Abstract
Alternative processing of precursor mRNAs (pre-mRNAs), including alternative transcription start sites, alternative splicing and alternative polyadenylation, is the major source of protein diversity and plays crucial roles in development, differentiation and diseases in higher eukaryotes. It is estimated from microarray analyses and deep sequencing of mRNAs from synchronized worms that up to 25% of protein-coding genes in Caenorhabditis elegans undergo alternative pre-mRNA processing and that many of them are subject to developmental regulation. Recent progress in visualizing the alternative pre-mRNA processing patterns in living worms with custom-designed fluorescence reporters has enabled genetic analyses of the regulatory mechanisms for alternative processing events of interest in vivo. Expression of the tissue-specific isoforms of actin depolymerising factor (ADF)/cofilin, UNC-60A and UNC-60B, is regulated by a combination of alternative splicing and alternative polyadenylation of pre-mRNA from a single gene unc-60. We recently found that muscle-specific splicing regulators ASD-2 and SUP-12 cooperatively switch the pre-mRNA processing patterns of the unc-60 gene in body wall muscles. Here I summarize the bichromatic fluorescence reporter system utilized for visualizing the tissue-specific alternative processing patterns of the unc-60 pre-mRNA. I also discuss the model for the coordinated regulation of the UNC-60B-type pre-mRNA processing in body wall muscles by ASD-2 and SUP-12.
Keywords: ADF, alternative splicing, ASD-2, fluorescence splicing reporter, body wall muscle, pre-mRNA processing, RBFOX, SUP-12, unc-60
Introduction
Newly synthesized pre-mRNAs are considered to be processed cotranscriptionally in eukaryotes.1 The sites of transcription initiation, splicing and polyadenylation should be tightly regulated for proper gene expression. Yet, a single gene can produce multiple mRNA and protein isoforms by the combination of alternative promoters, alternative splicing and alternative polyadenylation.2-4 A variety of cis-regulatory elements and tissue-specific trans-acting factors involved in the regulation of alternative splicing have been identified in higher eukaryotes by biochemical and bioinformatic approaches.5-7
In C. elegans, it is now estimated by a recent genome-wide analysis that up to 25% of the protein-coding genes undergo alternative splicing.8 Mutations mapped to isoform-specific exons or isoform-specific rescue experiments suggested isoform-specific functions for some genes.9-12 Some splicing regulators have been identified by their genetic interactions with mutations in other genes.13-22 But the tissue-specific splicing regulation was not subjected to genetic screening probably because specific morphological or behavioral phenotypes were not expected.23-25 The fluorescence alternative splicing reporter system, which is based on transgenic expression of multiple fluorescent proteins according to alternative pre-mRNA splicing patterns of reporter minigenes carrying genomic fragments of interest, has successfully visualized a variety of tissue-specific and developmentally regulated alternative splicing patterns in vivo and enabled forward and reverse genetic screenings of the regulators required for the splicing regulation.26-32 Here I discuss the findings regarding the muscle-specific regulation of the unc-60 pre-mRNA processing by ASD-2 and SUP-12.30
