparagine (ASN) and GLN stimulate ODC activity in a solution of physiological salts. More importantly, their presence is also required before growth factors and hormones such as EGF and insulin are able to increase ODC activity. ODC activity is inhibited by antizyme-1 (AZ) whose synthesis is stimulated by polyamines, thus, providing a negative feedback regulation of the enzyme. In the absence of amino acids mammalian target of rapamycin complex 1 (mTORC1) is inhibited, whereas, mTORC2 is stimulated leading to the inhibition of global protein synthesis but increasing the synthesis of AZ via a cap-independent mechanism. These data, therefore, explain why ASN or GLN is essential for the activation of ODC.
Interestingly, in a number of papers, AZ has been shown to inhibit cell proliferation, stimulate apoptosis or increase autophagy. Each of these activities results in decreased cellular growth. AZ binds to and accelerates the degradation of ODC and other proteins shown to regulate proliferation and cell death, such as Aurora-A, Cyclin D1 and Smad1. The correlation between the stimulation of ODC activity and the absence of AZ as influenced by amino acids is high. Not only do amino acids such as ASN and GLN stimulate ODC while inhibiting AZ synthesis, but also amino acids such as lysine, valine and ornithine, which inhibit ODC activity, increase the synthesis of AZ. The question remaining to be answered is whether AZ inhibits growth directly or whether it acts by decreasing the availability of polyamines to the dividing cells. In either case, evidence strongly suggests that the regulation of AZ synthesis is the mechanism through which amino acids influence the growth of intestinal mucosa.
This brief article reviews the experiments leading to the information presented above. We also present evidence from the literature that AZ acts directly to inhibit cell proliferation and increase the rate of apoptosis. Finally, we discuss future experiments that will determine the role of AZ in the regulation of intestinal mucosal growth by amino acids.
Keywords: asparagine, glutamine, polyamines, antizyme, mucosal cell proliferation, apoptosis
INTRODUCTION
The epithelial cells lining the intestine are exposed to more potential stimulants and inhibitors of growth than any tissue in the body (Johnson and McCormack, 1994). Not only are these cells affected by the normal classical growth-promoting hormones such as thyroxine and growth hormone, but they also respond to hormones originating from endocrine cells along the gastrointestinal (GI) tract such as gastrin, enteroglucagons and neurotensin. In addition, a host of purported trophic substances reach these cells from the lumen of the tract. These include agents, such as EGF, that are present in the secretions of the organs and cells of the digestive system, metabolic and secreted products of the gut microflora, ingested food and nutrient breakdown products. The latter of this group has frequently been referred to as luminal or local nutrition, meaning that the absorbing cells of the intestine are directly stimulated by specific nutrients in the lumen.
Whether local nutrients stimulate mucosal growth has been tested by infusing various substances into isolated gut loops. Liquid elemental diets have been reported to cause hyperplasia when infused into bypassed mucosa (Jacobs, et al, 1975). Various sugars, including glucose, have been shown to stimulate cell production when infused into isolated sections of intestine (Clarke RM, 1977). The strongest support for the stimulation of mucosal growth by enteral nutrients has come from experiments involving total parenteral nutrition (TPN), in which glutamine was supplemented enterally or added to the TPN mixture. Barrin et al (1994) showed that glutamine supplementation increases both jejunal height and surface area and attenuated most of the effects of TPN on the gut. Glutamine supplementation also increased protein synthesis in the intestinal mucosa of rats on TPN and decreased the tissue damage caused by sepsis (Yoshida S et al, 1992). Glutamine has also been shown to stimulate protein synthesis in isolated intestinal epithelial cells, and to be the only amino acid to do so of several tested (Higashiguchi T et al, 1993).
These types of results have lead to the suggestion that luminally derived amino acids might be important for nutrition and growth of small intestinal mucosa (Hirschfield and Kern, 1969). However, if one compares the protein distributions of orally administered amino acids with those administered parenterally, the patterns are different (Alpers 1972). After intravenous amino acid administration, proteins from cells of the crypt junctions are most heavily labeled. Following intraluminal administration, protein from cells near the villous tips is most heavily labeled with radioactive precursor. In other words, mature non-dividing enterocytes incorporate luminal amino acids into protein. The cells responsible for growth, the crypt cells, are supplied from the blood. This is strong evidence against the local nutrition concept, and to date there is no satisfactory explanation of the mechanism by which certain amino acids, or the lack thereof, affect the growth of the mucosa of the small intestine.
POLYAMINES
The polyamines, spermidine and spermine, and their precursor, putrescine, are found in virtually all cells of higher eukaryotes and are required for cell growth and proliferation (Pegg and McCann, 1982). Intracellular polyamine levels are highly regulated and are dependent primarily on the activity of ornithine decarboxylase (ODC), which catalyzes the first rate-limiting step in polyamine biosynthesis, the decarboxylation of ornithine to form the diamine putrescine (Russell, 1985). DL-α-difluoromethylornithine (DFMO) blocks ODC and depletes cells of polyamines. DFMO is a specific and irreversible inhibitor of ODC, having no effects except those caused by ODC inhibition and the subsequent depletion of polyamines (Pegg et al, 1987).
For a number of years, our laboratory has been interested in the role of polyamines in the growth and repair of the GI mucosa and has examined these processes in both rats and cultured normal intestinal cells. The mammalian GI epithelium is especially suited to study the control of proliferation because of its rapid and continuous renewal. Inhibition of ODC by DFMO depletes polyamines and inhibits the growth of cultured IEC-6 cells, a nontransformed line originally developed from rat crypt cells by Quaroni et al (1988), and prevents the normal repair of mucosal stress ulcers (Wang and Johnson 1990, Wang and Johnson 1991a, Zimmerman et al 1995).
Cells maintain intracellular polyamine levels at optimal concentrations by regulating synthesis and degradation and uptake and release. Basal levels of ODC are low, but enzyme activity increases rapidly in response to a variety of stimuli, including growth factors, insulin, stress, and certain amino acids (Wang and Johnson 1991, Rinehart and Canellakis 1985). ODC has one of the shortest half-lives of any mammalian enzyme and is feedback regulated by polyamines which induce the synthesis of an inhibitor called antizyme (Heller et al, 1976). The active form of ODC is a homodimer composed of two 53-kDa subunits. The active site is formed at the interface of the two monomers, the monomers having no enzymatic activity (Coleman et al 1994). Antizyme (AZ) inhibits ODC activity by binding to the monomers to prevent them from coming together (Mitchell and Chen, 1990; Li and Coffino, 1992). The second function of AZ is to feedback inhibit the uptake of polyamines by cells (Pegg 2006).
Amino Acids and ODC
Early studies by Fausto (1971) showed that some amino acids could stimulate ODC in regenerating mammalian liver. Similar results showing that amino acids induced ODC in cultured cells were obtained by Kay et al (1972) and Hogan and Murden (1974) using standard cell culture media. Important progress to understanding the role of amino acids occurred in 1977 when Chen and Canellakis developed the use of a salts-glucose solution to maintain cultured cells in the absence of complex growth media and serum. They determined that asparagine (ASN) and then glutamine (GLN) were the most effective amino acids in stimulating ODC activity in a neuroblastoma cell line. Since there were no other stimulants present, the induction of enzyme activity was a direct effect of the amino acids. Additional studies by the same group showed that the induction of ODC activity depended on Na+ but not on the incorporation of ASN into protein (Viceps-Madore et al, 1982). They also found that some non-metabolizable amino acids such as α-aminoisobutric acid (AIB) were effective stimulants of ODC activity. Although ASN was the most effective, other amino acids transported by one of the Na+-dependent systems could also induce ODC activity (Rinehart et al 1985). Of particular interest was the finding that, in a variety of cell types, the induction of ODC by growth factors and hormones required the presence of ASN, GLN or a similar amino acid (Rinehart and Canellakis, 1985). Neither epidermal growth factor (EGF), nerve growth factor, nor insulin could induce ODC activity unless ASN was present in the medium.
