Abstract
The claudin family of proteins are integral components of tight junctions and are responsible for determining the ion specificity and permeability of paracellular transport within epithelial and endothelial cell layers. Several members of the claudin family have been shown to be important during embryonic development and morphogenesis. However, detailed embryonic expression patterns have been described for only a few claudins. Here, we provide a phylogenetic analysis of the chicken claudins and a comprehensive analysis of their mRNA expression profiles. We found that claudin family members exhibit both overlapping and unique expression patterns throughout development. Especially striking were the distinct expression boundaries observed between neural and non-neural ectoderm, as well as within ectodermal derivatives. Claudins were also expressed in endodermally-derived tissues, including the anterior intestinal portal, pharynx, lung and pancreas and in mesodermally derived tissues such as the kidney, gonad and heart. The overlapping zones of claudin expression observed in the chick embryo may confer distinct domains of ion permeability within the early epiblast and in epithelial, mesodermal and endothelial derivatives that may ultimately influence embryonic patterning and morphogenesis during development.
Keywords: claudin, chick development, boundaries, tight junction, expression
Introduction
Tight junctions are required to maintain the integrity and function of epithelial and endothelial cell barriers, where they have two well-established roles in the cell. First, they act to restrict the movement of proteins between the apical and basolateral membrane compartments and, second, they regulate the passage of ions and small molecules in the paracellular space.1-4 Several proteins are positioned at the tight junction, including scaffolding, transmembrane and cytoplasmic proteins, which modulate the function of the junction. The integral component is the transmembrane claudin protein, which determines the ion specificity and permeability of the tight junction.5,6 The tightness of the paracellular barrier is determined by the combination of and interactions between claudin family members that are present within the cell.7
In vertebrates, over 20 claudin family members have been identified to date.8 All claudins contain four transmembrane domains, two extracellular loops and an intracellular C-terminal domain. The first extracellular loop contains several charged residues that establish a particular claudin’s ability to regulate the passage of charged ions. The second extracellular loop participates in homotypic, and in some cases heterotypic, interactions with other claudin family members.9 The greatest sequence heterogeneity between family members is within their cytoplasmic C-termini, which contain phosphorylation sites that regulate claudin localization and function and a PDZ-binding domain that interacts with scaffolding proteins that bridge the tight junction to the actin cytoskeleton.10-12
Recent studies investigating the role of claudins in morphogenesis and embryonic development have revealed that claudins are critical regulators of epithelial integrity during embryogenesis.13-16 For example, Claudin-4 and -6 are required in the mouse blastocyst to maintain the hydrostatic pressure that gives the blastocyst its shape,17 and Claudin E is required for epiboly movements during gastrulation in zebrafish.18 In addition, altering expression of the Xenopus claudin Xcla, or chicken Claudin-1, during gastrulation, randomizes the direction of heart-looping, the first conserved morphological sign of left-right patterning in vertebrates. However, in some instances targeted gene deletion of individual family members in the mouse have not yielded any detectable phenotype19 or resulted in only very restricted postnatal phenotypes,20-22 while overexpression of some claudins have severe effects. For example, the Claudin-6 knockout mice are viable and fertile,19 whereas overexpressing Claudin-6 in its endogenous location in the suprabasal layer of the epidermis leads to a skin barrier defect that is lethal within 48 h of birth.23 These gain- and loss-of-function studies suggest that some claudins are functionally redundant during the early stages of embryonic development, and the balance of claudin gene expression is critical. In order to explore these questions of redundancy and address this possibility it is essential to know the temporal and spatial expression patterns of the claudin family in the embryo.
We are using the chick embryo model system to explore the roles of claudins during early morphogenetic events in the embryo. The chick embryo is a powerful animal model that is amenable for gain-and loss-of-function studies to manipulate the expression of individual or multiple family members to investigate their roles in embryogenesis. In order to determine which family members should be studied in parallel it is essential to know where and when they are expressed in the developing embryo. Therefore, we performed a comprehensive temporal and spatial expression analysis of the claudin family in chick embryos. We have previously published detailed expression patterns of Claudin-1 and -5.24,25 Here, we present the expression patterns for 14 chick claudin family members during gastrulation, neurulation and the early stages of organogenesis. We observed that claudins are expressed in both unique and overlapping patterns, often with very distinct expression boundaries within a histologically uniform epithelial layer. These data suggest that the combinations of claudins expressed within a specific zone of the epiblast or surface ectoderm could demarcate regions with differing paracellular transport properties and ultimately influence the patterning of underlying mesodermal tissue.
Results and Discussion
The claudin gene family in the chicken
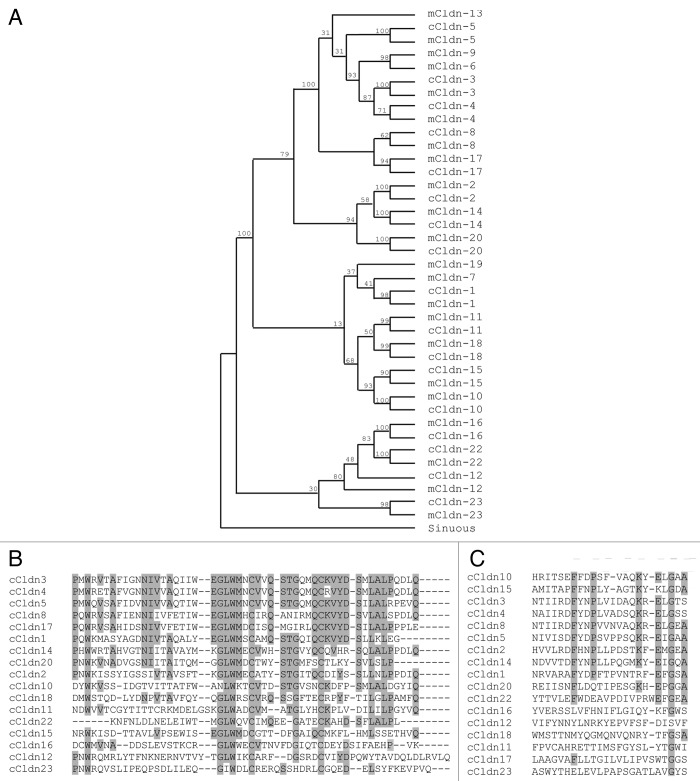
Seventeen claudin family members are annotated in the most recent build of the chicken genome (NCBI Gallus_gallus-4.0, GCA_000002315.2). To examine the relationship between these chicken claudins and their mouse orthologs, we aligned amino acid sequences and performed phylogenetic analyses using the Drosophila claudin-like molecule Sinuous (Fig. 1A) or C. elegans vab-9 (data not shown) as independent outgroups. A phylogenetic analysis that included the human claudin orthologs was performed in parallel (data not shown). All analyses gave the same clustering of orthologs, although the bootstrapping values were slightly higher when Sinuous was used as the outgroup. Each chicken claudin clustered more closely with its mouse or human ortholog than with other claudin family members. The relationship between individual chicken claudin family members determined in our phylogenetic analysis agrees with that shown for Takifugu rubripes, zebrafish, mouse and human claudins.26
Figure 1. Analysis of the relationship between chick claudins. (A) Phylogenetic tree of chick (c) claudin proteins compared with their mouse (m) claudin orthologs. Bootstrapping values are indicated on tree nodes. The Drosophila claudin-like molecule Sinuous was specified as the outgroup for analysis. (B) Alignment of the first extracellular loop of chick claudins. Grey highlighted residues are conserved in greater than half of the claudins. (C) Alignment of the second extracellular loop of chick claudins. Grey highlighted residues are conserved in greater than half of the claudins. Accession numbers of sequences used are provided in Table 2.
Our analyses demonstrate that claudin orthologs are highly conserved in vertebrate evolution and that the majority of family members were duplicated prior to the divergence of birds and mammals.
The genomic organization of claudins in chicken is similar to what has been observed in humans and mice. Claudins are distributed throughout the genome with the exception of three closely linked pairs: Claudin-3 and -4, Claudin-6 and -9 and Claudin-8 and -17. Claudin-3 and -4 are located 5 kb apart on chicken chromosome 19, and Claudin-8 and -17 are located 3 kb apart on chromosome 1. We noted that the chicken orthologs of Claudin-6 and -9 lie within a syntenic block that has not been annotated in the chicken genomic database.