Structure of the unc-60 Gene
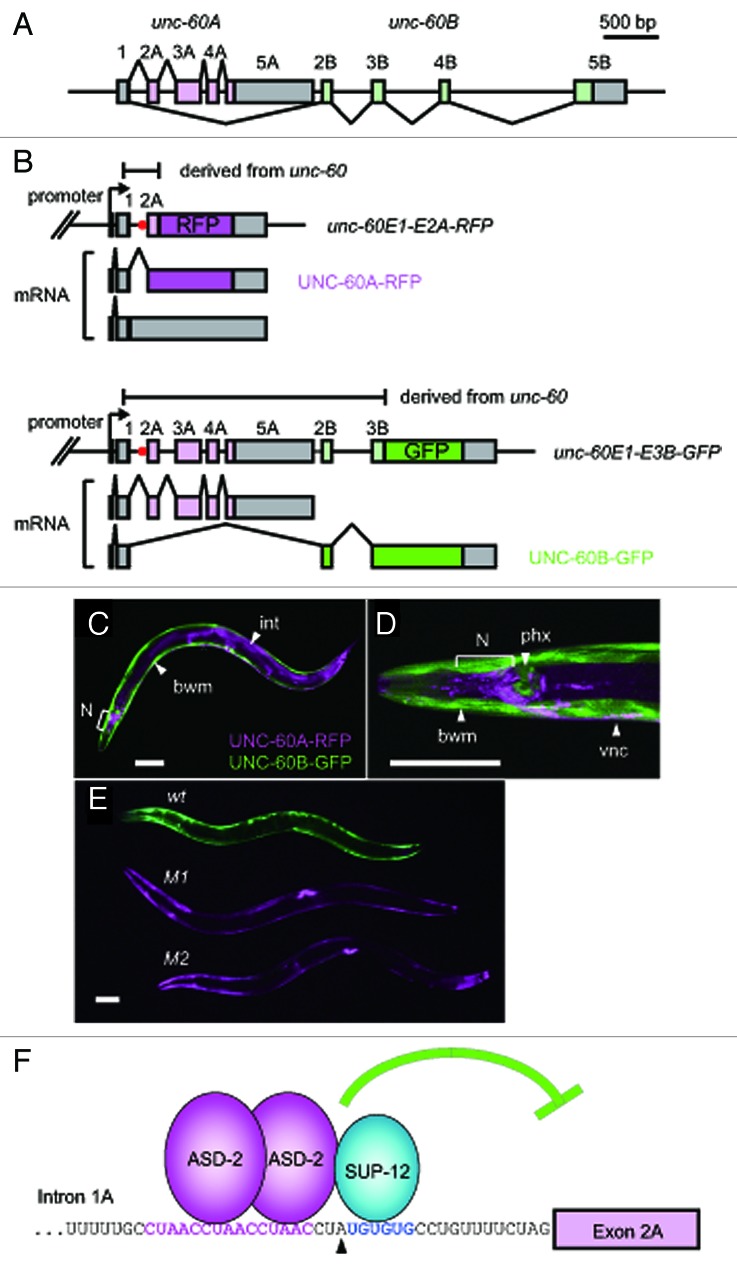
The unc-60 gene, generating two mRNA isoforms UNC-60A and UNC-60B, has a common first exon followed by two separate series of downstream exons, 2A through 5A for UNC-60A and 2B through 5B for UNC-60B (Fig. 1A).33 The unique structure of the unc-60 gene suggested that the choice between exons 2A-5A and exons 2B-5B in combination with alternative polyadenylation at the 3′ end of exon 5A or exon 5B determine the fate of the unc-60 pre-mRNA. The structure of the gene also suggested that the splice donor site for exon 1 should be preserved during the course of transcription until the splice acceptor site for exon 2B is available for splicing in situations where the UNC-60B mRNA is generated.
Figure 1. Fluorescence reporters reveal the tissue-specific selection patterns, the trans-acting regulatory factors and the cis-elements for the unc-60 pre-mRNA processing. (A) Schematic structure of the unc-60 gene. Numbered boxes indicate exons. The open reading frames (ORFs) for UNC-60A and UNC-60B are colored in light magenta and light green, respectively. (B) Schematic illustration of the pair of the unc-60 reporter minigenes and the UNC-60A- and UNC-60B-type mRNAs derived from them. The unc-60E1-E2A-RFP (top) and unc-60E1-E3B-GFP (bottom) cassettes carry the unc-60 genomic fragments from exon 1 through exon 2A and from exon 1 through exon 3B, respectively. The cDNA cassettes and predicted ORFs for RFP and GFP are colored in magenta and green, respectively. Red dots indicate the position of the CUAAC repeats and the UGUGUG stretch shown in (F). RFP is expressed only when intron 1A is excised from the unc-60E1-E2A-RFP cassette. GFP expression indicates the UNC-60B-type processing of the unc-60E1-E3B-GFP cassette. (C−E) Confocal images of the transgenic unc-60 reporter worms under the control of the unc-51 promoter (C, D) and a micrograph of the transgenic worms expressing the unc-60 reporter minigene pairs without (wt) or with the mutations in the CUAAC repeats (M1) or the UGUGUG stretch (M2) under the control of the myo-3 promoter (E). Anterior is to the left. The fluorescence images of UNC-60A-RFP and UNC-60B-GFP are pseudo-colored in magenta and green, respectively. bwm, body wall muscles; int, intestine; N, neurons in the head ganglia; phx, pharynx; vnc, ventral nerve cord. Scale bars, 50 μm. (F) Schematic illustration of the cooperative repression of the acceptor site for exon 2A by ASD-2 and SUP-12. The nucleotide sequence of the 3′ end region of intron 1A is indicated. The CUAAC repeats and the UGUGUG stretch are shown in magenta and blue, respectively. A black triangle indicates a putative branch site.45(C−E) are reproduced and modified from ref. 30.