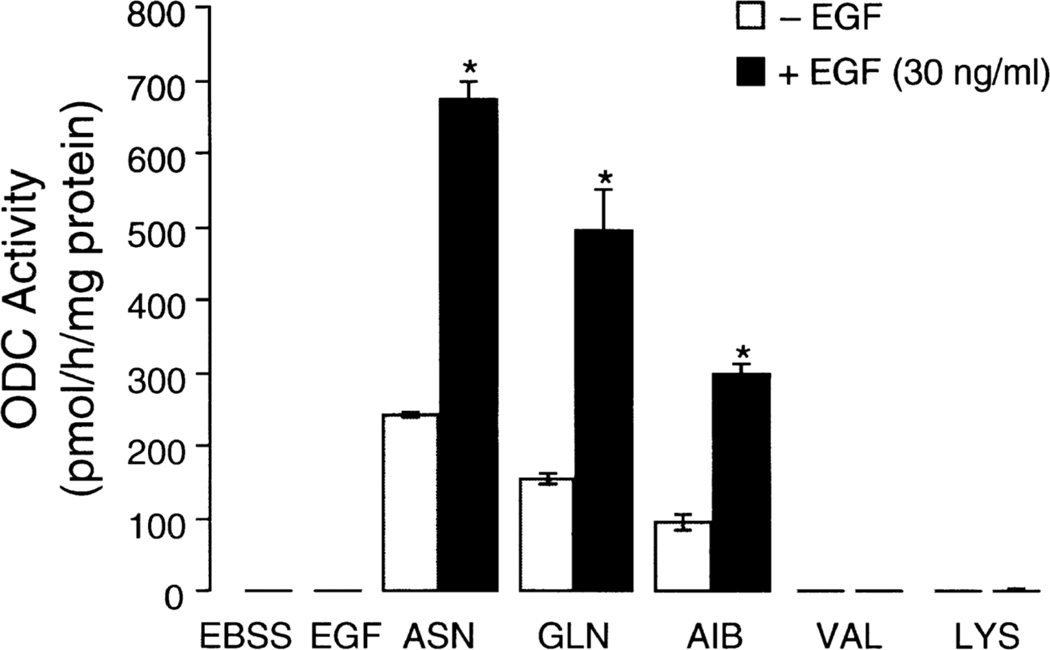
Our own laboratory examined the interaction of ASN and EGF in the regulation of ODC in intestinal epithelial (IEC-6) cells (Ray et al 1999). EGF was unable to increase ODC activity in cells maintained in a salt-glucose solution, but the addition of ASN (10mM) in the presence of EGF (30 ng/ml) increased ODC activity 3-fold over the response to ASN alone (Fig. 1). EGF was also able to induce ODC activity in the presence of GLN or AIB, but induction was maximum with ASN. Other amino acids, such as lysine and valine, had no effect alone or in combination with EGF. Thus, the results in intestinal epithelial cells are the same as those shown by the Canellakis group in other cell types.
Fig. 1.
Effect of asparagine (ASN), glutamine (GLN), α-aminoisobutyric acid (AIB), valine (VAL) and lysine (LYS) on the induction of ODC activity in the presence and absence of EGF (30 ug/ml). Amino acids were present at 10mM, and ODC was measured after a 3h incubation. Means ± SE; n=6 *P<0.5 compared to corresponding control. From: Ray et al 1999.
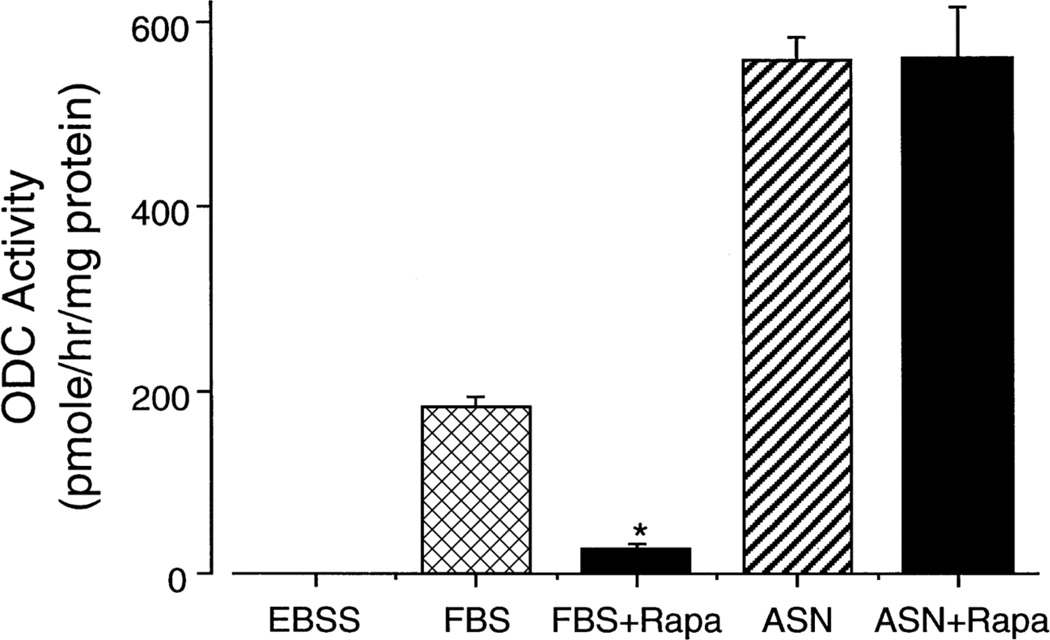
All of these studies taken together indicate that the mechanism of the interaction between ASN (and related amino acids) and EGF (and other growth factors and hormones) is important not only for understanding the regulation of ODC under physiological conditions but also for understanding the regulation of growth. Using the same conditions, Ray et al (1999) found that EGF alone increased the expression of the genes for c-Jun and c-Fos but not for ODC. ASN alone increased the expression of the ODC gene without effect on those for c-Jun and c-Fos. When ASN was added in the presence of EGF, ODC gene expression increased significantly over that seen with ASN alone. The addition of ASN had no effect on the response of the proto-oncogenes to EGF (Ray et al 1999). Interestingly, western analysis showed no significant difference in the levels of ODC protein in salt solution, ASN, EGF or ASN plus EGF, indicating that the synthesis and degradation of ODC were unaffected. The role of protein translation was examined using the mammalian target of Rapamycin (mTOR) 1 inhibitor rapamycin. mTOR1 is active in the presence of normal amounts of amino acids and phosphorylates 4EBP1, the binding protein which inhibits eIF-4E, a key factor in the translation of mRNA. Phosphorylation of 4EBP1 prevents its binding to eIF-4E and allows cap-dependent protein synthesis to take place (Wang and Proud, 2006). Figure 2 shows that rapamycin inhibited ODC activity stimulated by serum approximately 85 percent but had no effect on the much larger response to ASN. The failure of rapamycin to block the response to serum totally was probably due to the presence of amino acids in the serum as well as growth factors. These data rule out the involvement of eIF-4E in the induction of ODC activity by ASN and establish that the stimulation of ODC activity by ASN and similar amino acids is a distinct process from that of growth factors (Ray et al 1999). Further support for this conclusion is the original finding that the combination of ASN and EGF results in potentiation, which in itself requires separate mechanisms of action for the two agents.