The permeability properties of claudins have been determined primarily by overexpression studies in cell lines with assessment of the change in transepithelial resistance. Claudins that impart barrier properties are Claudin-1, -4, -5, -6, -8, -9, -11 and -19, whereas Claudin-2, -10, -15 and -16 are thought to create pores, making the junction leakier to ions.27 In our analysis of the chick claudin family, several of the barrier claudins are clustered together: Claudin-1, -4, -5 and -8. The remaining barrier claudins (Claudins-6, -9 and -19) are not annotated in the chick genome. Additionally, pore-forming Claudin-10 and -15 cluster closely together and are almost 36% identical at the amino acid level. Claudin-12 and -23 appear to be the least similar to other members of the family. These claudins are often described as “non-classic” claudins as their sequences show greater variation from other claudins and both lack PDZ-binding motifs in their C-terminal cytoplasmic tails, although the significance of these differences is not understood.
Functionally important charged residues located within the first extracellular loop determine the specific ion permeability properties of a claudin. We aligned these sequences to examine if common motifs were apparent in the chick family of claudins (Fig. 1B). A key motif that is found in the first extracellular loop is W-GLW-C-C. The two cysteine residues in this highly conserved motif are hypothesized to form a disulphide bond that enhances stability of the polymerized claudin molecule.6 The W-GLW-C-C motif is present in all chick claudins with the exception of Claudin-23, which also lacks the C-terminal PDZ-binding domain that is present in all other family members. As a consequence, Claudin-23 has lower identity to other family members and is the most distant member on the phylogenetic tree.
The second extracellular loop is thought to participate in side-to-side interactions between claudins in the same cell and in head-to-head interactions between claudins of an opposing cell. Heterotypic oligomerization is limited to certain combinations of claudin family members and little is known regarding what determines the ability of a particular claudin to participate in these interactions. We aligned the amino acids corresponding to the second extracellular loop of the chick claudins to determine if we could observe any conserved residues or motifs (Fig. 1C). The second extracellular loops clustered into two groups based on sequence homology: Claudin-1, -2, -3, -4, -5, -8, -10, -14 and -15 and Claudin-11, -12, -16, -18, -20, -22 and -23. These groups of claudins correspond to the “classic” and “non-classic” based on the sequence homology of the full-length mouse claudin proteins.28 While Claudin-17 is clustered with the “classic” claudins in the phylogenetic tree of the full length chick claudin proteins, it is an outlier in the second extracellular loop alignment as it clusters with the “non-classic” claudins, indicating that the sequence of the second extracellular loop is less conserved than the rest of the protein sequence.
Expression patterns of claudins during gastrulation and neurulation
To determine which of the claudin family members are expressed during embryonic development, we first performed RT-PCR analyses using poly A+ mRNA from whole gastrulation and neurulation-staged embryos (HH 4–8) (Fig. 2). All 17 members of the claudin family were detected by RT-PCR during these stages. The identity of all products was confirmed by sequencing. Single RT-PCR products were detected for all claudins except Claudin-4, -12, -15 and -16. Subcloning and sequencing of the multiple bands obtained for the RT-PCR reactions of Claudin-4, -12, -15 and -16 revealed that only one of the amplicons encoded the claudin of interest. Additionally, although the relative levels of claudin family members differ, the expression level of individual claudins did not appear to change significantly between HH 4 and HH 8.
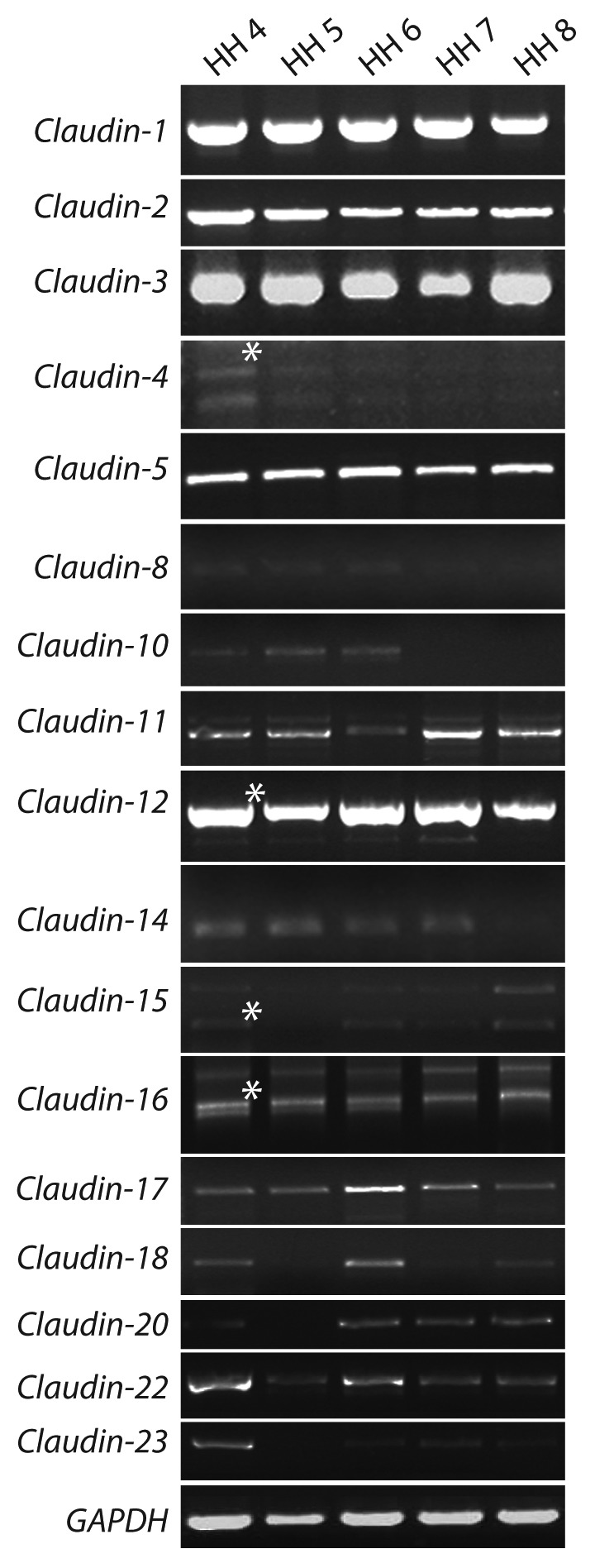
Figure 2. RT-PCR analysis of claudin family gene expression during chicken development. mRNA from gastrulation (HH 4–7) and neurulation (HH 8) were amplified by reverse transcriptase PCR. Multiple products for Claudin-4, -12, -15 and-16 were consistently amplified non-specifically with different oligonucleotide pairs. The correct band is indicated with an asterisk and was confirmed by sequencing. Gapdh was detected at all stages and in all tissues examined.
Next, we performed in situ hybridization analysis to characterize the spatial expression domains of claudins during early development, beginning at gastrulation. The results of these analyses are summarized in Table 1. We detected mRNA expression for all claudins amplified by RT-PCR except for Claudin-20. We have previously published the expression patterns for Claudin-124 and Claudin-525 and so these are discussed but not reported in detail here. In describing the expression patterns, we have primarily focused on tissues where distinct claudin expression domain are observed and those claudins that are most highly expressed. In contrast, several claudin family members were expressed at quite low levels and did not show significant variation in the different epithelial layers. We have included images of these embryos during gastrulation and neurulation, and their expression patterns are summarized in Table 1.