The UNC-60A and UNC-60B mRNAs share only the initiation codon harbored in the common first exon. Yet the UNC-60A and UNC-60B proteins share 38% amino acid sequence identity.33 The UNC-60 isoforms have distinct biochemical properties in the regulation of actin dynamics34,35 and different in vivo functions during development and in muscle organization.36,37 Immunohistochemical staining demonstrated that UNC-60A is predominantly expressed in non-muscle tissues, while UNC-60B is mainly detected in body wall muscles,36-38 suggesting that alternative processing of the unc-60 pre-mRNA is regulated in a tissue-specific manner.
Visualization of the Tissue-Specific Alternative Pre-mRNA Processing Patterns of the unc-60 Gene
In order to visualize the binary processing patterns of the unc-60 transcript in vivo, a pair of reporter minigenes was constructed (see Fig. 1B for details). The unique property of the unc-60 reporter is that the minigenes are asymmetric, i.e., the two minigenes carry distinct portions from the unc-60 gene. The bichromatic and trichromatic reporter minigenes utilized in other studies for visualizing in vivo selection patterns of mutually exclusive exons or cassette exons were symmetric pairs,28,31,32 a symmetric trio31 or a single construct.26 Because the UNC-60A- and UNC-60B-type mRNAs end at the distinct exons, the symmetric-pair type was not applicable and the asymmetric pair was designed. The unc-60E1-E2A-RFP cassette (Fig. 1B, top panel), carrying the short genomic fragment spanning from exon 1 through exon 2A, was designed to focus on the excision of the only intron between exon 1 and exon 2A (hereafter referred to as intron 1A) via expression of RFP-fusion protein (UNC-60A-RFP) based on the assumption that intron 1A should be retained in situations where UNC-60B is expressed. On the other hand, the unc-60E1-E3B-GFP cassette (Fig. 1B, bottom panel) was designed to monitor the UNC-60B-type processing via expression of GFP-fusion protein (UNC-60B-GFP).
When the unc-60 reporter minigene pair was expressed under the control of the unc-51 promoter, UNC-60A-RFP and UNC-60B-GFP were expressed in a mutually exclusive manner; UNC-60A-RFP was expressed in non-muscle tissues such as the nervous system, intestine and hypodermis, while UNC-60B-GFP was expressed in body wall muscles and pharyngeal muscles (Fig. 1C and D). The expression pattern indicated that the unc-60 reporter pre-mRNAs were processed in a tissue-specific manner that is consistent with the previous results of immunohistochemical staining of the endogenous UNC-60 proteins.36-38
The unc-60 reporter driven under the control of the myo-3 promoter (Fig. 1E) was utilized for further genetic analyses of the muscle-specific processing regulation. Reverse genetic screening of candidate RNA-binding proteins revealed that muscle-specific RNA-binding proteins ASD-2 and SUP-12 are required for the expression of UNC-60B-GFP in body wall muscles.30 Directed mutagenesis of short stretches in the pairs of the reporter minigenes revealed CUAAC repeats and a UGUGUG stretch residing near the putative branch site for intron 1A as the crucial cis-elements for the UNC-60B-type processing in body wall muscles (Fig. 1E−F). Electrophoretic mobility shift assays (EMSAs) revealed that ASD-2 and SUP-12 specifically recognize the CUAAC repeats and the UGUGUG stretch, respectively, to cooperatively bind to intron 1A.30 Thus, once the alternative processing patterns of the gene of interest is successfully visualized in vivo with the fluorescence reporter system, the standard genetic tools can be applied to elucidate the regulatory mechanisms in C. elegans.29
Models of the Muscle-Specific Switching of the unc-60 Pre-mRNA Processing Patterns by ASD-2 and SUP-12
Figure 2 illustrates the models of the tissue-specific pre-mRNA processing of the unc-60 gene. In non-muscle tissues (Fig. 2, top), the introns are excised during or after transcription and the UNC-60A mRNA is produced. The order of removal of the four introns does not appear to be strictly regulated.30 In muscles (Fig. 2, bottom), ASD-2 and SUP-12 cooperatively bind to the CUAAC repeats and the UGUGUG stretch, respectively, in intron 1A to repress its excision during the transcription and processing of the UNC-60A region. Intron 3A and intron 4A appear to be immediately removed upon transcription.30 When exon 2B is transcribed and become available, the preserved donor site for exon 1 is readily spliced to exon 2B and the pre-mRNA is committed to the UNC-60B isoform.