Fig. 2.
Effect of rapamycin (100 nM) on the induction of ODC by 10% fetal bovine serum (FBS) or ASN. Cells were incubated for 3h and ODC activity measured. Means ± SE; n=6 cultures; *P<0.05 compared to corresponding control. From: Ray et al 1999.
Amino Acids and Antizyme
As mentioned earlier, AZ is an important negative regulator of intracellular polyamine levels. AZ binds ODC monomers preventing the formation of the active dimers, eventually leading to ubiquitin-independent degradation by the 26 S proteosome. Over expression of AZ prevents tumor growth and inhibits cell proliferation (Feith et al 2001) analogous to polyamine depletion by DFMO. An ODC-like protein called antizyme inhibitor (AZin) increases ODC activity by binding to AZ. Serum and phorbol ester increase AZin expression prior to ODC activity and promote growth and tumor formation, respectively (Pegg et al 2003). Thus, AZ and AZin are important regulators of ODC activity, polyamine levels and, thereby, growth.
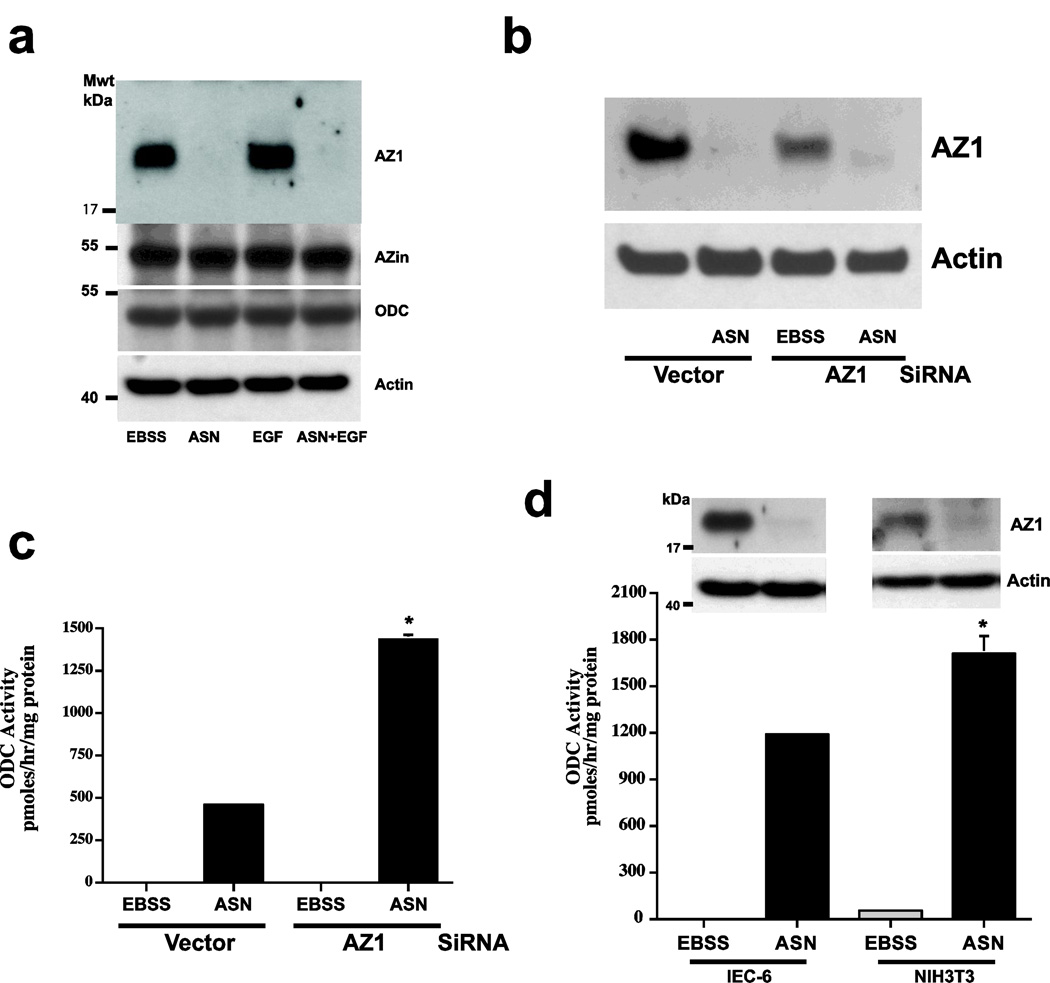
Since amino acids transported by A and N type systems are required for the induction of ODC, ASN might affect ODC activity by regulating AZ levels. As shown in Fig. 3a, amino acid starvation (EBSS) increased the expression of AZ in IEC-6 cells, and the addition of EGF to the balanced salt solution produced no change in AZ protein. In the presence of ASN, however, AZ decreased to undetectable levels in both cases. In no group was there a change in either AZin or ODC protein levels. Transfection of plasmid-based AZ siRNA decreased AZ expression in EBSS (Fig. 3b) and significantly increased ODC activity compared with that induced in vector-transfected cells in response to ASN (Fig. 3c). The siRNA data confirmed the finding that EBSS or the absence of amino acids induces the expression of AZ. Figure 4d shows that EBSS induced AZ expression in NIH3T3 fibroblasts as well as in IEC-6 cells, and that ASN prevents AZ expression in both cell types with similar increases in ODC activity. Despite differences in AZ protein levels and ODC activities, ASN had similar effects in both cell lines. The time courses for the activation of ODC and disappearance of AZ following the addition of ASN were similar. ASN when added after a 3h incubation in EBSS, ODC activity was detected within 60 min and continued to increase for at least 180 min, which correlated with the absence of AZ (Ray et al 2012).
Fig. 3.
A: ASN (10mM) was added to EBSS ± EGF (30 ug/ml) and incubated for 3h. IEC-6 extracts were analyzed by Western blot to determine levels of AZ, AZin, ODC and actin. B: Cells transfected with vector or AZ1 siRNA were incubated with EBSS ±ASN (10 mM) for 3h. Extracts were analyzed by western blot to determine levels of AZ and C: assay for ODC activity. D: IEC-6 and NIH 3T3 cells were incubated in EBSS ± ASN (10 mM) for 3h. ODC activity and AZ levels were determined. Means ± SE; n=3 cultures *P<0.05 compared to corresponding control. From: Ray et al (2012).