Table 1. Summary of Claudin expression patterns.
| 1* | 2 | 3 | 4 | 5* | 8 | 10 | 11 | 12 | 14 | 15 | 16 | 17 | 18 | 22 | 23 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gastrulation (HH4 - HH7) | ||||||||||||||||
| Epiblast | ++ | + | + | + | - | + | + | + | + | + | + | + | + | + | + | + |
| Mesoderm | - | + | - | + | - | - | + | - | - | - | + | - | + | - | - | + |
| Endoderm | + | + | - | + | - | + | + | + | - | + | + | + | + | + | - | + |
| Hensen's node | - | + | - | + | - | + | +a | - | - | + | + | + | + | + | - | + |
| Extraembryonic ectoderm | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Neurulation (HH8-HH12) | ||||||||||||||||
| Neural ectoderm | - | + | - | + | - | + | + | - | - | + | + | + | + | + | - | + |
| Non-neural ectoderm | + | + | ++ | + | - | + | ++ | ++ | + | + | + | + | + | + | ++ | + |
| Pharynx | + | + | ++ | ++ | - | + | - | - | - | ++ | + | + | ++ | + | + | + |
| Heart | - | ++ | - | - | - | - | + | - | - | + | - | - | - | - | - | - |
| Somites | - | + | - | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Vasculature | - | - | - | - | ++ | - | - | - | - | - | - | - | - | - | - | - |
| Organogenesis (E3-E10) | ||||||||||||||||
| Eye epithelium | + | +b | + | + | - | + | +b | - | - | + | + | + | + | + | + | - |
| Otic vesicle | +b | - | ++b | + | - | + | + | + | +b | + | +b | + | - | - | - | - |
| Nasal epithelium | + | + | + | - | - | +b | +b | +b | +b | +b | +b | +b | +b | +b | - | +b |
| Limb ectoderm | + | + | + | + | - | + | + | + | + | + | + | + | + | + | + | + |
| Apical ectodermal ridge | + | - | + | + | - | - | - | + | + | - | - | + | - | + | + | - |
| Surface ectoderm | + | + | ++b | + | - | + | +b | +b | +b | + | +b | + | + | + | +b | + |
| Feather buds | + | - | + | + | - | - | - | - | - | - | - | - | - | - | - | - |
| Pancreas | + | - | + | + | - | - | - | + | - | - | - | - | - | - | - | - |
| Lung | + | - | + | - | - | - | + | - | - | - | - | - | - | - | - | - |
| Kidney | + | - | + | + | - | - | ++ | - | - | - | - | - | - | - | - | - |
| Gonad | n/a | - | - | - | - | - | - | ++ | - | - | - | - | - | - | - | - |
| Heart | - | + | - | - | - | - | - | + | - | - | - | + | + | - | - | + |
, the expression patterns of Claudin-1 and Claudin-5 have been published previously; -, absent expression; +, positive expression; ++, robust expression ; +a, asymmetric expression; +b, expression with boundaries
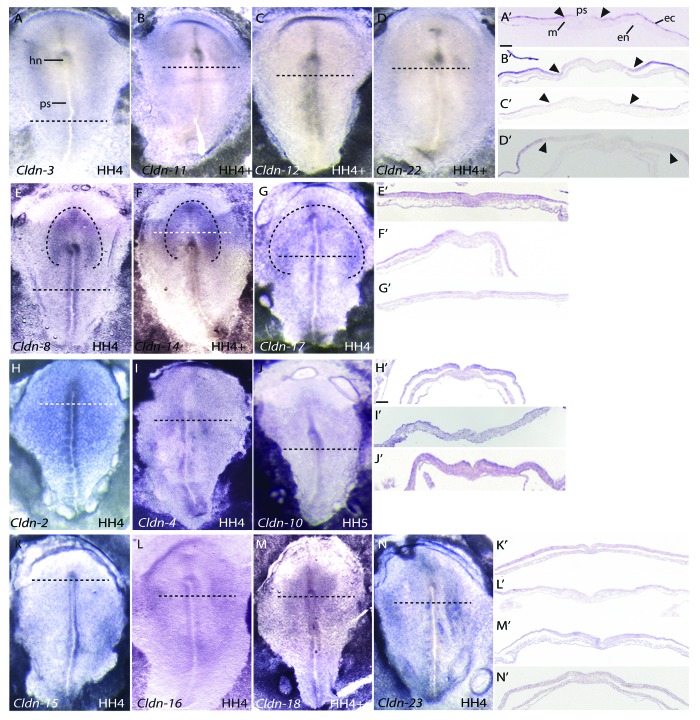
Gastrulation involves a series of highly coordinated cellular movements that generate the three germ layers of the embryo. During this process, cells from the epiblast undergo epithelial to mesenchymal transitions at the primitive streak to allow cells to ingress and give rise to the endoderm and mesoderm. The cells that remain in the epiblast become ectoderm, which gives rise to both the neuroectoderm and surface ectoderm. At HH 4, several claudins were expressed at Hensen’s node, the embryonic “organizer” in the chick that is a critical signaling site for axis formation. Claudin-2, -4, -8, -14, -15, -16, -17, -18 and -23 were bilaterally expressed at Hensen’s node (Fig. 3). Only Claudin-10 showed asymmetric expression on the right side of Hensen’s node (Fig. 3J).
Figure 3. Expression patterns of claudins during chicken gastrulation (HH 4–5) by RNA in situ hybridization. All images are dorsal views. Transverse sections were taken through the embryo at the position indicated by the horizontal dashed line. Dashed lines drawn in D, H and K outline the presumptive neural plate. Arrowheads mark boundaries of claudin expression in the epiblast. Scale bar in A’ represents 0.1 mm. Abbreviations: ec, ectoderm; en, endoderm; hn, Hensen’s node; m, mesoderm; ps, primitive streak.
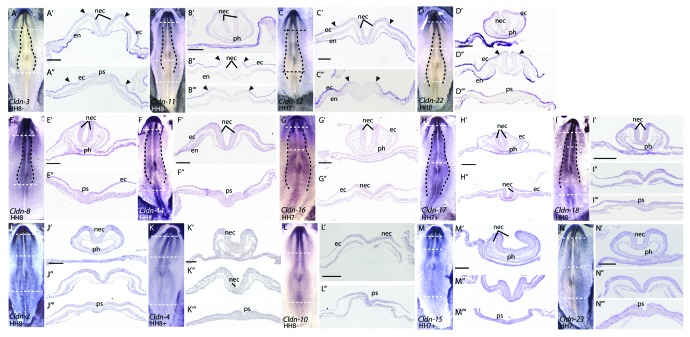
Throughout gastrulation and neurulation, Claudin-2, -4, -10, -15, -16, -18 and -23 were uniformly expressed in the epiblast/embryonic ectoderm (Figs. 3 and 4 H–N), and all appear to be expressed at lower levels than Claudin-1.24 Interestingly, a subset of claudins showed differential expression across the epiblast, corresponding to the boundary between the non-neural ectoderm and the presumptive neural plate cells, which are located in a horseshoe shape at the anterior end of the embryo.29-31 Four claudins, Claudin-3, -11, -12 and -22, were expressed in the lateral non-neural ectoderm and absent from the medial epiblast, which corresponds to the presumptive neural plate (Fig. 3A–D). Three claudins, Claudin-8, -14 and -17, exhibited enriched expression in the presumptive neural plate (Fig. 3E–G). During neurulation (HH 8–12), the neural plate cells thicken to give rise to the neural folds that move together at the midline and fuse to form the closed neural tube. Claudin-3, -11, -12 and -22 were absent from the neural folds (Fig. 4A–D) but were highly expressed in non-neural ectoderm, while Claudin-8, -14 and -17 continued to be more highly expressed in the open neural folds at HH 8 (Fig. 4E, F and H). In addition, expression of Claudin-16 and -18 also became enriched in the open neural folds at HH 8 (Fig. 4G and I). Similarly to the dramatic downregulation of Claudin-1 during neural tube closure, claudin expression was not detected in the closed neural tube (Fig. 5).
Figure 4. Expression patterns of claudins during early neurulation (HH 7–8) by RNA in situ hybridization. All images are dorsal views. Transverse sections were taken through the embryos at the position indicated by the horizontal dashed line. Dashed lines drawn in B, D, F, G, H, J, K, L and M outline the neural folds. Arrowheads mark boundaries of claudin expression in the epiblast. Scale bar represents 0.1 mm. Abbreviations: ec, ectoderm; en, endoderm; nec, neural ectoderm; ph, pharynx; ps, primitive streak.