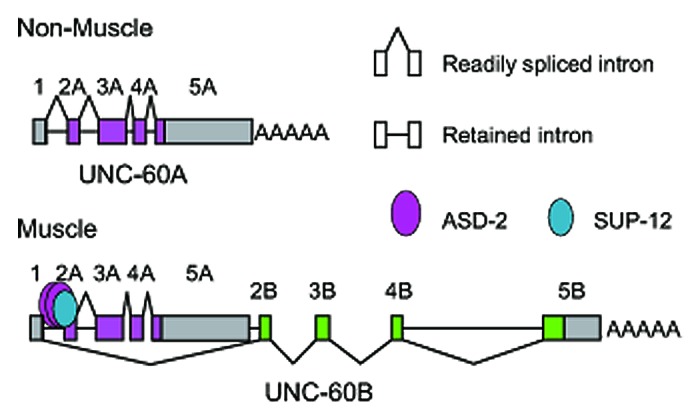
Figure 2. Models of the alternative processing of the unc-60 pre-mRNAs in non-muscle tissues (top) and muscles (bottom). See the main text and ref. 30 for details.
The orders of the intron excision in the models presented here and described in more detail in ref. 30 are suggested by comparison of the partially-spliced RNA species from the endogenous unc-60 gene in N2 and the sup-12 mutant. The detected partially-spliced RNAs in the steady-state may not necessarily be the processing intermediates for the mature mRNAs but instead be dead-end products of aberrant processing. Yet, the differences in the repertoires of the partially-spliced unc-60 RNAs were consistent with the differences in the amounts of the mature UNC-60 mRNAs between the wild type and the sup-12 mutant,30 suggesting that the RNA species are the actual processing intermediates.
Repression of the Acceptor Site for Exon 2A is the Key Event to Switch the Processing Patterns of the Entire Pre-mRNA
Expression of the UNC-60A protein instead of UNC-60B in body wall muscles in the asd-2 and sup-12 mutants has been demonstrated by immunohistochemical staining and suppression of the uncoordinated (Unc) phenotype of the unc-60B-specific mutant.19,30 The findings indicated that ASD-2 and SUP-12 switch the processing patterns of the entire unc-60 pre-mRNA from UNC-60A-type to UNC-60B-type in body wall muscles. This raises a question about whether ASD-2 and SUP-12 repress excision of only intron 1A as demonstrated by the unc-60E1-E2A-RFP reporter shown in Figure 1B (top) or they also bind to other unspecified site(s) and repress splicing of exons 3A, 4A and/or 5A to inhibit aberrant splicing between exon 1 and these exons. Disruption of the CUAAC repeats (M1) or the UGUGUG stretch (M2) in intron 1A of the unc-60E1-E3B-GFP reporter led to proper expression of the mature full-length UNC-60A mRNA (Fig. 1B, bottom, Fig. 1E), indicating that splicing of exons 3A, 4A and 5A is unaffected in body wall muscles in the wild-type background. Therefore, the repression of the acceptor site for exon 2A via the CUAAC repeats and the UGUGUG stretch in intron 1A is the crucial event to switch the processing patterns of the entire unc-60E1-E3B-GFP cassette from UNC-60A-type to UNC-60B-type as shown in Figure 2.
Does Polyadenylation at Exon 5A Need to Be Repressed in Muscles?
The remaining question is whether proper regulation of the alternative polyadenylation at exon 5A is crucial for the UNC-60B-type pre-mRNA processing; we do not have a conclusive answer.