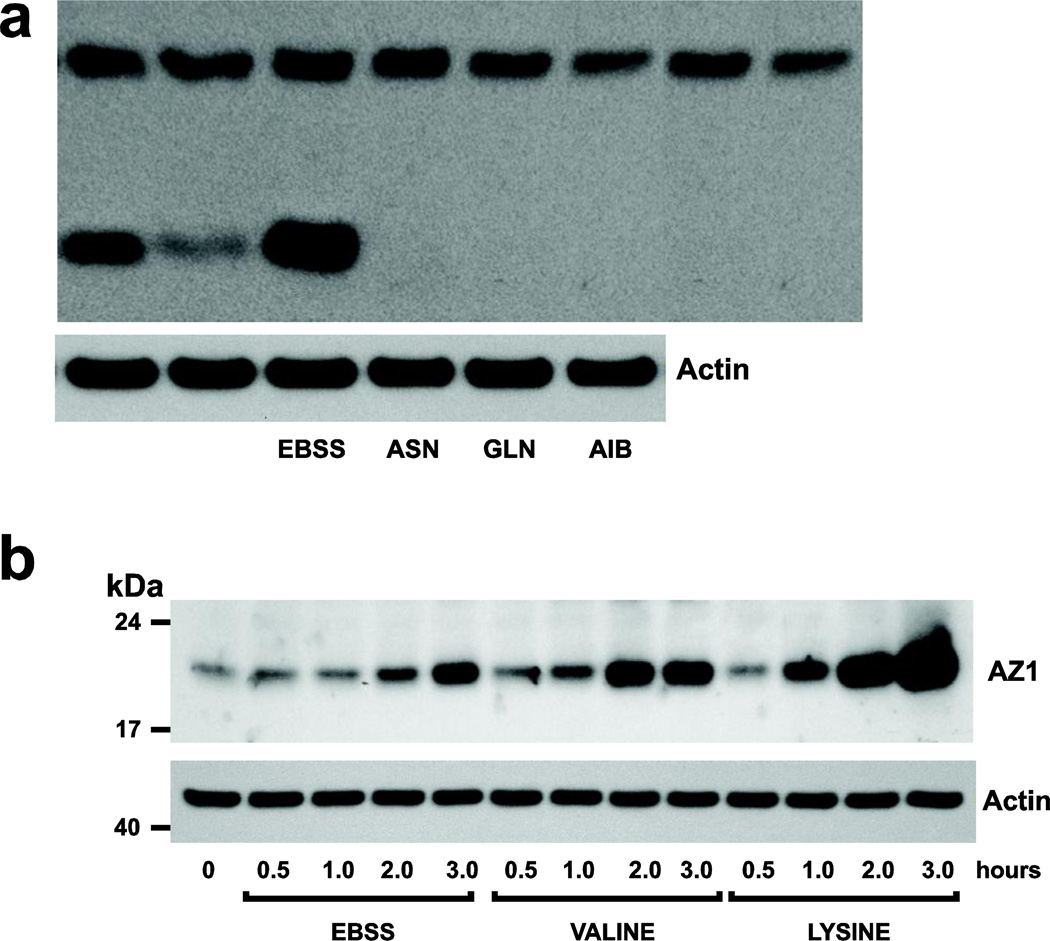
Fig. 4.
A: Confluent, serum starved cells were incubated in EBSS ± ASN, GLN, or AIB (10 mM) for 3h, and extracts were analyzed by Western blot for AZ1 and actin. B: Cells were incubated in EBSS ±VAL or LYS and at timed intervals, extracts were analyzed by Western blot to determine the levels of AZ1 and actin. Representative Western blots from 3 observations are shown. From: Ray et al 2012.
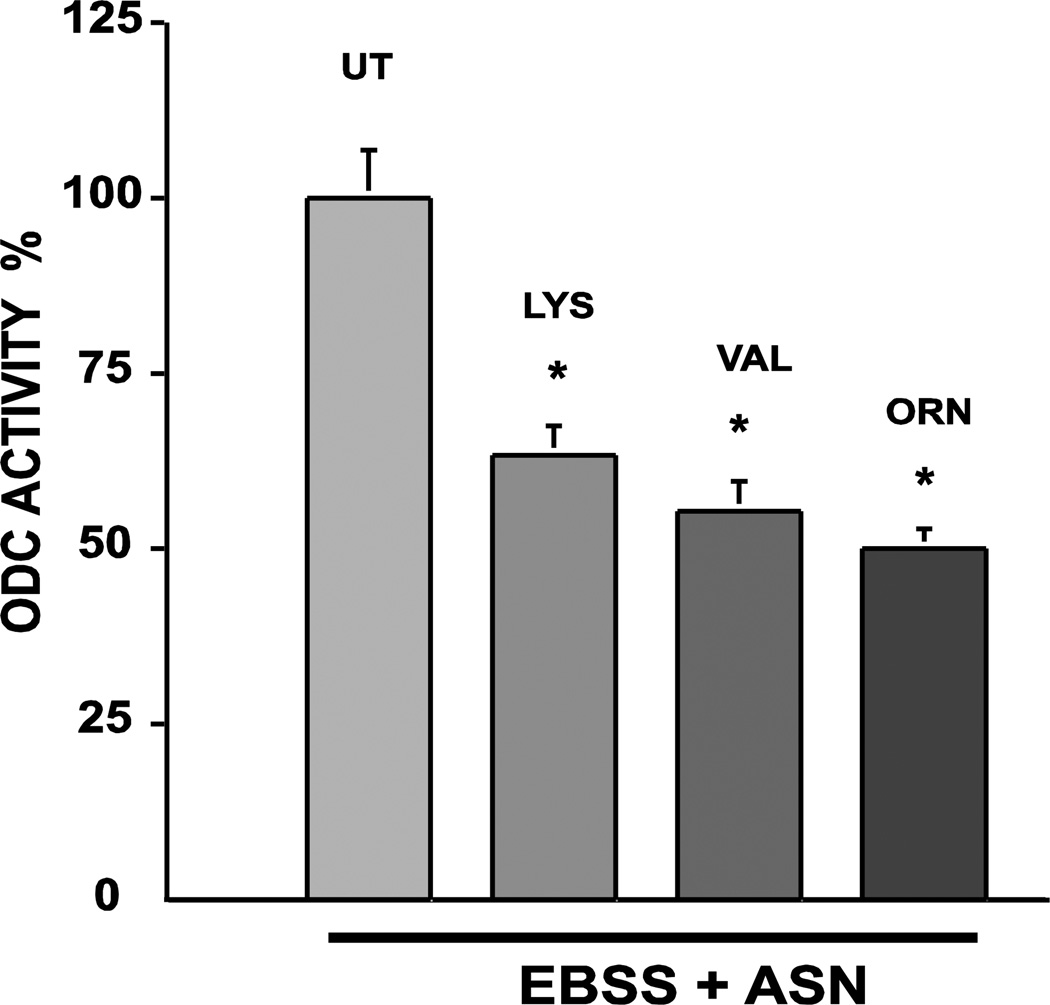
Previous studies already discussed (Rinehart et al 1985; Ray et al 1999) showed that, like ASN, glutamine (GLN) and α-aminoisobutyric acid (AIB) increased and lysine (LYS) and valine (VAL) failed to increase ODC activity in a balanced glucose-salt solution. When added to EBSS and incubated for 3h, GLN and AIB as well as ASN prevented the expression of AZ (Fig. 4a). Interestingly, both VAL and LYS, non-inducers of ODC activity increased AZ expression in a time dependent manner (Fig. 4b). These results predict that LYS and VAL should inhibit ODC activity induced by ASN (Ray et al, 2012). Figure 5 show that both LYS and VAL significantly inhibited the activation of ODC when incubated in salt solution with ASN. Ornithine, a known inducer of AZ, also inhibited the induction of ODC activity (Ray et al 2012).
Fig. 5.
Confluent serum starved IEC-6 cells were incubated in EBSS + 10 mM ASN in the presence or absence of either LYS or VAL and ODC activity was determined after 3h. Means ± SE; n=3 culture; *P<0.05 compared to UT (untreated). From: Ray et al 2012.