Figure 5. Expression patterns of claudins during late neurulation (HH12–14) by RNA in situ hybridization. All whole mount views are of the ventral surface. Transverse sections through the embryos are taken from the position indicated by the dashed line. Asterisks indicate trapped antisense riboprobe. Scale bars represent 0.1 mm. Abbreviations: aip, anterior intestinal portal; ec, ectoderm; en, endoderm; g, gut; h, heart; nf, neural folds; nt, neural tube; ph, pharynx; ps, primitive streak; s, somite; tb, tail bud.
Expression of claudins in the surface ectoderm
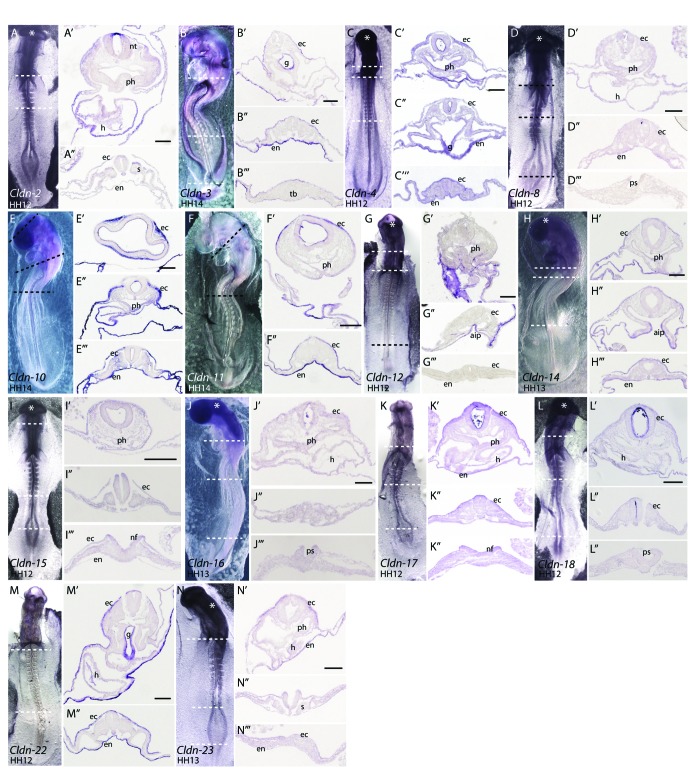
We next examined the expression pattern of each claudin between HH 12 and HH 14. A representative embryo from one of these stages for each claudin is shown in Figure 5. Sections through these embryos showed that Claudin-2, -3, -4, -10, -11, -14, 17 and -22 were most highly expressed in the surface ectoderm at these stages, while the remaining chicken claudin family members were expressed at low levels (Fig. 5).
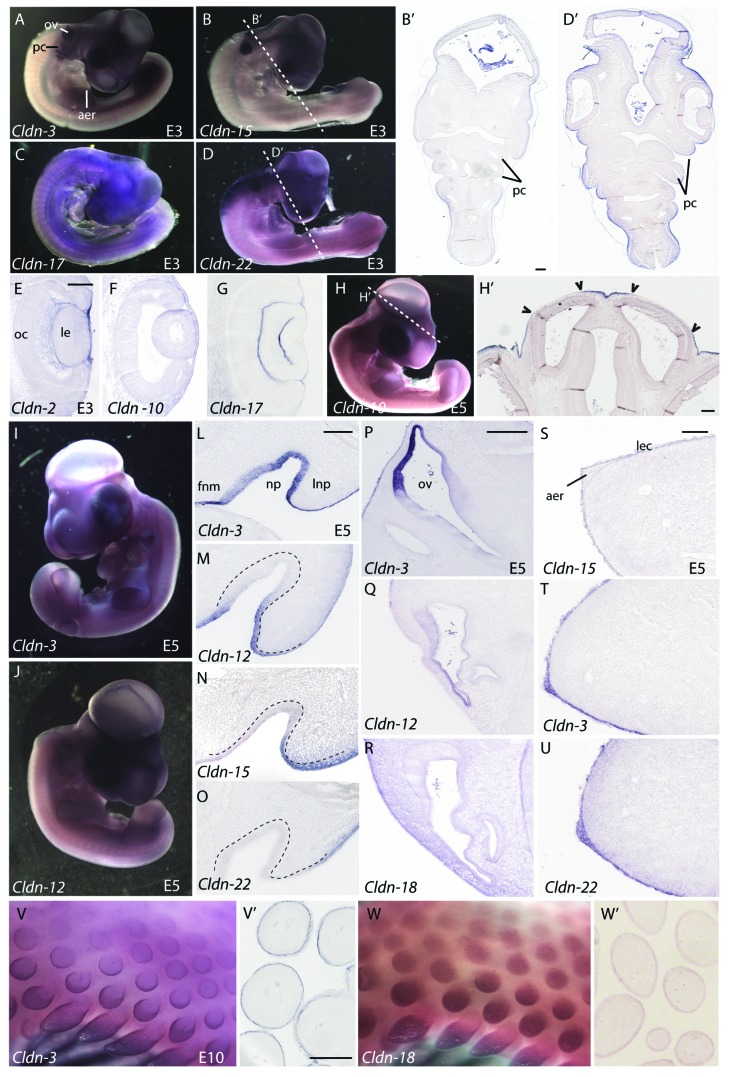
At embryonic day 3 (E3), Claudin-8, -14, -16, -18 and -23 were expressed evenly across the surface ectoderm and continued to be expressed at low levels at later stages examined (Fig. 5). In contrast, expression of Claudin-2, -3, -4, -10, -11, -12, -15, -17 and -22 was more restricted in the surface ectoderm; discrete claudin-specific “on-off” expression boundaries were observed. Some examples of these boundaries are described below. In the head region, all of these claudins showed reduced expression in the pharyngeal clefts and in the ectoderm overlying the anterior neural tube. In contrast, most claudins were expressed at higher levels in the surface ectoderm overlying the neural tube at more posterior regions of the embryo. Representative sections illustrating these boundaries in embryos analyzed for Claudin-15 and -22 expression are shown in Figure 6B,B’ and D,D’, respectively. The majority of claudins were expressed uniformly in the ectoderm overlying the lens, which is derived from an ectodermal placode; a representative section is shown for Claudin-10 (Fig. 6F). However, unique boundaries of expression in this region were observed for three claudins: Claudin-2 was highly expressed in the ectoderm overlying the lens (Fig. 6E), while Claudin-17 and -22 were absent (Fig. 6G and data not shown).
Figure 6. Expression of claudins in ectodermal derivatives. (A–D) Expression of claudins in E3 embryos. Whole mount views of E3 embryos showing expression of Claudin-3 (A), Claudin-15(B), Claudin-17(C) and Claudin-22(D). (B’, D’) Transverse sections taken at the level of the dashed lines indicated in B and D indicate boundaries of claudin expression observed in the surface ectoderm. (E–G) Claudin expression patterns in the eye. Sections through the head of Claudin-2 (E), Claudin-10(F) and Claudin-17(G) illustrate claudin boundaries in the surface ectoderm overlying the developing eye of E3 embryos. (H, H’) Expression of Claudin-10 in a whole-mount view and transverse section through the head at the level of the dashed line. Arrowheads indicate boundaries of expression in the surface ectoderm overlying brain ventricles. (I, J) Whole mount views of E5 embryos hybridized to a Claudin-3 or Claudin-12 riboprobe. (L–O) Claudin expression in the nasal pit of a E5 embryo. Representative examples of four expression patterns observed in the nasal pit. Dashed lines outline the position of the nasal epithelium. (P–R) Expression patterns of claudins the otic vesicle. (S–U) Claudin expression in the apical ectodermal ridge (AER). Claudin-15 is a representative example showing expression in the limb ectoderm, but no AER expression (S) as compared with Claudin-3 and -22, two examples of claudins highly expressed in the limb ectoderm and AER (T, U). (V, W) Expression of Claudin-3 and -18 in the feather buds of E10 embryos. Sections through the skin and feathers are shown in V’ and W’. All scale bars represent 0.1 mm. Abbreviations: aer, apical ectodermal ridge; fnm, frontonasal mass; le, lens; lec, limb ectoderm; lnp, lateral nasal process; np, nasal pit; oc, optic cup; ov, otic vesicle; pc, pharyngeal clefts.