If the unc-60 pre-mRNA is cleaved at the 3′ end of exon 5A before the acceptor site for exon 2B is committed to splicing, exon 1 would not be spliced to exon 2B. Although it is likely that ASD-2, SUP-12 or other muscle-specific factors may play roles in repressing the polyadenylation, the muscle-specific repression of the cleavage at exon 5A might be unnecessary considering the following situations. The polyadenylation signal (PAS) for exon 5A is not the canonical motif AAUAAA but appears to be a variant PAS; the cleavage and polyadenylation site is just about 90-nucleotide upstream of the acceptor site for exon 2B;39 exon 2B appears to be readily spliced to exon 1 when available.30
If the cleavage and polyadenylation at exon 5A is tightly repressed specifically in muscles, the mature mRNAs produced in muscles of the asd-2 and sup-12 mutants would include all the exons, i.e.,, exons 1 through 5A and 2B through 5B, where the region from exon 5A through exon 2B behaves as one exon because there left no available donor site in this region. This type of mRNAs may be retained in the nucleus due to the potential acceptor site for exon 2B or may be rapidly degraded by nonsense-mediated mRNA decay (NMD) due to its long 3′ untranslated region (UTR). However, such an mRNA isoform was not detected even in the NMD-deficient smg-2; sup-12 double mutant (unpublished observation), suggesting that exon 5A is consequently cleaved and polyadenylated even in muscles of the sup-12 mutant.
Cooperative Regulation of the unc-60 Pre-mRNA Processing by ASD-2 and SUP-12
The asd-2 gene has two tissue-specific promoters and therefore has two distinct first exons. Transcriptional fusion reporters revealed that transcription from exon 1a and exon 1b is driven in hypodermis and pharynx and in body wall muscles and pharynx, respectively.28 The ASD-2a and ASD-2b protein isoforms share most of the amino acid sequences including the evolutionarily conserved signal transduction and activation of RNA (STAR) domain.28 Ectopic expression of either ASD-2a or ASD-2b turned the let-2 alternative splicing reporter expression,28 indicating that both ASD-2a and ASD-2b are capable of regulating alternative splicing. Western blotting and immunohistochemical staining of the wild-type and asd-2b-specific allele of the asd-2 mutants revealed that ASD-2b is the major isoform in C. elegans and is specifically localized to the nuclei of body wall muscles.30
The mammalian ortholog of ASD-2, QKI, has been shown to form a homodimer40 and recognize a bipartite consensus sequence NACUAAY-N1–20-UAAY.41 The Drosophila ortholog HOW also forms a homodimer, which is enhanced upon phosphorylation.42 Considering the evolutionarily conserved amino acid sequences of the STAR domain that mediates the dimerization and RNA-binding28,42 as well as the conservation of the CUAAC repeats in intron 1A of the unc-60 gene in the genus Caenorhabditis,30 it is reasonable to suggest that ASD-2b also forms a homodimer when binding to the CUAAC repeats (Fig. 1F and 2, bottom).
SUP-12 represses the acceptor sites for unc-60 exon 2A30 and egl-15 exon 5B27 in a muscle-specific manner. In these cases, the presence of SUP-12 alone is not sufficient for repressing the splice sites but the partner regulators ASD-2b and the RBFOX family, respectively, are required for the proper regulation.27,30 Although SUP-12 has been shown to preferentially recognize the (U)GUGUG stretch in these cases,27,30 EMSAs revealed that SUP-12 can also bind to other unspecified site(s) in the unc-60 intron 1A probe.30 Therefore, the cooperation with the partner proteins is crucial for SUP-12 to specifically and efficiently regulate its target pre-mRNAs.
The full-length ASD-2b protein and the full-length SUP-12 protein cooperatively bind to unc-60 intron 1A.30 However, the STAR domain of ASD-2 and the RNA recognition motif (RRM) domain of SUP-12 did not exhibit cooperativity although they can specifically recognize the CUAAC repeats and the UGUGUG stretch, respectively, like the full-length proteins (unpublished observation). Considering that full-length ASD-2b and full-length SUP-12 can interact even in the absence of the target RNA,30 the portions other than the RNA-binding domains may contribute to the protein-protein interaction required for the cooperativity. Similarly, full-length SUP-12 can interact with full-length ASD-1 and FOX-1 in the absence of the target RNA27 and full-length SUP-12 and full-length ASD-1 or FOX-1 cooperatively bind to egl-15 intron 4.27 In contrast to the highly conserved RNA-binding domains, the N- and C-terminal portions of ASD-2, SUP-12, ASD-1 or FOX-1 are not conserved in the amino acid sequences among the orthologs from vertebrates, insects and nematodes19,28,43 but share similarities in the amino acid composition; they are rich in alanine (A) and glutamine (Q) residues (Fig. 3). As these A/Q-rich regions are hydrophobic, they may contribute to the specific protein-protein interaction in a certain or any combination in the cooperative recognition of the target RNAs.