The primary nutrient sensor in eukaryotes is target of rapamycin (TOR), a kinase that acts on a subset of proteins to regulate translation, autophagy and apoptosis (Zoncu et al 2011). Mammalian TOR (mTOR) is a PI3 kinase related enzyme that is the catalytic subunit of two separate complexes, distinguished by their accessory proteins. mTORC1 contains regulatory-associated protein of mTOR (RAPTOR) and is inhibited by rapamycin and activated by amino acids (Hara et al 2002). mTORC2 is associated with rapamycin-insensitive companion of TOR (RICTOR) (Sabassov DD et al 2004) and is activated by PI3K/AKT signaling (Tato et al 2011). Substrates of mTORC1, S6 kinase (S6K1) and eIF-4E binding protein (4EBP1) regulate mRNA translation initiation to control rates of protein synthesis and growth. mTORC1 integrates several major regulatory signals: nutrients (primarily amino acids), energy, and growth factors, which activate it, and stress, which inhibits. For example, in the absence of amino acids, mTORC1 is inhibited resulting in decreased cell growth and increased autophagy which provides amino acids for essential cell functions by degrading proteins (Zoncu et al 2011). Much less is known about mTORC2 than mTORC1. TORC2 was originally identified as a regulator of cytoskeletal reorganization (Jacinto et al 2004). mTORC2 also regulates cell cycle progression and cell survival by phosphorylating and activating AKT and protein kinase C (Sarbassov et al 2005). Few specifics are known regarding the upstream activation of mTORC2, although it is believed to be activated by growth factors, and insulin causes the phosphorylation of Ser 473 of AKT via TORC2 (Sarbassov et al 2005).
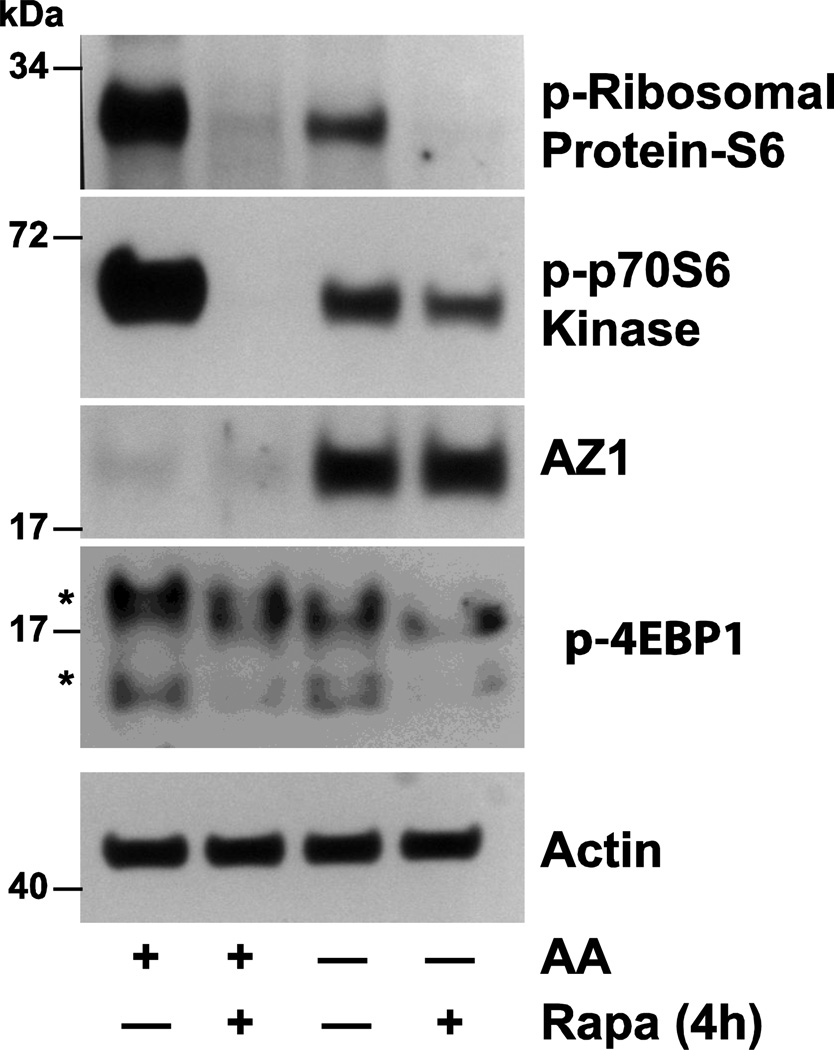
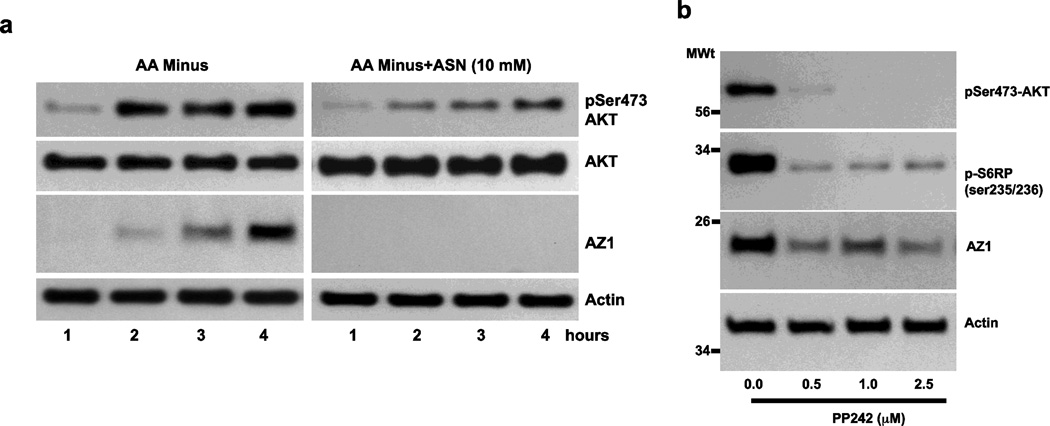
Figure 6 shows that following 4h incubation in the presence of amino acids mTORC1 is activated as evidenced by the phosphorylation of ribosomal protein S6, p70S6 kinase and 4EBP1, all of which is inhibited by rapamycin (Ray et al 2012). The right 2 columns of Fig. 6 show that 4h incubation in the absence of amino acids causes a dramatic increase in AZ protein and decreases in the phosphorylation of S6, p70S6 kinase and 4EBP1. Each of these responses except the increase in AZ are further inhibited by rapamycin. Since rapamycin insensitive mTOR signaling is mediated by mTORC2, these data suggest that mTORC2 is involved in the regulation of AZ expression. In the absence of amino acids mTORC2 activity increases as evidenced by increased AKT phosphorylation at the same time the level of AZ protein increases as well (Fig. 7a, left panel). Incubation in the presence of ASN results in undetectable levels of AZ after only 1hr and inhibition of the phosphorylation of AKT (right panel). Further evidence of the involvement of mTORC2 is shown in Fig. 7b. PP242, an inhibitor of mTORC2 prevents the phosphorylation of AKT and significantly decreases AZ levels after 4h incubation in the absence of amino acids.
Fig. 6.
Confluent serum starved cells were incubated in DMEM AA (amino acids) in the presence or absence of rapamycin (20nM) for 4h. Cell extracts were analyzed by Western blot to determine the levels of phosphoribosomal protein-S6, phospho-p7056K, AZ, and actin. Representative blots from three observations. From: Ray et al 2012.
Fig. 7.
A: Serum starved IEC-6 cells were incubated in DMEM in the absence of amino acids (AA-) in the absence and presence of ASN for the times indicated. Cell extracts were analyzed by Western blot to determine the levels of phosphoserine 473-AKT, AKT, AZ and actin. B: Cells were incubated with the indicated concentrations of PP242 for 4h, and extracts were analyzed for levels of phosphoserine 473-AKT, phosphoserine 235/236-S6 ribosomal protein, AZ and actin. Representative Western blots from three observations. From: Ray et al 2012.