Claudin-10 expression in the head ectoderm was distinct from other family members. At HH 14 (embryonic day 2, E2), Claudin-10 was absent from the ectoderm directly overlying the closed neural tube throughout most of the body (Fig. 5E”–E”’). At the anterior end of the embryo, sharp boundaries of Claudin-10 expression were observed over the brain ventricles and closed neural tube (Fig. 5E’). These boundaries remained through E5 (Fig. 6H and H’). Interestingly, Claudin-10 expression was absent from the surface ectoderm that is in direct contact with the neuroectoderm and present in the surface ectoderm that was separated from the neuroectoderm by a layer of mesoderm (Fig. 5E’ and 6H’).
Distinct expression boundaries were also observed in the ectoderm covering the craniofacial regions. Examples of the four different patterns that were observed in the nasal pit are shown (Fig. 6L–O). Claudin-2 and -3 were expressed uniformly across the epithelium outside and lining the nasal pit (Fig. 6L). Claudin-10, -11, -12, -16 and -18 were expressed in the frontonasal mass epithelium and lateral nasal prominence, but were absent in the epithelium at the base of the pit (Fig. 6M). Claudin-15 and -23 were absent from the frontonasal mass epithelium and nasal pit, but were expressed in the lateral nasal prominence (Fig. 6N). Finally, Claudin-4 and 22 were expressed at high levels around the lateral nasal prominence but absent from the frontonasal process epithelium and nasal pit (Fig. 6O).
In the otic vesicle at E5, most claudins were expressed at low levels throughout the epithelium (Claudin-2, -4, -8, -10, -11, -14, or -16) or absent (Claudin-17, -18, -22 and -23), as shown for Claudin-18 (Fig. 6R). However, distinct boundaries of gene expression were observed for Claudin-3, -12 and -15 in the region of the otic epithelium that is in closest proximity to the surface ectoderm (Fig. 6P and Q), similar to the pattern that we have previously reported for Claudin-1.24
Another region of specialized ectoderm that exhibited differential expression of claudin family members was the apical ectodermal ridge (AER), an ectodermal thickening at the distal tip of the limb bud that is a critical signaling center for limb development. Claudin-3, -4, -11 and -12 were expressed in the AER beginning at E3 (Fig. 6A and T and data not shown) and Claudin-16, -18 and -22 transcripts were detected in the AER beginning at E5 (Fig. 6U and data not shown). The remaining claudins were never detected in the AER (Fig. 6S and data not shown), despite the fact that all claudins were expressed throughout the limb ectoderm (Fig. 6S–U).
Feather buds form from the surface ectoderm beginning at day 6.5. We have previously shown that Claudin-1 is expressed in the periderm of the developing feather bud, initiating in a few cells at the tip of the bud and elongating down the sides of the bud during feather outgrowth.24 Claudin-3, -4 and -18 were highly expressed in the forming feather buds and showed two different patterns of expression (Fig. 6V and W). Claudin-3 and Claudin-4 were expressed most strongly in an outer ring around the developing feather bud (Fig. 6V and V’, data not shown). In contrast, expression of Claudin-18 (Fig. 6W) was uniform throughout the feather bud, similar to Claudin-1. Sections of these feather buds revealed that Claudin-3, -4 and -18 expression was restricted to the outer periderm layer (Fig. 6V’ and W’).
Expression of claudins in endoderm-derived tissues
Although none of the claudins were expressed in the newly formed endoderm at gastrulation (Fig. 3), most family members were expressed in the endoderm beginning at neurulation. At HH 7–8, Claudin-3, -12 and -22 were mostly highly expressed in the endoderm (Fig. 4A, C and D) and all three were expressed more strongly at the anterior end of the embryo. Claudin-22 expression was highest in the lateral endoderm (Fig. 4D”), while Claudin-3, - 8, -11, -12, -16, -17, -18 and -22 had higher levels of expression in the endoderm underlying the head folds (Fig. 4A–E and G–I) as compared with the posterior end of the embryo. In contrast, Claudin-2, -4 -10, -14, -15, -17 and -23 were detected uniformly in the endoderm along the anterior-posterior axis (Fig. 4F’, H’ and J–N’).
A primary derivative of the endoderm is the gut. At HH 8, Claudin-2, -3, -8, -10, -14, -17, -18, -22 and -23 transcripts were detected in the ventral pharynx (Fig. 4A’, D’-F’, H’-J’, L’ and N’). However, by HH 12–14, only Claudin-10, -14 and -22 were expressed in the pharynx (Fig. 5E,” H’ and M’). By HH 14, there was a marked increase in the level of Claudin-10 and -11 in the endoderm along the entire anterior-posterior axis. Claudin-3, -4, -11, -12, -14 and -22 were expressed in the anterior intestinal portal (AIP) (Fig. 5B, C, F, G, H and M). A strong boundary of Claudin-14 expression was observed in the AIP (Fig. 5H”), at the level at which the two points will fuse to form a closed gut tube. Only Claudin-3, -4 and 22 were observed in the closed gut tube (Fig. 5B’, C’ and M’ and Fig. 6J’).
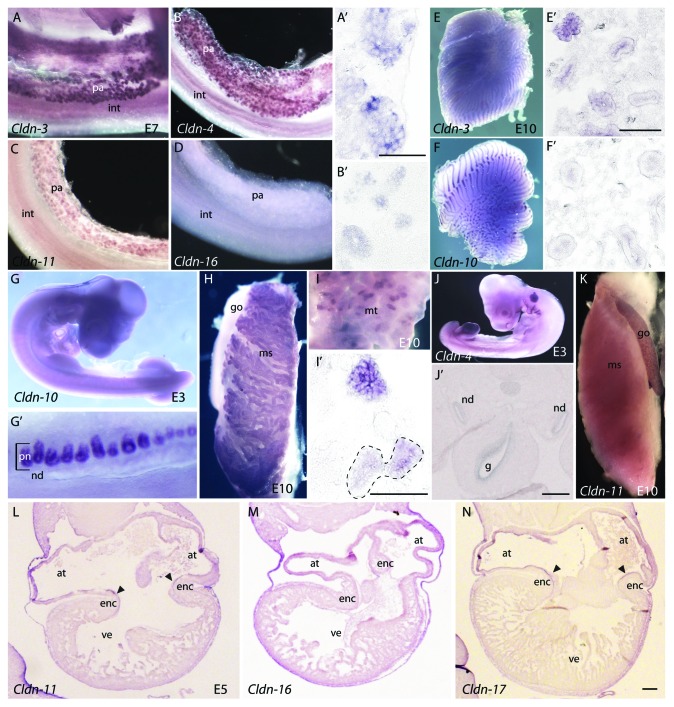
We also examined the endodermally-derived organs at E5 and E10, including the pancreas and lungs, for claudin expression. The pancreas is formed from two buds that develop from the foregut. These buds fuse to form a single organ by E7. Claudin-3 and -4 were detected in primitive buds at E5 (data not shown) and were highly expressed at E7 in the epithelial cells of the pancreatic ducts (Fig. 7A and B). The level of expression is equivalent in all positive cells (Fig. 7A’ and B’). Faint expression of Claudin-11 was observed in the pancreas starting at E7 (Fig. 7C). No other claudins were expressed in the pancreas (Fig. 7D and data not shown).
Figure 7. Claudin mRNA expression patterns in organs derived from the endoderm and mesoderm. (A–C)Claudin-3, -4 and -11 are expressed in the pancreas. Shown for comparison is Claudin-16, which is not expressed in the pancreas (D). Sections through the pancreas showing expression of Claudin-3 and -4 in epithelialized ductal cells are shown in A’ and B’. (E, F) Expression of Claudin-3 and -10 in the E10 lung. Frontal sections through the lung show differences in the localization of Claudin-3(E’) and uniform expression of Claudin-10(F’) in the bronchi. (G–J) Expression of claudins in the developing kidney. Expression of Claudin-10 in a E3 embryo (G) and pronephros (G’), in the E10 mesonephros (H) and metanephros (I, I’). Frontal sections through the metanephric kidney show localization of expression to duct cells (outlined in black dashed line). (J) Whole-mount view of the expression of Claudin-4 in a E3 embryo. (J’) Transverse sections through the E3 embryo show expression in the nephric duct. (K) Expression of Claudin-11 in the gonad of a E10 embryo. (L–N) Expression of claudins in the E5 heart. Arrowheads indicate expression in the endocardial cushions. All scale bars represent 0.1 mm. Abbreviations: at, atria; enc, endocardial cushions; g, gut; go, gonad; int, intestine; ms, mesonephros; mt, metanephros; nd, nephric duct; pa, pancreas; pn, pronephric tubules; ve, ventricle.