Figure 3. The N-terminal and C-terminal portions of ASD-2b, SUP-12, ASD-1 and FOX-1 are rich in the alanine and glutamine residues. The protein structures are schematically shown with boxes. The STAR domain and the RRM domains are colored in magenta and orange, respectively. The total contents of the alanine (A) and glutamine (Q) residues in the N-terminal and C-terminal portions of each protein are indicated.
Pre-mRNA Processing of the Cholinergic Gene Locus
Another example of a locus with a structure similar to the unc-60 gene is the cholinergic gene locus consisting of unc-17, encoding vesicular acetylcholine transporter (VAChT), and cha-1, encoding choline acetyltransferase (ChAT).44 The two genes share a common 5′ untranslated exon, and the other three exons of the unc-17 gene reside in the first intron of the cha-1 gene.44 The structure of the locus suggests the switch-like regulation of the pre-mRNA processing like the unc-60 gene. Interestingly, the products of the two genes function in sequential steps in the metabolism of the neurotransmitter acetylcholine. In contrast to the unc-60 gene that produce the UNC-60A and UNC-60B isoforms in distinct tissues in a mutually exclusive manner, both the UNC-17 and CHA-1 mRNAs and proteins should be produced in the same cholinergic neurons. Therefore, not only the binary pre-mRNA processing patterns but the ratio of the amounts of the mature mRNAs should be somehow properly regulated for the cholinergic gene locus.
Acknowledgments
I thank Hiroaki Iwasa of Tokyo Medical and Dental University for fruitful discussion.
Glossary
Abbreviations:
- A
alanine
- ADF
actin depolymerising factor
- ChAT
choline acetyltransferase
- EMSA
electrophoretic mobility shift assay
- GFP
green fluorescent protein
- NMD
nonsense-mediated mRNA decay
- ORF
open reading frame
- PAS
polyadenylation signal
- pre-mRNA
precursor messenger RNA
- Q
glutamine
- RFP
red fluorescent protein
- RRM
RNA recognition motif
- STAR
signal transduction and activation of RNA
- Unc
uncoordinated
- UTR
untranslated region
- VAChT
vesicular acetylcholine transporter
Submitted
01/02/2013
Revised
01/27/2013
Accepted
01/30/2013
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/worm/article/23834
References
- 1.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 3.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–98. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 4.Li Q, Lee JA, Black DL. Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci. 2007;8:819–31. doi: 10.1038/nrn2237. [DOI] [PubMed] [Google Scholar]
- 5.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10:741–54. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McManus CJ, Graveley BR. RNA structure and the mechanisms of alternative splicing. Curr Opin Genet Dev. 2011;21:373–9. doi: 10.1016/j.gde.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Witten JT, Ule J. Understanding splicing regulation through RNA splicing maps. Trends Genet. 2011;27:89–97. doi: 10.1016/j.tig.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramani AK, Calarco JA, Pan Q, Mavandadi S, Wang Y, Nelson AC, et al. Genome-wide analysis of alternative splicing in Caenorhabditis elegans. Genome Res. 2011;21:342–8. doi: 10.1101/gr.114645.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rogalski TM, Gilchrist EJ, Mullen GP, Moerman DG. Mutations in the unc-52 gene responsible for body wall muscle defects in adult Caenorhabditis elegans are located in alternatively spliced exons. Genetics. 1995;139:159–69. doi: 10.1093/genetics/139.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pujol N, Bonnerot C, Ewbank JJ, Kohara Y, Thierry-Mieg D. The Caenorhabditis elegans unc-32 gene encodes alternative forms of a vacuolar ATPase a subunit. J Biol Chem. 2001;276:11913–21. doi: 10.1074/jbc.M009451200. [DOI] [PubMed] [Google Scholar]
- 11.Goodman SJ, Branda CS, Robinson MK, Burdine RD, Stern MJ. Alternative splicing affecting a novel domain in the C. elegans EGL-15 FGF receptor confers functional specificity. Development. 2003;130:3757–66. doi: 10.1242/dev.00604. [DOI] [PubMed] [Google Scholar]