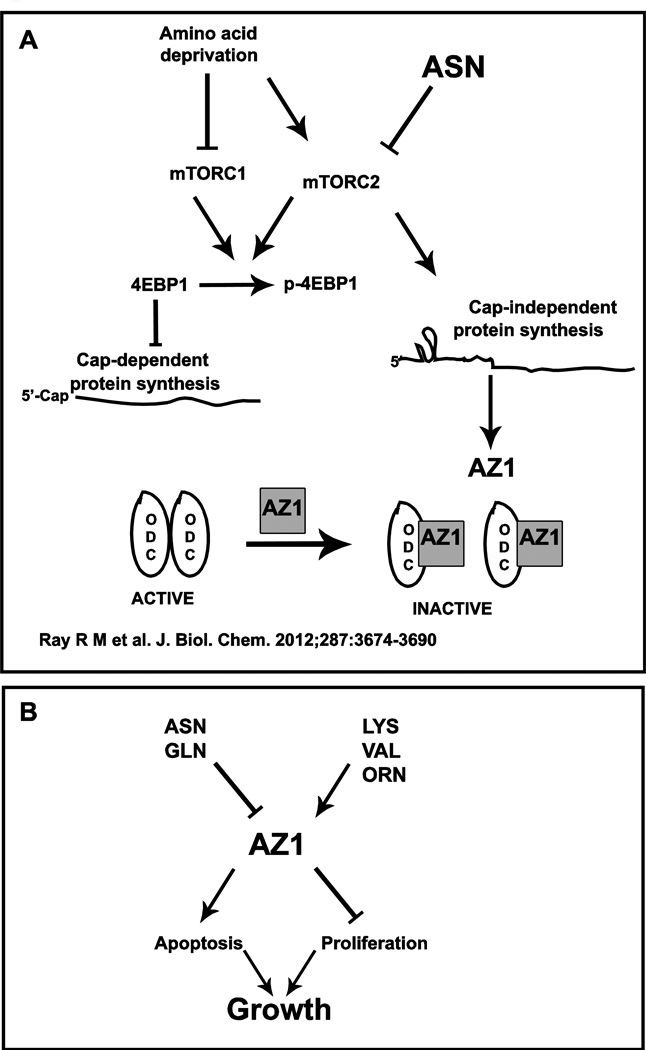
Taken together, these data indicate that in the absence of amino acids, mTORC1 is inhibited which leads to a decrease in cap-dependent protein synthesis by phosphorylating 4EBP-1. At the same time, mTORC2 is activated, and AZ is synthesized by cap-independent translation leading to the inhibition of ODC. ASN or GLN block mTORC2 resulting in a rapid decrease in AZ levels and in the activation of ODC, which can then be potentiated by growth factors (Fig. 8A).
Fig. 8.
In the absence of amino acids, mTORC1 is inhibited decreasing the phosphorylation of 4EBP1, leading to the inhibition of cap-dependent mRNA translation. At the same time mTORC2 activity is stimulated which increases cap-independent translation and the synthesis of AZ protein, which inhibits ODC. ASN inhibits mTORC2 activity and AZ synthesis allowing ODC monomers to form the active dimeric enzyme. From: Ray et al 2012. (b) Amino acids regulate growth via AZ1.
Antizyme and Growth
Tissue growth, or lack thereof, is determined by the balance between cell proliferation and cell death, in most cases apoptosis. The regulation of AZ levels may turn out to be the mechanism whereby amino acids, especially ASN and GLN, regulate gastrointestinal mucosal growth. There are several recent reports that AZ directly binds regulators of apoptosis and proliferation, targeting them for degradation via the proteosome in the same way it accelerates the degradation of ODC; these include DNp73 (Dulloo et al, 2010), cyclin D1 (Newman et al 2004), Smad1 (Lin et al 2002), and Aurora-A proteins (Lim and Gopalan 2007).
DNp73 is an antiapoptotic protein which acts as a dominant negative inhibitor of both TAp73 and p53, and TAp73 has an obligatory role in regulating apoptosis in response to DNA damage (Lin et al 2004). Dulloo et al (2010) showed that DNp73 was degraded in a c-Jun dependent pathway that was mediated by direct binding to AZ, which targeted the protein for degradation. AZ is also a potent tumor suppressor and is necessary for the conversion of pancreatic tumor cells into glucagon-producing differentiated cells (Suzuki et al 2009).
Proliferation is regulated by cyclin dependent kinases (CDKs), which phosphorylate and activate cyclins to regulate the progression of cells from the G0/G1 phase into the S phase of the cell cycle (Woo and Poon 2003). AZ binds cyclin D1 in a noncovalent association causing its degradation via the proteosome. Newman et al (2004) referred to this as a “novel mechanism for cell growth regression.” Recently Kusbek et al (2010) reported that AZ binds monopolar spindle-1 kinase which is involved in cell cycle regulation, leading to its degradation and decreasing the formation of centrosomes.
Despite these reports of AZ binding directly to proteins that regulate growth, the question remains as to whether AZ exerts its effects directly or whether they are due to its control over polyamine levels. Fong et al (2003) found that transgenic mice over expressing AZ in the basal cell layer of the forestomach had decreased cell proliferation and increased levels of apoptosis, both of which decrease tissue growth. In addition, AZ inhibited the induction of tumors by N-nitrosomethylbenzylamine. The authors found that DFMO, and the resulting depletion of polyamines, had effects similar to AZ and attributed their results to the ability of AZ to decrease the levels of polyamines. However, the same group has reported that excessive accumulation of polyamines induced apoptosis in L1210 cells that over produced ODC (Pouline et al 1995).
Additional reports provide considerable evidence that AZ directly increases apoptosis. Packham and Cleveland (1994) concluded that ODC and polyamines are mediators of c-myc induced apoptosis. Kahana and Tobias (1995) found that excessive accumulation of putrescine led to cell death in mouse myeloma cells that over expressed ODC. Our laboratory has published a number of studies in which we examined the effects of DFMO on apoptosis. Since both DFMO and AZ inhibit ODC and lead to polyamine depletion, these experiments relate to the question of whether AZ acts directly. In each case we found that DFMO and polyamine depletion decrease apoptosis, the opposite effect one would expect if AZ caused apoptosis by decreasing polyamine levels. Polyamine depletion significantly protected IEC-6 cells from apoptosis induced by the topoisomerase 1 inhibitor, camptothecin, and decreased basal levels of apoptosis as well. When putrescine was added to the DFMO containing medium, the levels of apoptosis increased to normal (Ray et al 2000). Pretreatment of IEC-6 cells with DFMO also significantly reduced radiation-induced apoptosis as evidenced by caspase-3 activity and DNA fragmentation (Deng et al 2005). In the same report, pretreatment of mice with 2% DFMO in drinking water reduced apoptotic cells from 2.75 to 1.61 per crypt-villus unit, accompanied by significant decreases in caspase-3 activity. Crypt cell survival was increased more than 2-fold by polyamine depletion.
The issue regarding whether amino acids and AZ act independently of polyamines to stimulate proliferation is more complicated, since AZ decreases polyamine levels, and polyamine depletion inhibits growth. This question will only be settled by experiments designed carefully to address it. We have preliminary data indicating that spermidine, a potent inducer of AZ synthesis, inhibits proliferation in the presence of DFMO. When ASN was added to the incubation mixture AZ became undetectable and spermidine was then able to restore proliferation to control levels that were present prior to giving DFMO.