At E5, no claudins were observed in the esophagus and only faint expression of Claudin-3 was observed in the early lung bronchus (data not shown). The lung bud endoderm subsequently undergoes several rounds of budding and branching. By E7, we observed Claudin-3 and -10 expression in the branched lung (Fig. 7E and F). Frontal sections through the E10 lung showed that Claudin-10 expression was uniform throughout the branching bronchi (Fig. 7F and F’). In contrast, Claudin-3 was preferentially expressed at the bud termini (Fig. 7E and E’) and exhibited variable expression along the length of the lung epithelium. This pattern is similar to what we observed for Claudin-1 expression in the lung.24
Claudin expression in mesodermally-derived tissues
Much of our knowledge about claudins and their physiological role in regulating ion permeability comes from functions described in the adult mammalian kidney, where well-defined boundaries of claudin expression along the nephron render particular segments permeable to different ions.7 Additionally, the functional relevance of these boundaries of claudin expression are reflected in human mutations in claudins that affect resorption of calcium and magnesium ions,32-34 as well as in mouse transgenic models.20,35-37
We examined the expression of claudins during three stages of kidney development: development of the pronephros, mesonephros and metanephros. In addition to Claudin-1, which is expressed in the nephric duct, and Claudin-3, whose expression in the nephric duct and kidney has been described previously,38 the only other claudin detected in the nephric duct was Claudin-4 (Fig. 7J and J’). Claudin-4 expression was more temporally restricted, and it was not detected in the pronephric, mesonephric, or metanephric kidney (data not shown). In contrast, Claudin-10 was not expressed in the nephric duct but was highly expressed in the pronephric tubules at E3 (Fig. 7G and G’) and in the mesonephric and metanephric kidneys at E10 (Fig. 7H and I). Sections through the metanephric kidney show that Claudin-10 expression is restricted to the epithelial cells of the kidney tubules (Fig. 7I’). We did not detect expression of other claudins in the mesonephric or metanephric kidneys by in situ hybridization at the stages we examined.
In addition to giving rise to the kidney, the intermediate mesoderm will also differentiate to form the gonad, which is positioned against the mesonephric kidney. We only detected expression of Claudin-11 in this tissue (Fig. 7K), which was expressed bilaterally in the gonads, but absent from the kidney. Interestingly, Claudin-11 has been implicated in male fertility in mice. The Claudin-11 null mouse line exhibits Sertoli cell dissociation from the basement membrane, resulting in abnormally localized germ cells, increased apoptosis in germ cells and halted spermatogenesis at meiosis.39
The embryonic heart is derived from lateral plate mesoderm tissue, cardiac fields and neural crest cells. We did not detect any claudins in those tissues during gastrulation and early neurulation. However, at E5 we observed expression of Claudin-2, -11, -16, -17 and -23 in the endocardial lining of the atria (Fig. 7L–N, data not shown). With the exception Claudin-16, all of these claudins were also expressed in the forming endocardial cushions.
Experimental Procedures
Phylogenetic analysis of chicken claudins
Alignment of chicken claudin amino acid sequences was performed using ClustalX 2.0.12.41 Multiple and pairwise alignment parameters were kept at default values with the exception of using the PAM series protein weight matrix for both alignment criteria. Sequences were trimmed at the N-terminal end to the first conserved aligned amino acid. Aligned sequences were processed using PHYLIP version 3.69 programs SEQBOOT, PROTDIST, NEIGHBOR and CONSENSE.42 Briefly, 100 bootstrapped data sets were created from the original alignment using SEQBOOT and used in PROTDIST to generate a distance matrix using the Dayhoff PAM matrix method. Distance matrices were then used as randomized input in NEIGHBOR to generate a neighbor-joining tree, and the Drosophila claudin-like molecule Sinuous (Fig. 1A) or C. elegans vab-9 was specified as the outgroup. The final tree was generated using CONSENSE. Unless specified, default parameters were used in all programs described. Accession numbers of claudin protein sequences used for analysis are described in Table 2.
Table 2. Accession numbers of claudin amino acid sequences used for phylogenetic analysis.
| Gene | Accession number |
|---|---|
| cClaudin-1 | NP_001013629 |
| cClaudin-2 | XP_420271 |
| cClaudin-3 | NP_989533 |
| cClaudin-4 | XP_003642430 |
| cClaudin-5 | NP_989532 |
| cClaudin-8 | XP_425544 |
| cClaudin-10 | XP_001232227.2 |
| cClaudin-11 | ENSGALP00000015219 |
| cClaudin-12 | XP_001234158 |
| cClaudin-14 | XP_425552 |
| cClaudin-15 | ENSGALP00000003668 |
| cClaudin-16 | ENSGALP00000011742 |
| cClaudin-17 | XP_425543 |
| cClaudin-18 | XP_426691 |
| cClaudin-20 | XP_001232003 |
| cClaudin-22 | XP_425804 |
| cClaudin-23 | ENSGALP00000031534 |
| mClaudin-1 | NM_016674 |
| mClaudin-2 | NP_057884 |
| mClaudin-3 | NM_009902 |
| mClaudin-4 | NP_034033 |
| mClaudin-5 | AAH83341 |
| mClaudin-6 | AAH50138 |
| mClaudin-7 | AAH50007 |
| mClaudin-8 | AAH03868 |
| mClaudin-9 | AAH58186 |
| mClaudin-10 | AAH29019 |
| mClaudin-11 | NM_008770 |
| mClaudin-12 | AAH247057 |
| mClaudin-13 | AAN03863 |
| mClaudin-14 | AAG0051 |
| mClaudin-15 | NM_021719 |
| mClaudin-16 | NP_444471 |
| mClaudin-17 | NM_181490 |
| mClaudin-18 | NP_062789 |
| mClaudin-19 | AAM12735 |
| mClaudin-20 | NP_001095030 |
| mClaudin-22 | NM_029383 |
| mClaudin-23 | AAH85262 |
| Sinuous | NP_647971 |
| vab-9 | NM_063435 |
RT-PCR and cDNA cloning of chicken claudins
Fertilized eggs were obtained from Couvoir Simetin Hatchery (Mirabel) and incubated at 38.5°C until the appropriate age was reached. Embryos were staged using Hamburger and Hamilton (HH) staging criteria.43 Embryos were handled according to the Canadian Council on Animal Care guidelines. Whole HH 4–8 embryos (excluding extra-embryonic membranes) were dissected and flash frozen in liquid nitrogen. Poly A+ mRNA was isolated using the MicroFast Track 2.0 kit (Invitrogen) and RT-PCR was performed using the QIAGEN One-Step RT-PCR kit.. Oligonucleotide sequences and fragment sizes are described in Table 3. Full length or partial sequences were cloned into the pSC-A vector using the Strataclone kit (Stratagene) and clones were sequenced to confirm their identity using the BigDye Terminator v3.1 Cycle Sequencing kit (Applied Biosystems).