- 12.Van Buskirk C, Sternberg PW. Epidermal growth factor signaling induces behavioral quiescence in Caenorhabditis elegans. Nat Neurosci. 2007;10:1300–7. doi: 10.1038/nn1981. [DOI] [PubMed] [Google Scholar]
- 13.Lundquist EA, Herman RK. The mec-8 gene of Caenorhabditis elegans affects muscle and sensory neuron function and interacts with three other genes: unc-52, smu-1 and smu-2. Genetics. 1994;138:83–101. doi: 10.1093/genetics/138.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lundquist EA, Herman RK, Rogalski TM, Mullen GP, Moerman DG, Shaw JE. The mec-8 gene of C. elegans encodes a protein with two RNA recognition motifs and regulates alternative splicing of unc-52 transcripts. Development. 1996;122:1601–10. doi: 10.1242/dev.122.5.1601. [DOI] [PubMed] [Google Scholar]
- 15.Nicoll M, Akerib CC, Meyer BJ. X-chromosome-counting mechanisms that determine nematode sex. Nature. 1997;388:200–4. doi: 10.1038/40669. [DOI] [PubMed] [Google Scholar]
- 16.Davies AG, Spike CA, Shaw JE, Herman RK. Functional overlap between the mec-8 gene and five sym genes in Caenorhabditis elegans. Genetics. 1999;153:117–34. doi: 10.1093/genetics/153.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spike CA, Shaw JE, Herman RK. Analysis of smu-1, a gene that regulates the alternative splicing of unc-52 pre-mRNA in Caenorhabditis elegans. Mol Cell Biol. 2001;21:4985–95. doi: 10.1128/MCB.21.15.4985-4995.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spike CA, Davies AG, Shaw JE, Herman RK. MEC-8 regulates alternative splicing of unc-52 transcripts in C. elegans hypodermal cells. Development. 2002;129:4999–5008. doi: 10.1242/dev.129.21.4999. [DOI] [PubMed] [Google Scholar]
- 19.Anyanful A, Ono K, Johnsen RC, Ly H, Jensen V, Baillie DL, et al. The RNA-binding protein SUP-12 controls muscle-specific splicing of the ADF/cofilin pre-mRNA in C. elegans. J Cell Biol. 2004;167:639–47. doi: 10.1083/jcb.200407085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Spartz AK, Herman RK, Shaw JE. SMU-2 and SMU-1, Caenorhabditis elegans homologs of mammalian spliceosome-associated proteins RED and fSAP57, work together to affect splice site choice. Mol Cell Biol. 2004;24:6811–23. doi: 10.1128/MCB.24.15.6811-6823.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yochem J, Bell LR, Herman RK. The identities of sym-2, sym-3 and sym-4, three genes that are synthetically lethal with mec-8 in Caenorhabditis elegans. Genetics. 2004;168:1293–306. doi: 10.1534/genetics.104.029827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calixto A, Ma C, Chalfie M. Conditional gene expression and RNAi using MEC-8-dependent splicing in C. elegans. Nat Methods. 2010;7:407–11. doi: 10.1038/nmeth.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.David CJ, Manley JL. The search for alternative splicing regulators: new approaches offer a path to a splicing code. Genes Dev. 2008;22:279–85. doi: 10.1101/gad.1643108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zahler AM. Alternative splicing in C. elegans In: The C. elegans Research Community, ed. WormBook - Molecular biology -: http://www.wormbook.org, 2005:1-13. [DOI] [PMC free article] [PubMed]
- 25.Zahler AM. Pre-mRNA splicing and its regulation in Caenorhabditis elegans In: The C. elegans Research Community, ed. WormBook - Molecular biology -: http://www.wormbook.org, 2012:1-21. [DOI] [PMC free article] [PubMed]
- 26.Kuroyanagi H, Kobayashi T, Mitani S, Hagiwara M. Transgenic alternative-splicing reporters reveal tissue-specific expression profiles and regulation mechanisms in vivo. Nat Methods. 2006;3:909–15. doi: 10.1038/nmeth944. [DOI] [PubMed] [Google Scholar]
- 27.Kuroyanagi H, Ohno G, Mitani S, Hagiwara M. The Fox-1 family and SUP-12 coordinately regulate tissue-specific alternative splicing in vivo. Mol Cell Biol. 2007;27:8612–21. doi: 10.1128/MCB.01508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohno G, Hagiwara M, Kuroyanagi H. STAR family RNA-binding protein ASD-2 regulates developmental switching of mutually exclusive alternative splicing in vivo. Genes Dev. 2008;22:360–74. doi: 10.1101/gad.1620608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuroyanagi H, Ohno G, Sakane H, Maruoka H, Hagiwara M. Visualization and genetic analysis of alternative splicing regulation in vivo using fluorescence reporters in transgenic Caenorhabditis elegans. Nat Protoc. 2010;5:1495–517. doi: 10.1038/nprot.2010.107. [DOI] [PubMed] [Google Scholar]