Summary and Future Studies
A recent article has pointed out that circulating products derived from food act as hormones stimulating receptors and signaling pathways to regulate growth (Ryan and Seeley, 2013). Among these are omega-3 fatty acids and a variety of short chain fatty acids that bind peroxisome proliferation-activated receptor γ (PPARγ) or modify the binding of the hormone ghrelin. Branched chain amino acids bind to and activate mTORC1, and are believed to play a role in cell-cycle progression and insulin activity.
The studies summarized in this brief review suggest that amino acids like ASN and GLN may also act as hormones. Known for years to be essential for the activation of ODC by growth factors, the finding that they act by inhibiting AZ synthesis provides a mechanism for their effects. Given the strong evidence that AZ directly stimulates apoptosis and that its synthesis can be regulated through the mTOR pathway, these amino acids may have direct effects on tissue growth independent of their nutrient value (Fig. 8b).
Crucial experiments need to be carried out to examine whether changes in AZ levels induced by ASN and GLN correlate with cell proliferation and apoptosis, and whether the effects can be shown to be independent of polyamines. To date all studies have been performed with cultured cells. Determining whether these amino acids regulate AZ levels in whole animals will be a significant step in deciding the importance of these findings.
ACKNOWLEDGEMENTS
This publication was made possible by Grants (Number DK -16505 and DK-052784) from the National Institute of Diabetes and Digestive and Kidney Disease (NIDDK) and by support from the Thomas A. Gerwin endowment. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institute of Health.
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
REFERENCES
- Alpers DH. Protein synthesis in intestinal mucosa: the effect of route of administration of precursor amino acids. J Clin Invest. 1972;51:167–173. doi: 10.1172/JCI106788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrin DG, Shulman RJ, Langston C, Storm MC. Supplemental alanyl glutamine, organ growth and nitrogen metabolism in neonatal pigs fed by total parenteral nutrition. J Parenter Enteral Nutr. 1994;18:313–319. doi: 10.1177/014860719401800406. [DOI] [PubMed] [Google Scholar]
- Chen KY, Canellakis ES. Enzyme regulation in neuroblastoma cells in a salts-glucose medium: Induction of ornithine decarboxylase by asparagine and glutamine. Proc Natl Acad Sci USA. 1977;74:3791–3795. doi: 10.1073/pnas.74.9.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke RM. “Luminal nutrition” versus “functional work-load” as controllers of mucosal morphology and epithelial replacement in the rat small intestine. Digestion. 1977;15:411–412. doi: 10.1159/000198029. [DOI] [PubMed] [Google Scholar]
- Coleman CS, Stanley BA, Viswanath R, Pegg AE. Rapid exchange of subunits of mammalian ornithine decarboxylase. J Biol Chem. 1994;269:3155–3158. [PubMed] [Google Scholar]
- Deng W, Viar MJ, Johnson LR. Polyamine depletion inhibits irradiation-induced apoptosis in intestinal epithelia. Am J Physiol. 2005;289:G599–G606. doi: 10.1152/ajpgi.00564.2004. [DOI] [PubMed] [Google Scholar]
- Dulloo I, Gopalan G, Melino G, Sabapathy K. The antiapototic delta Np73 is degraded in a c-Jun-dependent manner upon genotoxic stress through the antizyme-mediated pathway. Proc Natl Acad Sci USA. 2010;107:4902–4917. doi: 10.1073/pnas.0906782107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fausto N. The control of ornithine decarboxylase activity during liver regeneration. Biochim Biophys Acta. 1971;238:116–128. doi: 10.1016/0005-2787(71)90015-3. [DOI] [PubMed] [Google Scholar]
- Feith DJ, Shantz LM, Pegg AE. Targeted antizyme expression in the skin of transgenic mice reduces tumor promoter induction of ornithine decarboxylase and decreases sensitivity to chemical carcinogenesis. Cancer Res. 2001;61:6073–6081. [PubMed] [Google Scholar]
- Fong LYY, Feith DJ, Pegg AE. Antizyme over expression in transgenic mice reduces cell proliferation, increases apoptosis, and reduces N-nitrosomethylbenzylamide-induced forestomach carcinogenesis. Cancer Res. 2003;63:3945–3954. [PubMed] [Google Scholar]
- Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell. 2002;110:177–189. doi: 10.1016/s0092-8674(02)00833-4. [DOI] [PubMed] [Google Scholar]
- Heller JS, Fong WF, Canellakis ES. Induction of a protein inhibitor to ornithine decarboxylase by the end products of its reaction. Proc Natl Acad Sci USA. 1976;73:1858–1862. doi: 10.1073/pnas.73.6.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashiguchi T, Hasselgren P-O, Wagner K, Fischer JE. Effect of glutamine on protein synthesis in isolated intestinal epithelial cells. J Parenter Enteral Nutr. 1993;17:307–314. doi: 10.1177/0148607193017004307. [DOI] [PubMed] [Google Scholar]
- Hirschfield JS, Kern F. Protein starvation and the small intestine. III. Incorporation of orally and intraperitoneally administered l-leucine 4,55-3H into intestinal protein of protein-deprived rats. J Clin Invest. 1969;48:1224–1229. doi: 10.1172/JCI106086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan BLM, Murden S. Effect of growth conditions on the activity of ornithine decarboxylase in cultured hepatoma cells: I. Effect of amino acid supply. J Cell Physiol. 1974;83:345–352. doi: 10.1002/jcp.1040830304. [DOI] [PubMed] [Google Scholar]
- Jacinto E, Loewith R, Schmidt A, Liu S, Ruegg MA, Hall A, Hall MN. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat Cell Biol. 2004;6:1122–1128. doi: 10.1038/ncb1183. [DOI] [PubMed] [Google Scholar]
- Jacobs LR, Taylor BR, Dowling RH. Effect of luminal nutrition on intestinal adaptation following Thiry-Vella bypass in the dog. Clin Sci Mol Med. 1975;49:26. [Google Scholar]
- Johnson LR, McCormack SA. Regulation of gastrointestinal mucosal growth. In: Johnson LR, editor. Physiology of the Gastrointestinal Tract. 3rd ed. New York: Raven; 1994. pp. 611–642. [Google Scholar]
- Kasbek C, Yang CH, Fisk HA. Antizyme restrains centrosome amplification by regulating the accumulation of Mps1 at centrosomes. Mol Biol Cell. 2010;21:3878–3889. doi: 10.1091/mbc.E10-04-0281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JE, Lindsay VJ, Cooke A. Ornithine decarboxylase in phytohaemagglutinin stimulated lymphocytes: Control of degradation by amino acids. FEBS Lett. 1972;21:123–126. doi: 10.1016/0014-5793(72)80118-2. [DOI] [PubMed] [Google Scholar]