Table 3. Oligonucleotide primers used for RT-PCR.
| Gene | Gene ID | Primer Sequence | Product Size |
|---|---|---|---|
| cClaudin-2 | GI:118089520 | Forward 5′-ccatggtctctatgggactcc-3′ | 684 |
| Reverse 5′-gcttctacacgtatcccgtc-3′ | |||
| cClaudin-3 | GI:46049088 | Forward 5′-gcccatgtggagggtgac-3′ | 376 |
| Reverse 5′-caccagcgggttgtagaaat-3′ | |||
| cClaudin-4 | GI: 363741048 | Forward 5′-cgggatccgatggcctccatggggct-3′ | 622 |
| Reverse 5′-gtggaattccttacacgtagttgctg-3′ | |||
| cClaudin-8 | GI:50730032 | Forward 5′-atgatcagtggtgcgtgccaaattgc-3′ | 675 |
| Reverse 5′-ctagacgtactgactcttagagtagg-3′ | |||
| cClaudin-10 | GI:118084678 | Forward 5′-catggcgagcacgtcggcggag-3′ | 687 |
| Reverse 5′-ttaaacgtaagcgttcttgtc-3′ | |||
| cClaudin-11 | GI:118095299 | Forward 5′-catgggtggccacctgcctgca-3′ | 608 |
| Reverse 5′-ttcagacgtgggcgctctta-3′ | |||
| cClaudin-12 | GI:118085731 | Forward 5′-atgggctgcagggatgttcat-3′ | 738 |
| Reverse 5′-cttaagatgtgtgtgtcacaaca-3′ | |||
| cClaudin-14 | GI:118083818 | Forward 5′-atggcaagcacggccgttcagttac-3′ | 708 |
| Reverse 5′-tcacacatagtcattcaatctgtagc-3′ | |||
| cClaudin-15 | GI:50751942 | Forward 5′-atggcttcatcttctctacag-3′ | 675 |
| Reverse 5′-ttagatgtactccttg-3′ | |||
| cClaudin-16 | GI:50752418 | Forward 5′-atgccactggatgtaccaacacc-3′ | 744 |
| Reverse 5′-tcacactcttgtatccacggc-3′ | |||
| cClaudin-17 | GI:118083832 | Forward 5′-catggcttgctgtgcattac-3′ | 669 |
| Reverse 5′-gccttctatacgtatgcctgc-3′ | |||
| cClaudin-18 | GI:118094873 | Forward 5′-atgtccacgaccctctggc-3′ | 780 |
| Reverse 5′-ctagacgtactgactcttagagtagg-3′ | |||
| cClaudin-20 | GI:118088400 | Forward 5′-ccgaactggaaggtaaatgc-3′ | 522 |
| Reverse 5′-catgctccttgctgctttttg-3′ | |||
| cClaudin-22 | GI:118101949 | Forward 5′-atgctactggcgctcttcgg-3′ | 654 |
| Reverse 5′-cagattaccaggtcagcg-3′ | |||
| cClaudin-23 | GI: 50746852 | Forward 5′-atgcggacaccggccgtgat-3′ | 702 |
| Reverse 5′-tcatcttggggtctgg-3′ |
Whole-mount in situ hybridization
Antisense digoxygenin-labeled riboprobes for chicken claudins were made from linearized cDNA constructs and transcribed using the appropriate RNA polymerase according to standard protocols. Whole-mount in situ hybridization was performed as previously described.44 Proteinase K (10μg/ml) treatment was performed for 2 min for embryos younger than embryonic day 2, 5 min for E3–4 embryos, 10 min for E5–6 embryos and 20 min for E10 embryos. Embryos were photographed under a Leica MZFLIII dissecting microscope with SPOT Advanced digital image capture software. Prior to sectioning, embryos were either dehydrated through graded ethanol washes, cleared in xylene and embedded in paraffin or were cryoprotected in sucrose and embedded in OCT (Tissue-Tek). Blocks containing whole embryos were sectioned at 10 μm, and limbs were sectioned at 7 μm. Paraffin sections were collected on Fisherbrand Superfrost Plus microscope slides, dried overnight, rinsed twice in xylene to remove paraffin and coverslipped with Permount. Cryosections were air-dried for a minimum of two hours, washed in PBS and coverslipped using glycerol gelatin (Sigma). Images were captured using a Zeiss Axiophot compound microscope and AxioCamMRc camera with Axiovision v4.7.1.0 software.
Summary
Our analysis of the claudin family of integral tight junction proteins revealed that chicken claudins cluster more closely with their mouse and human orthologs than with other claudin family members. We found that the genomic organization of two claudin pairs is conserved between chicken and mouse, suggesting these gene duplication events occurred early during chordate evolution. Despite the observation that the chicken genome has an increased rate of loss compared with other genomes,40 the majority of the claudin family is present in the chicken suggesting that these genes are part of the “core” vertebrate genome. Spatial and temporal investigation of the mRNA expression patterns of the claudins investigated in this manuscript and those that we have previously published revealed that many claudins are co-expressed, suggesting that their transcription may be co-regulated by a similar set of transcription factors. We also identified tissues that express only a subset of claudins and examined tissues that have boundaries of claudin expression, including epithelial derivatives in the craniofacial region, feathers and internal organs. These results suggest that a combinatorial code of claudins in a particular tissue may be involved with the establishment of microenvironments necessary for normal development or affect the morphogenesis or patterning of an apparently uniform epithelium at these early stages of embryogenesis.
Acknowledgments
We would like to thank members of the Ryan lab, K. Dewar, I. Gupta and L. Jerome-Majewska for helpful discussions and comments. M.M.C. is the recipient of a doctoral studentship from the Fonds de la Recherche en Santé du Quebec (FRSQ) and A.I.B. is the recipient of a studentship from the Foundation of Stars. This work was funded by a Natural Sciences and Engineering Research of Canada Discovery Grant and a Canadian Institutes of Health Research Operating Grant to A.K.R. A.K.R. is a member of the Research Institute of the McGill University Health Centre, which is supported in part by the FRSQ.
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/tissuebarriers/article/24517
References
- 1.Steed E, Balda MS, Matter K. Dynamics and functions of tight junctions. Trends Cell Biol. 2010;20:142–9. doi: 10.1016/j.tcb.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 2.González-Mariscal L, Betanzos A, Nava P, Jaramillo BE. Tight junction proteins. Prog Biophys Mol Biol. 2003;81:1–44. doi: 10.1016/S0079-6107(02)00037-8. [DOI] [PubMed] [Google Scholar]
- 3.Matter K, Balda MS. Signalling to and from tight junctions. Nat Rev Mol Cell Biol. 2003;4:225–36. doi: 10.1038/nrm1055. [DOI] [PubMed] [Google Scholar]
- 4.Turksen K, Troy TC. Barriers built on claudins. J Cell Sci. 2004;117:2435–47. doi: 10.1242/jcs.01235. [DOI] [PubMed] [Google Scholar]
- 5.Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J Cell Biol. 1998;141:1539–50. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol. 2006;68:403–29. doi: 10.1146/annurev.physiol.68.040104.131404. [DOI] [PubMed] [Google Scholar]
- 7.Angelow S, Ahlstrom R, Yu AS. Biology of claudins. Am J Physiol Renal Physiol. 2008;295:F867–76. doi: 10.1152/ajprenal.90264.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lal-Nag M, Morin PJ. The claudins. Genome Biol. 2009;10:235. doi: 10.1186/gb-2009-10-8-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furuse M, Sasaki H, Tsukita S. Manner of interaction of heterogeneous claudin species within and between tight junction strands. J Cell Biol. 1999;147:891–903. doi: 10.1083/jcb.147.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamazaki Y, Itoh M, Sasaki H, Furuse M, Tsukita S. Multi-PDZ domain protein 1 (MUPP1) is concentrated at tight junctions through its possible interaction with claudin-1 and junctional adhesion molecule. J Biol Chem. 2002;277:455–61. doi: 10.1074/jbc.M109005200. [DOI] [PubMed] [Google Scholar]