- 30.Ohno G, Ono K, Togo M, Watanabe Y, Ono S, Hagiwara M, et al. Muscle-specific splicing factors ASD-2 and SUP-12 cooperatively switch alternative pre-mRNA processing patterns of the ADF/cofilin gene in Caenorhabditis elegans. PLoS Genet. 2012;8:e1002991. doi: 10.1371/journal.pgen.1002991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuroyanagi H, Watanabe Y, Hagiwara M. CELF family RNA-binding protein UNC-75 regulates two sets of mutually exclusive exons of the unc-32 gene in neuron-specific manners in Caenorhabditis elegans. PLoS Genet. doi: 10.1371/journal.pgen.1003337. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuroyanagi H, Watanabe Y, Suzuki Y, Hagiwara M. Position-dependent and neuron-specific splicing regulation by the CELF family RNA-binding protein UNC-75 in Caenorhabditis elegans. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKim KS, Matheson C, Marra MA, Wakarchuk MF, Baillie DL. The Caenorhabditis elegans unc-60 gene encodes proteins homologous to a family of actin-binding proteins. Mol Gen Genet. 1994;242:346–57. doi: 10.1007/BF00280425. [DOI] [PubMed] [Google Scholar]
- 34.Ono S, Benian GM. Two Caenorhabditis elegans actin depolymerizing factor/cofilin proteins, encoded by the unc-60 gene, differentially regulate actin filament dynamics. J Biol Chem. 1998;273:3778–83. doi: 10.1074/jbc.273.6.3778. [DOI] [PubMed] [Google Scholar]
- 35.Yamashiro S, Mohri K, Ono S. The two Caenorhabditis elegans actin-depolymerizing factor/cofilin proteins differently enhance actin filament severing and depolymerization. Biochemistry. 2005;44:14238–47. doi: 10.1021/bi050933d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ono K, Parast M, Alberico C, Benian GM, Ono S. Specific requirement for two ADF/cofilin isoforms in distinct actin-dependent processes in Caenorhabditis elegans. J Cell Sci. 2003;116:2073–85. doi: 10.1242/jcs.00421. [DOI] [PubMed] [Google Scholar]
- 37.Ono K, Yamashiro S, Ono S. Essential role of ADF/cofilin for assembly of contractile actin networks in the C. elegans somatic gonad. J Cell Sci. 2008;121:2662–70. doi: 10.1242/jcs.034215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ono S, Ono K. Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J Cell Biol. 2002;156:1065–76. doi: 10.1083/jcb.200110013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, et al. The landscape of C. elegans 3’UTRs. Science. 2010;329:432–5. doi: 10.1126/science.1191244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen T, Richard S. Structure-function analysis of Qk1: a lethal point mutation in mouse quaking prevents homodimerization. Mol Cell Biol. 1998;18:4863–71. doi: 10.1128/mcb.18.8.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galarneau A, Richard S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat Struct Mol Biol. 2005;12:691–8. doi: 10.1038/nsmb963. [DOI] [PubMed] [Google Scholar]
- 42.Nir R, Grossman R, Paroush Z, Volk T. Phosphorylation of the Drosophila melanogaster RNA-binding protein HOW by MAPK/ERK enhances its dimerization and activity. PLoS Genet. 2012;8:e1002632. doi: 10.1371/journal.pgen.1002632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuroyanagi H. Fox-1 family of RNA-binding proteins. Cell Mol Life Sci. 2009;66:3895–907. doi: 10.1007/s00018-009-0120-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alfonso A, Grundahl K, McManus JR, Asbury JM, Rand JB. Alternative splicing leads to two cholinergic proteins in Caenorhabditis elegans. J Mol Biol. 1994;241:627–30. doi: 10.1006/jmbi.1994.1538. [DOI] [PubMed] [Google Scholar]
- 45.Blumenthal T, Steward K. RNA processing and gene structure. In: D. Riddle, T. Blumenthal, B. Meyer, J. Priess, eds. C elegans II. N.Y.: Cold Spring Harbor Laboratory Press, 1997:117–45. [PubMed] [Google Scholar]