- Li X, Coffino P. Regulated degradation of ornithine decarboxylase requires interaction with the polyamine-inducible protein antizyme. Mol Cell Biol. 1992;12:3556–3562. doi: 10.1128/mcb.12.8.3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Martin J, Gruendler C, Farley J, Meng X, Li BY, Lechleider R, Huff C, Kim RH, Grasser WA, Paralker V, Wang T. A novel link between the proteasome pathway and the signal transduction pathway of the bone morphogenetic proteins (BMPs) BMC Cell Biol. 2002;3:15. doi: 10.1186/1471-2121-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KW, Nam SY, Toh WH, Dulloo I, Sabapathy K. Multiple stress signals induce p73 beta accumulation. Neoplasia. 2004;6:546–557. doi: 10.1593/neo.04205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim SK, Gopalan G. Antizyme mediates AURKA1P1-dependent degradation of Aurora-A. Oncogene. 2007;26:6593–6603. doi: 10.1038/sj.onc.1210482. [DOI] [PubMed] [Google Scholar]
- Mitchell JL, Chen HJ. Conformational changes in ornithine decarboxylase enable recognition by antizyme. Biochim Biophys Acta. 1990;1037:115–121. doi: 10.1016/0167-4838(90)90109-s. [DOI] [PubMed] [Google Scholar]
- Newman RM, Mosbacher A, Mangold U, Koike C, Diah S, Schmidt M, Finley D, Zetter BR. Antizyme targets cyclin DI for degradation. A novel mechanism for cell growth repression. J Biol Chem. 2004;279:41504–41511. doi: 10.1074/jbc.M407349200. [DOI] [PubMed] [Google Scholar]
- Packham G, Cleveland JL. Ornithine decarboxylase is a mediator of c-Myc induced apoptosis. Mol Cell Biol. 1994;14:5741–5747. doi: 10.1128/mcb.14.9.5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE, McCann PP. Polyamine metabolism and function. Am J Physiol. 1982;243:C212–C221. doi: 10.1152/ajpcell.1982.243.5.C212. [DOI] [PubMed] [Google Scholar]
- Pegg AE, McGovern KA, Weist L. Decarboxylation of alpha-difluoromethylamine by ornithine decarboxylase. Biochem J. 1987;241:305–307. doi: 10.1042/bj2410305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg AE, Feith DJ, Fong LY, Coleman CS, O’Brien TG, Shantz LM. Transgenic mouse models for studies of the role of polyamines in normal, hypertropic and neoplastic growth. Biochem Soc. Trans. 2003;31:356–360. doi: 10.1042/bst0310356. [DOI] [PubMed] [Google Scholar]
- Pegg AE. Regulation of ornithine decarboxylase. J Biol Chem. 2006;281:14529–14532. doi: 10.1074/jbc.R500031200. [DOI] [PubMed] [Google Scholar]
- Pouline R, Pelletier G, Pegg AE. Induction of apoptosis by excessive polyamine accumulation in ornithine decarboxylase-overproducing L1210 cells. Biochem J. 1995;311:723–727. doi: 10.1042/bj3110723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaroni A, Wands J, Trelstad RL, Isselbacher KJ. Epithelioid cell culture from rat small intestine. Characterization by morphologic and immunologic criteria. J Cell Biol. 1979;80:248–265. doi: 10.1083/jcb.80.2.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray RM, Viar MJ, Patel TB, Johnson LR. Interaction of asparagine and EGF in the regulation of ornithine decarboxylase in IEC-6 cells. Am J Physiol. 1999;276:G773–G780. doi: 10.1152/ajpgi.1999.276.3.G773. [DOI] [PubMed] [Google Scholar]
- Ray RM, Viar MJ, Yuan Q, Johnson LR. Polyamine depletion delays apoptosis of rat intestinal epithelial cells. Am J Physiol. 2000;278:C400–C489. doi: 10.1152/ajpcell.2000.278.3.C480. [DOI] [PubMed] [Google Scholar]
- Ray RM, Viar MJ, Johnson LR. Amino acids regulate the expression of antizyme-1 to modulate ornithine decarboxylase activity. J Biol Chem. 2012;287:3674–3690. doi: 10.1074/jbc.M111.232561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinehart CA, Jr, Viceps-Madore D, Fong WF, Ortiz JG, Canellakis ES. The effect of transport system A and N amino acids and nerve and epidermal growth factors on the induction of ornithine decarboxylase activity. J Cell Physiol. 1985;123:435–441. doi: 10.1002/jcp.1041230321. [DOI] [PubMed] [Google Scholar]
- Rinehart CA, Jr, Canellakis ES. Induction of ornithine decarboxylase activity by insulin and growth factors is mediated by amino acids. Proc Natl Acad Sci USA. 1985;82:4365–4368. doi: 10.1073/pnas.82.13.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell DH. Ornithine decarboxylase: A key regulatory enzyme in normal and neoplastic growth. Drug Met Rev. 1985;16:1–88. doi: 10.3109/03602538508991430. [DOI] [PubMed] [Google Scholar]
- Ryan KK, Seeley RJ. Food as a hormone. Science. 2013;339:918–919. doi: 10.1126/science.1234062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Kim DH, Guertin DA, Latek RR, Erdjument-Bromage H, Tempst P, Sabitini DM. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr Biol. 2004;14:1296–1302. doi: 10.1016/j.cub.2004.06.054. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by Rictor mTOR complex. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- Suzuki J, Murakami Y, Samejima K, Ontani KK, Oka T. Antizyme is necessary for conversion of pancreatic tumor cells into glucagon-producing differentiated cells. Endocr Relat Cancer. 2009;16:649–659. doi: 10.1677/ERC-09-0004. [DOI] [PubMed] [Google Scholar]
- Tato I, Bartons R, Ventura F, Rosa JL. Amino acids activate mammalian target of rapamycin complex 2 (mTORC2) via P13K/Akt signaling. J Biol Chem. 2011;272:26457–26463. doi: 10.1074/jbc.M110.166991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias KE, Kahana C. Exposure to ornithine results in excessive accumulation of putrescine and apoptotic cell death in ornithine decarboxylase overproducing mouse myeloma cells. Cell Growth Differ. 1995;10:1279–1285. [PubMed] [Google Scholar]
- Viceps-Madore D, Chen KY, Tsou HR, Canellakis ES. Studies on the role of protein synthesis and of sodium on the regulation of ornithine decarboxylase activity. Biochim Biophys Acta. 1982;717:305–315. doi: 10.1016/0304-4165(82)90184-2. [DOI] [PubMed] [Google Scholar]
- Wang X, Proud CG. The mTOR pathway in the control of protein synthesis. Physiology. 2006;21:362–369. doi: 10.1152/physiol.00024.2006. [DOI] [PubMed] [Google Scholar]
- Wang J-Y, Johnson LR. Role of ornithine decarboxylase in the repair process of gastric mucosal stress ulcers. Am J Physiol. 1990;258:G78–G85. doi: 10.1152/ajpgi.1990.258.1.G78. [DOI] [PubMed] [Google Scholar]
- Wang J-Y, Johnson LR. Polyamines and ornithine decarboxylase during repair of duodenal mucosa after stress in rats. Gastroenterology. 1991a;100:333–343. doi: 10.1016/0016-5085(91)90200-5. [DOI] [PubMed] [Google Scholar]
- Woo RA, Poon RY. Cyclin-dependent kinases and S phase control in mammalian cells. Cell Cycle. 2003;2:316–324. [PubMed] [Google Scholar]
- Yoshida S, Leskiw MJ, Schulter MD, et al. Effect of total parenteral nutrition, systemic sepsis, and glutamine on gut mucosa in rats. Am J Physiol. 1992;263:E368–E373. doi: 10.1152/ajpendo.1992.263.2.E368. [DOI] [PubMed] [Google Scholar]
- Zimmerman BJ, McCormack SA, Israel M, Johnson LR. Polyamine-mediated cell migration and growth: structural requirement for diamine analogues to substitute for putrescine. Cell Pharmacol. 1995;2:109–113. [Google Scholar]
- Zoncu R, Efeyan A, Sabitini DM. From growth signal integration to cancer, diabetes and aging. Nature Rev: Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]