- 11.Simard A, Di Pietro E, Young CR, Plaza S, Ryan AK. Alterations in heart looping induced by overexpression of the tight junction protein Claudin-1 are dependent on its C-terminal cytoplasmic tail. Mech Dev. 2006;123:210–27. doi: 10.1016/j.mod.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. J Cell Biol. 1999;147:1351–63. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheung ID, Bagnat M, Ma TP, Datta A, Evason K, Moore JC, et al. Regulation of intrahepatic biliary duct morphogenesis by Claudin 15-like b. Dev Biol. 2012;361:68–78. doi: 10.1016/j.ydbio.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta IR, Ryan AK. Claudins: unlocking the code to tight junction function during embryogenesis and in disease. Clin Genet. 2010;77:314–25. doi: 10.1111/j.1399-0004.2010.01397.x. [DOI] [PubMed] [Google Scholar]
- 15.Furuse M. Knockout animals and natural mutations as experimental and diagnostic tool for studying tight junction functions in vivo. Biochim Biophys Acta. 2009;1788:813–9. doi: 10.1016/j.bbamem.2008.07.017. [DOI] [PubMed] [Google Scholar]
- 16.Bagnat M, Cheung ID, Mostov KE, Stainier DY. Genetic control of single lumen formation in the zebrafish gut. Nat Cell Biol. 2007;9:954–60. doi: 10.1038/ncb1621. [DOI] [PubMed] [Google Scholar]
- 17.Moriwaki K, Tsukita S, Furuse M. Tight junctions containing claudin 4 and 6 are essential for blastocyst formation in preimplantation mouse embryos. Dev Biol. 2007;312:509–22. doi: 10.1016/j.ydbio.2007.09.049. [DOI] [PubMed] [Google Scholar]
- 18.Siddiqui M, Sheikh H, Tran C, Bruce AE. The tight junction component Claudin E is required for zebrafish epiboly. Dev Dyn. 2010;239:715–22. doi: 10.1002/dvdy.22172. [DOI] [PubMed] [Google Scholar]
- 19.Anderson WJ, Zhou Q, Alcalde V, Kaneko OF, Blank LJ, Sherwood RI, et al. Genetic targeting of the endoderm with claudin-6CreER. Dev Dyn. 2008;237:504–12. doi: 10.1002/dvdy.21437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyamoto T, Morita K, Takemoto D, Takeuchi K, Kitano Y, Miyakawa T, et al. Tight junctions in Schwann cells of peripheral myelinated axons: a lesson from claudin-19-deficient mice. J Cell Biol. 2005;169:527–38. doi: 10.1083/jcb.200501154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi D, Tamura A, Tanaka H, Yamazaki Y, Watanabe S, Suzuki K, et al. Deficiency of claudin-18 causes paracellular H+ leakage, up-regulation of interleukin-1β, and atrophic gastritis in mice. Gastroenterology. 2012;142:292–304. doi: 10.1053/j.gastro.2011.10.040. [DOI] [PubMed] [Google Scholar]
- 22.Ben-Yosef T, Belyantseva IA, Saunders TL, Hughes ED, Kawamoto K, Van Itallie CM, et al. Claudin 14 knockout mice, a model for autosomal recessive deafness DFNB29, are deaf due to cochlear hair cell degeneration. Hum Mol Genet. 2003;12:2049–61. doi: 10.1093/hmg/ddg210. [DOI] [PubMed] [Google Scholar]
- 23.Troy TC, Turksen K. ES cell differentiation into the hair follicle lineage in vitro. Methods Mol Biol. 2002;185:255–60. doi: 10.1385/1-59259-241-4:255. [DOI] [PubMed] [Google Scholar]
- 24.Simard A, Di Pietro E, Ryan AK. Gene expression pattern of Claudin-1 during chick embryogenesis. Gene Expr Patterns. 2005;5:553–60. doi: 10.1016/j.modgep.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 25.Collins MM, Baumholtz AI, Ryan AK. Claudin-5 expression in the vasculature of the developing chick embryo. Gene Expr Patterns. 2012 doi: 10.1016/j.gep.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Loh YH, Christoffels A, Brenner S, Hunziker W, Venkatesh B. Extensive expansion of the claudin gene family in the teleost fish, Fugu rubripes. Genome Res. 2004;14:1248–57. doi: 10.1101/gr.2400004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou J, Rajagopal M, Yu AS. Claudins and the Kidney. Annu Rev Physiol. 2013;75:479–501. doi: 10.1146/annurev-physiol-030212-183705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krause G, Winkler L, Piehl C, Blasig I, Piontek J, Müller SL. Structure and function of extracellular claudin domains. Ann N Y Acad Sci. 2009;1165:34–43. doi: 10.1111/j.1749-6632.2009.04057.x. [DOI] [PubMed] [Google Scholar]
- 29.Hatada Y, Stern CD. A fate map of the epiblast of the early chick embryo. Development. 1994;120:2879–89. doi: 10.1242/dev.120.10.2879. [DOI] [PubMed] [Google Scholar]
- 30.Chapman SC, Schubert FR, Schoenwolf GC, Lumsden A. Analysis of spatial and temporal gene expression patterns in blastula and gastrula stage chick embryos. Dev Biol. 2002;245:187–99. doi: 10.1006/dbio.2002.0641. [DOI] [PubMed] [Google Scholar]
- 31.Puelles L, Fernández-Garre P, Sánchez-Arrones L, García-Calero E, Rodríguez-Gallardo L. Correlation of a chicken stage 4 neural plate fate map with early gene expression patterns. Brain Res Brain Res Rev. 2005;49:167–78. doi: 10.1016/j.brainresrev.2004.12.036. [DOI] [PubMed] [Google Scholar]
- 32.Simon DB, Lu Y, Choate KA, Velazquez H, Al-Sabban E, Praga M, et al. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science. 1999;285:103–6. doi: 10.1126/science.285.5424.103. [DOI] [PubMed] [Google Scholar]
- 33.Weber S, Hoffmann K, Jeck N, Saar K, Boeswald M, Kuwertz-Broeking E, et al. Familial hypomagnesaemia with hypercalciuria and nephrocalcinosis maps to chromosome 3q27 and is associated with mutations in the PCLN-1 gene. Eur J Hum Genet. 2000;8:414–22. doi: 10.1038/sj.ejhg.5200475. [DOI] [PubMed] [Google Scholar]
- 34.Konrad M, Schaller A, Seelow D, Pandey AV, Waldegger S, Lesslauer A, et al. Mutations in the tight-junction gene claudin 19 (CLDN19) are associated with renal magnesium wasting, renal failure, and severe ocular involvement. Am J Hum Genet. 2006;79:949–57. doi: 10.1086/508617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatum R, Zhang Y, Salleng K, Lu Z, Lin JJ, Lu Q, et al. Renal salt wasting and chronic dehydration in claudin-7-deficient mice. Am J Physiol Renal Physiol. 2010;298:F24–34. doi: 10.1152/ajprenal.00450.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou J, Shan Q, Wang T, Gomes AS, Yan Q, Paul DL, et al. Transgenic RNAi depletion of claudin-16 and the renal handling of magnesium. J Biol Chem. 2007;282:17114–22. doi: 10.1074/jbc.M700632200. [DOI] [PubMed] [Google Scholar]
- 37.Breiderhoff T, Himmerkus N, Stuiver M, Mutig K, Will C, Meij IC, et al. Deletion of claudin-10 (Cldn10) in the thick ascending limb impairs paracellular sodium permeability and leads to hypermagnesemia and nephrocalcinosis. Proc Natl Acad Sci U S A. 2012;109:14241–6. doi: 10.1073/pnas.1203834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haworth KE, El-Hanfy A, Prayag S, Healy C, Dietrich S, Sharpe P. Expression of Claudin-3 during chick development. Gene Expr Patterns. 2005;6:40–4. doi: 10.1016/j.modgep.2005.05.002. [DOI] [PubMed] [Google Scholar]
- 39.Mazaud-Guittot S, Meugnier E, Pesenti S, Wu X, Vidal H, Gow A, et al. Claudin 11 deficiency in mice results in loss of the Sertoli cell epithelial phenotype in the testis. Biol Reprod. 2010;82:202–13. doi: 10.1095/biolreprod.109.078907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Consortium ICGS, International Chicken Genome Sequencing Consortium Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 41.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 42.Felsenstein J. Mathematics vs. Evolution: Mathematical Evolutionary Theory. Science. 1989;246:941–2. doi: 10.1126/science.246.4932.941. [DOI] [PubMed] [Google Scholar]
- 43.Hamburger V, Hamilton HL. A Series of Normal Stages in the Development of the Chick Embryo. J Morphol. 1951;88:49. doi: 10.1002/jmor.1050880104. [DOI] [PubMed] [Google Scholar]
- 44.Nieto MA, Patel K, Wilkinson DG. In situ hybridization analysis of chick embryos in whole mount and tissue sections. Methods Cell Biol. 1996;51:219–35. doi: 10.1016/S0091-679X(08)60630-5. [DOI] [PubMed] [Google Scholar]